Introduction
Plesiomonas shigelloides (P.
shigelloides) is the only species member of the
Plesiomonas genus, and the only oxidase-positive member of
the Enterobacteriaceae family (1). P. shigelloides is present
worldwide, primarily in aquatic environments, including freshwater,
estuarine and marine environments (2–4).
P. shigelloides induces various types of gastroenteritis,
including acute secretory gastroenteritis, invasive
shigellosis-like disease and cholera-like illness, infections
increasing in recent years (5–7).
In humans, although extra-intestinal diseases of
P. shigelloides are rare, it has been associated with
secondary infections in immunocompromised states, including
malignancy, blood disorders and hepatobiliary disease (8–10).
P. shigelloides may often be missed in stool samples due to
its small colony size and relatively low prevalence in
gastrointestinal samples. The lack of a routine assay for P.
shigelloides in cases of gastroenteritis means that this
bacterium is identified only occasionally (11). Xia et al (12) reported a case of
meningoencephalitis caused by P. shigelloides with a fatal
outcome in a Chinese neonate. Therefore, its earlier and accurate
identification, and the prescription of the correct antibiotic
therapy may be critical for patient prognosis.
The traditional culture-based approaches for
detection of P. shigelloides involve enrichment in liquid
media and isolation of colonies on selective media. Although
extensively used, these assays are time-consuming and laborious,
taking more than three days. In addition, the isolates of P.
shigelloides appear as green colonies on Hektoen enteric agar,
with an appearance similar to that of Shigella (13).
Molecular-based techniques, including polymerase
chain reaction (PCR) and quantitative PCR assays, have been
established for the detection of P. shigelloides, producing
reliable results. However, PCR-based techniques rely on expensive
thermal cycler or complex sample-handling procedures, limiting its
application (14). Therefore,
there is an urgent requirement to devise a novel strategy for
rapid, robust and sensitive identification of P.
shigelloides using simple equipment.
Loop-mediated isothermal amplification (LAMP), as a
rapid, specific and sensitive detection methodology, has been used
to detect various pathogens, including parasites, fungi, bacteria
and viruses (15). However, primer
design for LAMP techniques is complicated, requiring a specific,
long, highly conserved fragment. This limits the application of
LAMP for the detection of pathogens (16). A novel technology, cross-priming
amplification (CPA), overcomes the technical difficulties posed by
current LAMP approaches, which contains five specially designed
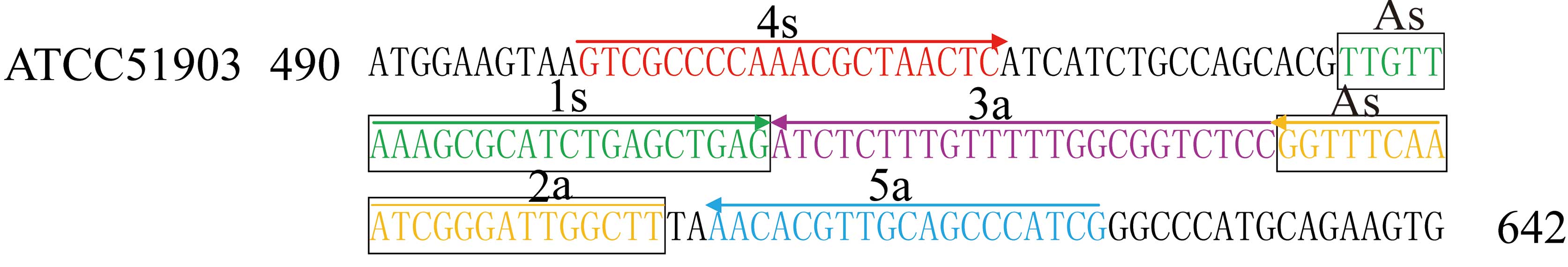
primers (1s, 2a, 3a, 4s and 5a) that recognise five conserved
regions on the target sequence. Each cross primer contains 5′ tail
sequences identical to each other's priming site and thus
introduces additional priming sites in each round of extension. The
primers are designed to accomplish the basic goal of isothermal
generation of single-stranded DNA (ssDNA) using a strand-displacing
polymerase such as Bst, and the DNA target sequence may be
amplified without an initial denaturation step or addition of a
nicking enzyme (17–20). The CPA products may be detected by
an increase in turbidity, agarose gel electrophoresis of amplicons
or by visualization of a colour alteration in the presence of
Loopamp® Fluorescent Detection Reagent.
Several potential virulence factors of P.
shigelloides have been described, however, the pathogenesis of
P. shigelloides-associated gastroenteritis remains to be
elucidated (21,22). Acquisition of iron has been
demonstrated to be involved in the virulence of a variety of
bacterial pathogens (23,24). Heme is the primary source of iron
within the body, and numerous pathogenic bacteria carry heme
transport systems (25). The
strains of P. shigelloides express highly specific outer
membrane receptors that bind, extract and transport heme into the
bacterial periplasm (26). The
hugA genes (heme iron utilization locuaccession no.
AY008342.1) encoding the heme iron utilization system of P.
shigelloides have been isolated and characterized, and are
essential for the growth of P. shigelloides.
The present study aimed to develop a rapid,
cost-effective and efficient CPA method for detecting P.
shigelloides, and evaluating the assay performance with
pathogen-simulated human stool. In addition, the CPA method was
compared with PCR to determine the sensitivity and evaluate the
practical application in clinical samples.
Materials and methods
Ethics statement
Stool specimens were acquired from 70 patients with
diarrhoea, aged from 18 to 50 years old, and written informed
consent was obtained from all participants. The study was reviewed
and approved by the ethics committee of the National Institute for
Communicable Disease Control and Prevention, Chinese Center for
Disease Control and Prevention (Beijing, China), according to the
medical research regulations of the Ministry of Health (Beijing,
China; approval no. ICDC-2014003).
