Introduction
As the most frequent primary malignant bone tumor,
osteosarcoma occurs most commonly in children and adolescents, and
presents with a high risk of metastasis (1). It predominantly occurs around regions
involved in the processes of bone growth and repair, including the
knee joint, lower femur and upper tibia. Osteosarcoma presents with
a low 5-year survival rate, with pulmonary metastases as a common
cause of death of patients (2).
Therefore, in addition to the traditional surgical cytoreduction of
the primary tumor and chemotherapy, the critical strategy for
improvement of the prognosis of patients carrying osteosarcoma is
to prevent the pulmonary metastases. MALAT-1 has been previously
demonstrated to be involved in the novel epigenetic regulatory
mechanism (3).
MALAT-1 is a novel large, noncoding RNA, which is
highly abundant and is expressed predominantly in healthy organs,
and localizes to the nucleus (4).
In addition its 3′ end can be processed to yield a tRNA-like
cytoplasmic RNA (5). In addition
to its presence in in healthy organs, MALAT-1 has been demonstrated
to be a potential marker for epithelial carcionmas (6–8) and
is significantly upregulated in lung adenocarcinoma metastasis
(5), endometrial stromal sarcoma
of the uterus (8), nonhepatic
human carcinomas (4) and placenta
previa in trophoblasts (9).
Overexpression of MALAT-1 has been observed to predict unfavorable
outcomes of drug therapy in patients with osteosarcoma (6). Metastasis has been associated with
upregulation of MALAT-1 by functioning as an epigenetic regulator
(10), however, the regulatory
mechanism of MALAT-1 expression levels remains to be fully
elucidated.
Specificity protein 1 (Sp1) binds to GC-rich DNA
regions through three C2H2-type zinc fingers in the C-terminal
domain (11). When compared with
adjacent normal tissues, tumor tissues exhibit significantly
increased expression levels of Sp1, including in breast cancer,
gastric tumors and lung cancer (12–14),
indicating a critical role in tumorigenesis. Increasing evidence
indicates that the Sp1 protein promotes the metastasis of numerous
tumor types via an unknown mechanism. In a recent study, it was
been demonstrated that Sp1 activates the promoter of MALAT-1 gene
by binding to its promoter region. Knockdown of Sp1 markedly
reduced the MALAT-1 expression levels and thus caused the
inhibition