Bacterial strains
A total of 53 strains (20 P. shigelloides
strains and 33 non-P. shigelloides strains, listed in
Table I) were used for specificity
testing. The bacterial load of the strains used for specificity
evaluation was 105 pg/ml, which is high enough to
prevent false-negative amplifications. P. shigelloides ATCC
51903 (GenBank accession number AY008342.1) was selected as the
positive control for the assay optimisation, sensitivity
evaluation, and to spike human stool samples. All strains were
cultured overnight at 37°C on brain heart infusion (BHI) agar (BD
Biosciences, Franklin Lakes, NJ, USA).
 | Table I.Bacterial strains used in the present
study. |
Table I.
Bacterial strains used in the present
study.
| Latin name | Strain no. (source
of strain) | No. of strains |
|---|
| Plesiomonas
shigelloides | ATCC 51903 | 1 |
|
| Isolated strains
(ICDC) | 19 |
| Enteropathogenic
Escherichia coli | Isolated strain
(ICDC) | 1 |
| Enterotoxigenic
Escherichia coli | Isolated strain
(ICDC) | 1 |
| Enteroinvasive
Escherichia coli | Isolated strain
(ICDC) | 1 |
| Enterohaemorrhagic
Escherichia coli | EDL 933 (isolated
previously in our laboratory) | 1 |
| Enteroaggregative
Escherichia coli | Isolated strain
(ICDC) | 1 |
| Salmonella
enterica | ATCC 14028 | 1 |
| Shigella
flexneri | Isolated strain
(ICDC) | 1 |
| Shigella
sonnei | ATCC 25931 | 1 |
| Proteus
vulgaris | Isolated strain
(ICDC) | 1 |
| Aeromonas
veronii | ATCC 35622 | 1 |
| Aeromonas
salmonicida | ATCC 7965 | 1 |
| Aeromonas
caviae | ATCC 15468 | 1 |
| Aeromonas
media | ATCC 33907 | 1 |
| Clostridium
perfringens | Isolated strain
(ICDC) | 1 |
| Enterobacter
cloacae | Isolated strain
(ICDC) | 1 |
| Serratia
marcescens | Isolated strain
(ICDC) | 1 |
| Vibrio
parahaemolyticus | ATCC 17802 | 1 |
| Staphylococcus
aureus | ATCC 6538 | 1 |
| Streptococcus
pneumoniae | Isolated strain
(ICDC) | 1 |
| Streptococcus
pyogenes | Isolated strain
(ICDC) | 1 |
| Streptococcus
sanguis | Isolated strain
(ICDC) | 1 |
| Streptococcus
salivarius | Isolated strain
(ICDC) | 1 |
| Streptococcus
bovis | Isolated strain
(ICDC) | 1 |
| Enterococcus
faecalis | ATCC 35667 | 1 |
| Yersinia
enterocolitica | ATCC 23715 | 1 |
| Pseudomonas
aeruginosa | ATCC 15442 | 1 |
| Aeromonas
hydrophila | ATCC 7966 | 1 |
| Listeria
monocytogenes | ATCC 54003 | 2 |
| Enterobacter
sakazakii | ATCC 51329 | 1 |
| Campylobacter
jejuni | ATCC 33291 | 1 |
| Vibrio
minicus | Isolated strain
(ICDC) | 1 |
| Vibrio
vulnificus | Isolated strain
(ICDC) | 1 |
Genomic DNA extraction
Bacterial genomic DNA was extracted from all
cultured strains using DNA extraction kits (QIAamp DNA
minikitQiagen, Hilden, Germany) according to the manufacturer's
instructions.
P. shigelloides hugA CPA primers and
reaction conditions
A set of five primers was manually designed to
target the nucleotide sequence of P. shigelloides ATCC
51903, based on the mechanism of CPA (27). The sequences and locations of the
primers within hugA are presented in Table II and Fig. 1. CPA reactions were performed using
the Loopamp kit (Eiken Chemical Co., Ltd., Tokyo, Japan) in a final
volume of 20 µl containing 2.4 mM cross primer As, 1.44 mM each of
primers 2a and 3a, 0.3 mM each of displacement primers 4s and 5a,
20 mM Tris-HCl (pH 8.8), 10 mM KCl, 4 mM MgSO4, 10 mM (NH4)2SO4,
0.1% Tween 20, 0.8 M betaine, 1.4 mM deoxynucleoside triphosphates
(dNTPs), 1 µl of Bst DNA polymerase (8 U µl−1), 1
µl Loopamp Fluorescent Detection Reagent (Eiken Chemical Co., Ltd.)
and 1 µl DNA template. The reaction mixture was incubated in an
LA320 Real-Time Turbidimeter (Teramecs Co., Ltd., Kyoto, Japan) at
63°C for 60 min, and then heated at 95°C for 5 min to terminate the
reaction. Amplified products were directly detected by observing a
colour change from orange to green by the naked eye, or by
electrophoresis on 2% agarose gels using staining with GoldenView
reagent. Furthermore, real-time monitoring of the CPA reaction was
performed by recording the optical density at 650 nm every 6 sec
using the LA-320C Real-Time Turbidimeter. A positive reaction was
defined as a turbidity cut-off value of >0.1 within 60 min.
 | Table II.CPA and PCR primers used to detect
Plesiomonas shigelloides. |
Table II.
CPA and PCR primers used to detect
Plesiomonas shigelloides.
| Assay type | Primer/probe
name | Sequence
(5′-3′) | Length (nt) |
|---|
|
hugA-CPA | AS (2a+1s) |
AAGCCAATCCCGATTTGAAACCTTTTGTTAAAGCGCATCTGAGCTGAG | 48 |
|
| 3a |
GGAGACCGCCAAAAACAAAGAGAT | 24 |
|
| 2a |
AAGCCAATCCCGATTTGAAACC | 22 |
|
| 4s |
GTCGCCCCAAACGCTAACTC | 20 |
|
| 5a |
CGATGGGCTGCAACGTGTT | 19 |
|
hugA-PCR | F |
GCGAGCGGGAAGGGAAGAACC | 21 |
|
| R |
GTCGCCCCAAACGCTAACTCATCA | 24 |
Evaluation of the specificity,
sensitivity and reproducibility of the P. shigelloides hugA CPA
assay
To determine the specificity of the CPA assay, the
CPA reaction was performed under the conditions described above
with DNA templates from 20 P. shigelloides and 33 non-P.
shigelloides strains (Table
I). All detection assays were performed in triplicate.
To assess the analytical sensitivity of CPA assay,
CPA assays were performed using serial dilutions (20, 2 ng, 200,
20, 2 pg, 200, 100 and 50 fg per µl) of P. shigelloides
genomic DNA. The genomic templates (1 µl) were added into the CPA
mixture and at least 3 replicates of each dilution were assessed to
define the limit of detection (LoD) of the CPA approach. Mixtures
without DNA templates served as a negative control. The sensitivity
of the CPA assay on P. shigelloides was determined by
analyzing the amplifications produced from the serial dilutions of
the P. shigelloides genomic DNA.
To compare the sensitivities of the CPA and PCR
assay in pure culture, template DNA from P. shigelloides
(ATCC 51903) was serially diluted (20, 2.0 ng, 200, 20, 2.0 pg, 200
fg, 100 and 50 fg per µl). The LoD of CPA and PCR was ascertained
using the two assays.
To evaluate the reproducibility of the CPA assay,
different concentrations (20 ng, 200 and 2.0 pg) of template DNA
from P. shigelloides (ATCC 51903) were amplified two ways
(10 times on one day and once each on 10 different days). The
intra-assay and inter-assay variation were analysed at the time of
precipitation, as measured by turbidity on the Real-Time
Turbidimeter. The coefficient of variation (CV) is equal to the
standard deviation (SD) divided by the mean average, multiplied by
100. Statistical analyses were conducted using SPSS software
(version, 19.0; IBM SPSS, Armonk, NY, USA).
PCR amplifications were performed in a final volume
of 20 µl containing 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 0.001%
gelatin, 1.5 mM MgCl2, 0.2 µM each of hugA forward and
hugA reverse primers, 0.2 mM each of dNTPs, 0.5 units of Ex
Taq DNA polymerase (Takara Bio, Inc., Otsu, Japan) and 1 µl DNA
template. The program consisted of an initial denaturation step of
5 min at 95°C, 35 cycles of 30 sec at 95°C, 30 sec at 60°C, and 30
sec at 72°C, and a final 5 min extension at 72°C. The PCR products
were visualised by 2% agarose gel electrophoresis to verify the
presence of the expected 435-bp fragment.
P. shigelloides hugA CPA application
in simulated human stools specimens
Human stool specimens were obtained from a healthy
donor with the written informed consent. The human stool specimens
were confirmed to be P. shigelloides-negative using a
traditional culture assay and PCR amplification (Table III). To determine the LoD of CPA
in human stool, 10-fold serial dilutions of a mid-log phase culture
of P. shigelloides grown in BHI broth at 37°C were prepared
in PBS, quantified using the standard plating method, and added to
the stool samples at 3×101-3×106 CFU/g.
Aliquots (0.2 g) of the stools were used for DNA extraction with a
QIAamp DNA Mini kit. This experiment was performed in triplicate
independently, and the supernatants (2 µl) were used for CPA and
PCR.
 | Table III.Reproducibility of the Plesiomonas
shigelloides hugA cross-priming amplification assay. |
Table III.
Reproducibility of the Plesiomonas
shigelloides hugA cross-priming amplification assay.
|
Reproducibility | Template DNA
(pg/reaction) | Number of
detections | Mean time of
precipitation (mins) | Standard
deviation | Coefficient of
variation (%) |
|---|
| Intra-assay |
2×104 | 10 | 23.4 | 0.21 | 0.90 |
|
|
2×102 | 10 | 27.3 | 0.37 | 1.36 |
|
| 2 | 10 | 38.4 | 0.49 | 1.28 |
| Inter-assay |
2×104 | 10 | 23.3 | 0.23 | 0.99 |
|
|
2×102 | 10 | 27.5 | 0.33 | 1.20 |
|
| 2 | 10 | 38.7 | 0.66 | 1.71 |
Practical application of the P.
shigelloides hugA CPA assay
To estimate the feasibility of the CPA assay to
detect P. shigelloides in clinical samples, 100 samples (70
stool specimens from patients with diarrhoea and 30 water samples
from the environment) were analysed using the CPA method, and
compared with the results from the traditional culture and PCR
methods. Culture-based detection of stool samples was performed by
enriching 2 g stool specimens in 20 ml tetrathionate broth without
iodine (Oxoid; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
for 10 h at 37°C, and then streaking on inositol brilliant green
bile salts (IBB) agar (Oxoid; Thermo Fisher Scientific, Inc.) and
the plates incubated at 35°C for 24 h. Pink colonies suspected to
be P. shigelloides were Gram stained, picked onto BHI agar
at 37°C for 18 h and subjected to biochemical tests using the API
20E system (BioMérieux, Marcy-l'Étoile, France) (25).
Water samples (500 ml) were filtered through sterile
analytical filters (NalgenThermo Fisher Scientific, Inc.) with pore
sizes of 0.45 µm, within 30 h of sample collection. The filters
were enriched in 20 ml tetrathionate broth without iodine for 10 h
at 37°C, streaked on IBB agar and the plates were incubated at 35°C
for 24 h (25). Pink colonies
suspected to be P. shigelloides were Gram stained, picked
onto BHI agar at 37°C for 18 h and subjected to biochemical tests
using the API 20E system.
DNA was extracted from 1 ml aliquots of the
enrichment broth using the QIAamp DNA Mini kit, and 2 µl of each
DNA extract was used as the template in the CPA and PCR assays.
P. shigelloides (ATCC 51903) genomic DNA was used as the
positive control template, and sterile water was used as the
negative control template.
Results
Primer design for the P. shigelloides
hugA CPA assay
For the P. shigelloides-specific hugA
gene, a set of 5 primers, which targeted 5 distinct regions, was
designed for the CPA assay by sequence alignment and primer
software Primer Premier 5.0 (Premier Biosoft International, Palo
Alto, CA, USA). These included the amplification primer 2a and 1s,
designated as the cross primer (As) and two amplification primers
(3a and 2a). The specificity of the CPA primers was confirmed using
the NCBI Basic Local Alignment Search Tool (National Institutes of
Health, Bethesda, MD, USA). The details of the primers are
presented in Table II and
Fig. 1.
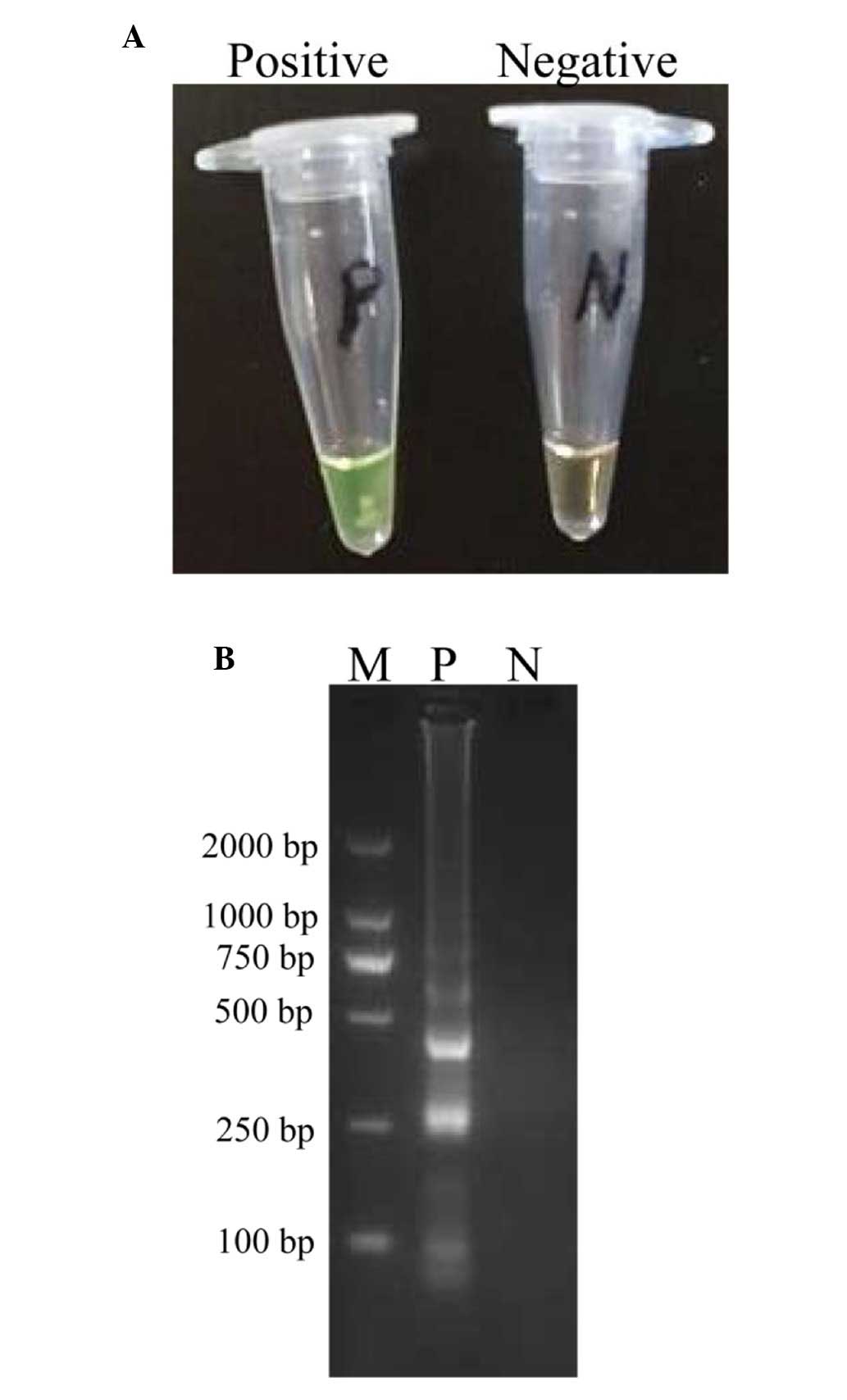
Confirmation and detection of P.
shigelloides CPA products
The amplification products were examined by visual
inspection using Loopamp Fluorescent Detection Reagent and the
positive amplifications were directly observed due to the colour
change from the original orange to green (Fig. 2A). In addition, the conventional
CPA products were assessed by 2% agarose gel electrophoresis, and
positive results demonstrated a typical ladder-like pattern
(Fig. 2B).
Specificity of the P. shigelloides
hugA CPA assay
The specificity of the CPA assay towards the P.
shigelloides hugA gene was examined by performing the assay
with DNA from 53 bacterial strains from 29 different species as the
template (Table I). The 20 P.
shigelloides strains were correctly identified, whereas no
amplification was observed in the 33 non-P. shigelloides
strains. The results demonstrated that the specificity of the CPA
assay was 100%, and the sequence revealed no cross-reaction with
different pathogens.
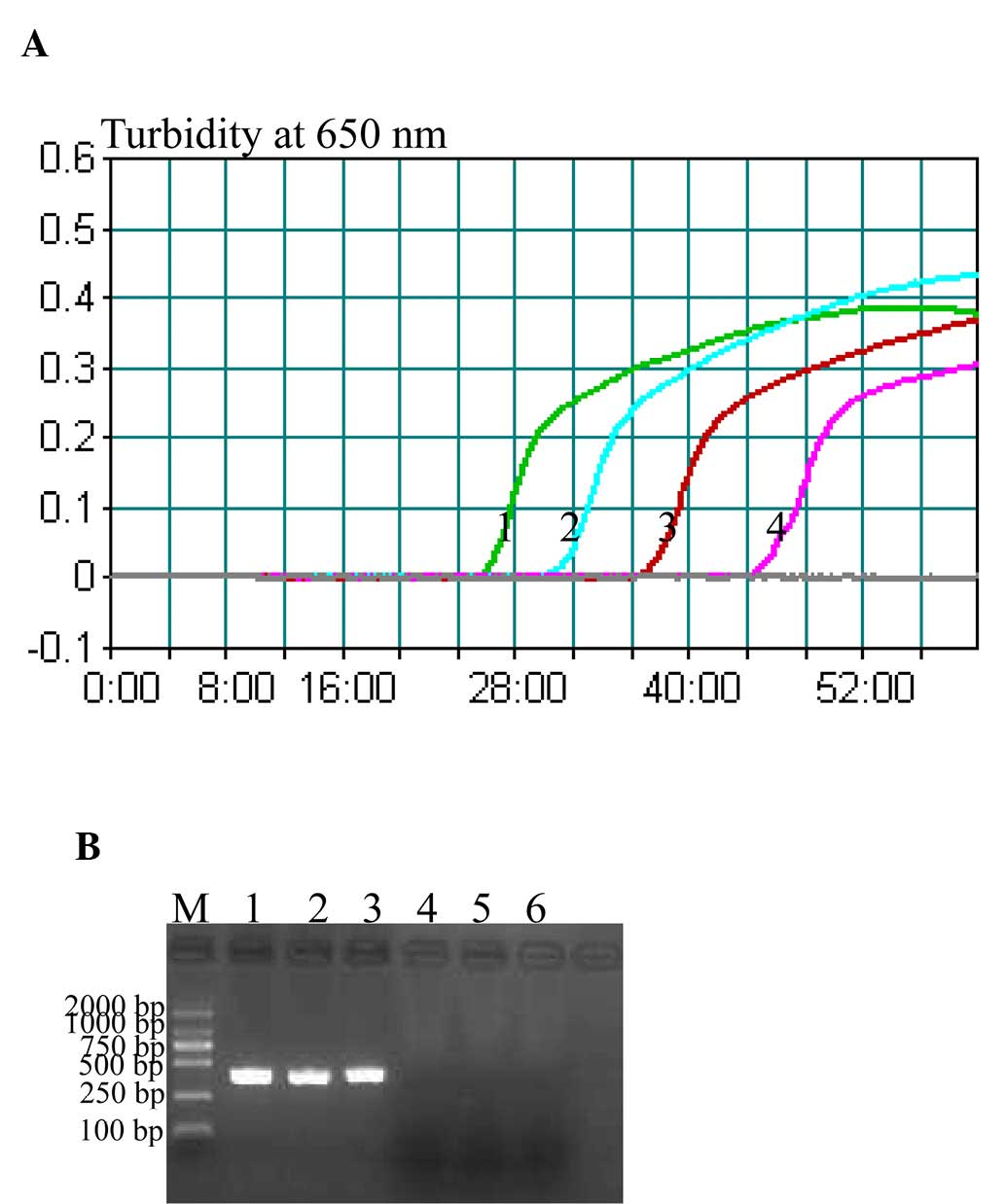
Sensitivity of the P. shigelloides
hugA CPA assay
The sensitivity of CPA assay towards P.
shigelloides was examined by determining the LoD of reactions
performed with serial dilutions of P. shigelloides genomic
DNA (20, 2 ng, 200, 20, 2 pg, 200, 100 and 50 fg per µl). The LoD
of CPA (Fig. 3A) was 200 fg
DNA/tube, whereas the LoD of PCR assay was 20 pg DNA/tube (Fig. 3B). These results indicated that the
CPA assay was 100-fold more sensitive than the PCR assay for
detecting P. shigelloides genomic DNA.
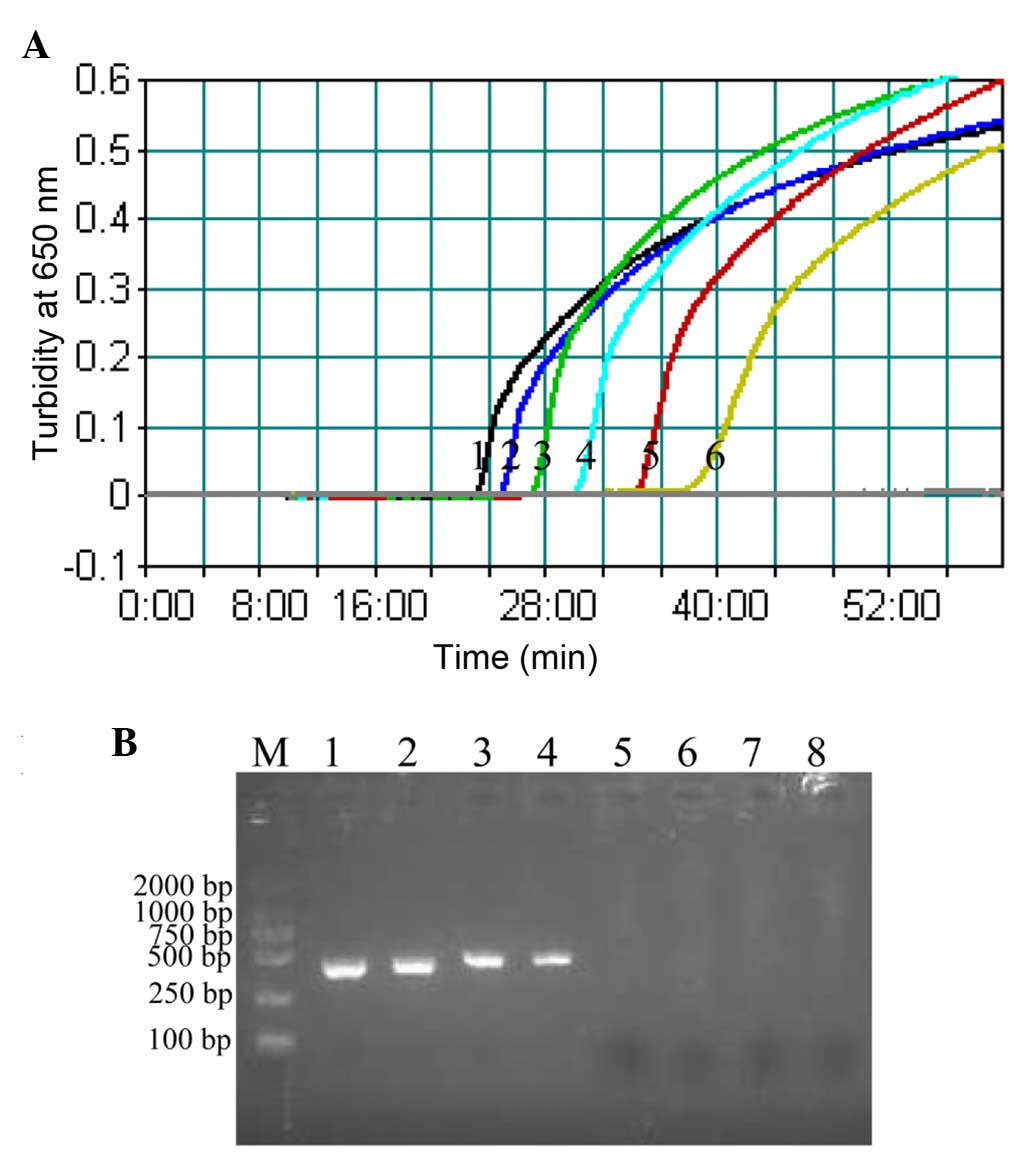 | Figure 3.Sensitivity of the CPA and PCR
methods. (A) Sensitivity of the CPA assay was assessed by measuring
the turbidity (optical density at 650 nm) of reactions over the
course of 60 min, using serial dilutions of Plesiomonas
shigelloides ATCC 51903 genomic DNA as template (1, 20 ng; 2, 2
ng; 3, 200 pg; 4, 20 pg; 5, 2 pg; and 6, 200 fg per µl,
respectively). A turbidity value of >0.1 within 60 min indicated
a positive reaction. (B) Sensitivity of the PCR method was
evaluated by detection of a 435-bp single target band by agarose
gel electrophoresis, using serial dilutions of Plesiomonas
shigelloides ATCC 51903 genomic DNA as template (1, 20 ng; 2, 2
ng; 3, 200 pg; 4, 20 pg; 5, 2 pg; 6, 200 fg; 7, 100 fg; and 8, 50
fg per µl). CPA, cross-priming amplification; PCR, polymerase chain
reaction; M, DL 2,000 bp DNA marker. |
Reproducibility of the P. shigelloides
hugA CPA assay
The intra-assay coefficient of variation (CV) was
determined using various quantities of template DNA (20 ng, 200 and
2.0 pg) 10 times in a single run. The inter-assay CV was determined
by performing the CPA assay using the same templates in 10 separate
runs. The intra-assay CV ranged from 0.9 to 1.36%, and the
inter-assay CV ranged from 0.99 to 1.71% (Table III). The reproducibility of the
P. shigelloides hugA CPA assay was, therefore, good.
P. shigelloides hugA CPA efficacy in
human stool specimens
The LoD of the P. shigelloides hugA CPA assay
on human stools containing measured concentrations of P.
shigelloides was examined. The CPA assay identified the
presence of P. shigelloides in stools containing as little
as 3×103 CFU per g stool (Fig. 4A), whereas PCR had a LoD of
3×104 CFU/g stool (Fig.
4B).
Utility of the P. shigelloides hugA
CPA assay for detection in clinical and environmental samples
The P. shigelloides hugA CPA assay, PCR and
culture-based detection were used to detect P. shigelloides
in 100 clinical and environmental specimens (70 stool samples from
patients with diarrhoea and 30 environmental water samples). The
P. shigelloides hugA CPA assay and PCR method detected P.
shigelloides in 11 (15.7%) and 8 (11.4%) stool specimens,
respectively (Table IV). In the
case of water samples, 4 (13.3%) and 3 (10.0%) water samples were
P. shigelloides positive by CPA and PCR, respectively
(Table IV). The samples that were
positive by PCR were also positive by CPA. P. shigelloides
strains were successfully isolated from all the CPA positive
samples. The CPA detection accuracy was 100% compared with the
traditional culture method. All samples were subjected to
culture-based detection. The P. shigelloides hugA CPA assay,
therefore, appears to be more sensitive for the detection of P.
shigelloides in clinical and environmental samples than
conventional PCR.
 | Table IV.Practical application of the
Plesiomonas shigelloides hugA cross-priming amplification
assay. |
Table IV.
Practical application of the
Plesiomonas shigelloides hugA cross-priming amplification
assay.
|
| Diarrhoea patient
specimens (n=70) | Environmental water
samples (n=30) |
|---|
|
|
|
|
|---|
| Detection
method | Positive | Negative | Positive | Negative |
|---|
| Polymerase chain
reaction | 8 | 62 | 3 | 27 |
| Culture | 11 | 59 | 4 | 26 |
| Cross-priming
amplification | 11 | 59 | 4 | 26 |
Discussion
In the present study, a CPA assay was developed for
the rapid detection of P. shigelloides as a potential
on-site and point-of-care test in clinics. P. shigelloides
is an important pathogen, which may contaminate food or aquatic
environment and causes gastrointestinal illness (6–8).
However, the current lack of a rapid and sensitive diagnostic
method can result in inappropriate antimicrobial therapies being
administered, potentially leading to further complications and
fatal outcomes (12,28). Therefore, a rapid, sensitive,
specific and economical detection method is urgently required.
The conventional methods for the isolation and
identification of P. shigelloides involve enrichment in
fluid media and subsequent isolation of colonies on selective
media. Although extensively performed, the methods are
labor-intensive and time-consuming, making it unsuitable for the
rapid detection of causative pathogens associated with sporadic and
outbreaks cases (25). As an
alternative, various PCR-based assays have been developed for the
detection of P. shigelloides. However, PCR-based methods
require a high-precision thermal cycler, which restricts their
widespread application and mean that these techniques are not
suited to diagnosis of P. shigelloides in basic clinical and
field laboratories in rural areas. Several isothermal amplification
methods have been developed for the rapid diagnosis of infectious
pathogens, including LAMP, which is a promising low-cost method for
detecting various infectious pathogens (17–20).
To date, the LAMP technique has been used to detect P.
shigelloides in stool and environment specimens. However, LAMP
assays require primers with high stringency, for which primer
design is complicated and requires specific software (Primer
Explorer V4 softwarEiken Chemical Co. Ltd.), therefore, posing an
obstacle for clinical application (16). Moreover, in LAMP, an additional
step of DNA template denaturation is required (29). The CPA assay reported in the study
does not require a denaturation step, does not require specific
software for primer design, and as the gene target sequence used
for primer design in the P. shigelloides hugA CPA assay is
shorter than required for the LAMP assay, the subsequently reduced
detection time is conducive to clinical application, as described
by Fang et al (30) for the
detection of M. tuberculosis in sputum samples. CPA is a
powerful innovative gene amplification technique, which has been
described as an easy and rapid diagnostic tool for the detection of
pathogens (27). The equipment
requirements for the CPA assay are also limited to a heat block or
water bath, maintaining a constant temperature of 63°C for 1 h. The
measurement of CPA products is possible by measuring turbidity,
electrophoresis of amplicons or visual observation when using the
Loopamp Fluorescent Detection Reagent. These features establish the
CPA assay as a suitable method for P. shigelloides detection
in basic clinical and field laboratories.
A 128-nucleotide fragment of the hugA gene
was selected as the target for the CPA assay primers, as this gene
is highly conserved in P. shigelloides strains (25). Primer specificity was determined by
subjecting 33 non-P. shigelloides strains (causing similar
clinical syndromes to P. shigelloides) to the P.
shigelloides hugA CPA assay, revealing 100% specificity of the
CPA assay for P. shigelloides. Positive amplification was
completed by visual inspection, and no positive reactions were
observed in the assays of non-P. shigelloides strains. The
results of the present study suggested that the CPA assay for the
detection of the gene that encodes the HugA outer membrane receptor
required for heme iron utilization by P. shigelloides may be
a reliable method to detect P. shigelloides. This procedure
combined with an enrichment step allows P. shigelloides
detection in clinical and environment specimens.
To the best of our knowledge, the present study is
the first to use CPA technology to detect P. shigelloides in
clinical and environmental specimens. The P. shigelloides
hugA CPA method was 100-fold more sensitive than conventional
PCR methods, detecting as little as 200 fg DNA per reaction.
Several previous studies have also demonstrated that CPA has
greater sensitivity than PCR for pathogen detection (17,29,31–34).
Thus, the P. shigelloides hugA CPA assay is more appropriate
than PCR for simple, rapid and sensitive detection of P.
shigelloides.
To evaluate the practical application of the P.
shigelloides hugA CPA assay for detection of P.
shigelloides in clinical samples, 100 specimens of clinical and
environmental origins were analysed using conventional
culture-based detection detection, PCR, and the P. shigelloides
hugA CPA assay. The P. shigelloides hugA CPA assay
exhibited greater P. shigelloides detection capability than
PCR, which was supported by several previous studies (17,29,31–34).
The conventional PCR method also led to false negative results that
were detected by the P. shigelloides hugA CPA assay; 3 stool
specimens and 1 water sample were positive by culture and CPA, but
PCR did not detect P. shigelloides in these samples. The
reduced detection rate of PCR may be due to copy numbers of the
P. shigelloides template that were less than the LoD, or the
presence of PCR-specific inhibitors that may have affected the
reaction sensitivity.
In conclusion, to the best of our knowledge, this is
the first report of a CPA assay for the rapid detection of P.
shigelloides. Compared with currently existing PCR methods, the
P. shigelloides hugA CPA assay offers the advantages of
improved sensitivity, rapidity, detection capability and ease of
operation. In general, the CPA assay provides increased flexibility
for clinical applications, and the isothermal amplification feature
provides a potential method for the simple and rapid detection of
P. shigelloides in basic clinical and field laboratories
with limited resources.
Acknowledgements
This work was supported by the Mega Project of
Research on The Prevention and Control of HIV/AIDS, Viral Hepatitis
Infectious Diseases (grant nos. 2011ZX10004-001 and
2013ZX10004-101) from the Ministry of Science and Technology and
the State Key Laboratory of Infectious Disease Prevention and
Control, Chinese Center for Disease Control and Prevention (grant
no. 2015SKLID507).
References
|
1
|
Garrity GM, Bell JA and Lilburn TG:
Taxonomic outline of the procaryotes. Bergey's Manual of Systematic
Bacteriology. 2nd edition. Release 4.0. http://141.150.157,2003.80/bergeysoutline/main.html
|
|
2
|
Bodhidatta L, McDaniel P, Sornsakrin S,
Srijan A, Serichantalergs O and Mason CJ: Case-control study of
diarrheal disease etiology in a remote rural area in Western
Thailand. Am J Trop Med Hyg. 83:1106–1109. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Krovacek K, Eriksson LM, González-Rey C,
Rosinsky J and Ciznar I: Isolation, biochemical and serological
characterisation of Plesiomonas shigelloides from freshwater in
Northern Europe. Comp Immunol Microbiol Infect Dis. 23:45–51. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Reinhardt JF and George WL: Plesiomonas
shigelloides-associated diarrhea. Jama. 253:3294–3295. 1985.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Aquilini E, Merino S, Regué M and Tomás
JM: Genomic and proteomic studies on Plesiomonas shigelloides
lipopolysaccharide core biosynthesis. J Bacteriol. 196:556–567.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
González-Rey C, Svenson SB, Bravo L,
Rosinsky J, Ciznar I and Krovacek K: Specific detection of
Plesiomonas shigelloides isolated from aquatic environments,
animals and human diarrhoeal cases by PCR based on 23S rRNA gene.
FEMS Immunol Med Microbiol. 29:107–113. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen X, Chen Y, Yang Q, Kong H, Yu F, Han
D, Zheng S, Cui D and Li L: Plesiomonas shigelloides infection in
Southeast China. PloS One. 8:e778772013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Schneider F, Lang N, Reibke R, Michaely
HJ, Hiddemann W and Ostermann H: Plesiomonas shigelloides
pneumonia. Med Mal Infect. 39:397–400. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ozdemir O, Sari S, Terzioglu S and
Zenciroglu A: Plesiomonas shigelloides sepsis and
meningoencephalitis in a surviving neonate. J Microbiol Immunol
Infect. 43:344–346. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Auxiliadora-Martins M,
Bellissimo-Rodrigues F, Viana JM, Teixeira GC, Nicolini EA,
Cordeiro KS, Colozza G, Martinez R, Martins-Filho OA and
Basile-Filho A: Septic shock caused by Plesiomonas shigelloides in
a patient with sickle beta-zero thalassemia. Heart Lung.
39:335–339. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chan SS, Ng KC, Lyon DJ, Cheung WL, Cheng
AF and Rainer TH: Acute bacterial gastroenteritis: A study of adult
patients with positive stool cultures treated in the emergency
department. Emerg Med J. 20:335–338. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xia FQ, Liu PN and Zhou YH:
Meningoencephalitis caused by Plesiomonas shigelloides in a Chinese
neonate: Case report and literature review. Ital J Pediatr.
41:32015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pence MA: The brief case: Wound infection
with Plesiomonas shigelloides following a freshwater injury. J Clin
Microbiol. 54:1180–1182. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Meng S, Xu J, Xiong Y and Ye C: Rapid and
sensitive detection of Plesiomonas shigelloides by loop-mediated
isothermal amplification of the hugA gene. PloS One. 7:e419782012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mori Y and Notomi T: Loop-mediated
isothermal amplification (LAMP): A rapid, accurate and
cost-effective diagnostic method for infectious diseases. J Infect
Chemother. 15:62–69. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Parida M, Sannarangaiah S, Dash PK, Rao PV
and Morita K: Loop mediated isothermal amplification (LAMP): A new
generation of innovative gene amplification techniquperspectives in
clinical diagnosis of infectious diseases. Rev Med Virol.
18:407–421. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang Y, Wang Y, Ma A, Li D and Ye C: Rapid
and sensitive detection of Listeria monocytogenes by cross-priming
amplification of lmo0733 gene. FEMS Microbiol Lett. Oct
1–2014.(Epub ahead of print). View Article : Google Scholar
|
|
18
|
Yang HL, Huang J, Yang B, Liu F and Zhang
QL: The establishment and application of isothermal cross-priming
amplification techniques in detecting penaeid shrimp white spot
syndrome virus. Lett Appl Microbiol. 59:200–206. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang F, Wang L, Fan K, Wu J and Ying Y:
The detection of T-Nos, a genetic element present in GMOs, by
cross-priming isothermal amplification with real-time fluorescence.
Anal Bioanal Chem. 406:3069–3078. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bai Z, Xie H, You Q, Pickerill S, Zhang Y,
Li T, Geng J, Hu L, Shan H and Di B: Isothermal cross-priming
amplification implementation study. Lett Appl Microbiol.
60:205–209. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Janda JM and Abbott SL: Expression of
hemolytic activity by Plesiomonas shigelloides. J Clin Microbiol.
31:1206–1208. 1993.PubMed/NCBI
|
|
22
|
Santos JA, González CJ, López TM, Otero A
and García-López ML: Hemolytic and elastolytic activities
influenced by iron in Plesiomonas shigelloides. J Food Prot.
62:1475–1477. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Villarreal DM, Phillips CL, Kelley AM,
Villarreal S, Villaloboz A, Hernandez P, Olson JS and Henderson DP:
Enhancement of recombinant hemoglobin production in Escherichia
coli BL21(DE3) containing the Plesiomonas shigelloides heme
transport system. Appl Environ Microbiol. 74:5854–5856. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Oldham AL, Wood TA and Henderson DP:
Plesiomonas shigelloides hugZ encodes an iron-regulated heme
binding protein required for heme iron utilization. Can J
Microbiol. 54:97–102. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Herrera FC, Santos JA, Otero A and
García-López ML: Occurrence of Plesiomonas shigelloides in
displayed portions of saltwater fish determined by a PCR assay
based on the hugA gene. Int J Food Microbiol. 108:233–238. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wandersman C and Stojiljkovic I: Bacterial
heme sources: The role of heme, hemoprotein receptors and
hemophores. Curr Opin Microbiol. 3:215–220. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xu G, Hu L, Zhong H, Wang H, Yusa S, Weiss
TC, Romaniuk PJ, Pickerill S and You Q: Cross priming
amplification: Mechanism and optimization for isothermal DNA
amplification. Sci Rep. 2:2462012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Okon E, Bishburg E, Ugras S, Chan T and
Wang H: Clostridium perfringens meningitis, Plesiomonas
shigelloides sepsis: A lethal combination. Am J Case Rep. 14:70–72.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wozniakowski G, Niczyporuk JS,
Samorek-Salamonowicz E and Gaweł A: The development and evaluation
of cross-priming amplification for the detection of avian reovirus.
J Appl Microbiol. 118:528–536. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fang R, Li X, Hu L, You Q, Li J, Wu J, Xu
P, Zhong H, Luo Y, Mei J and Gao Q: Cross-priming amplification for
rapid detection of Mycobacterium tuberculosis in sputum specimens.
J Clin Microbiol. 47:845–847. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang H, Feng S, Zhao Y, Wang S and Lu X:
Detection of Yersinia enterocolitica in milk powders by
cross-priming amplification combined with immunoblotting analysis.
Int J Food Microbiol. 214:77–82. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang X, Du XJ, Guan C, Li P, Zheng WJ and
Wang S: Detection of Vibrio cholerae by isothermal cross-priming
amplification combined with nucleic acid detection strip analysis.
Mol Cell Probes. 29:208–214. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Qiao B, Cui JY, Sun L, Yang S and Zhao YL:
Cross-priming amplification targeting the coagulase gene for rapid
detection of coagulase-positive Staphylococci. J Appl Microbiol.
119:188–195. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ke Y, Wang Y, Wang Z, Du X, Huang L and
Chen Z: Sensitive and rapid detection of blaNDM-1 in clinical
samples by isothermal cross-priming amplification. J Microbiol
Methods. 95:215–217. 2013. View Article : Google Scholar : PubMed/NCBI
|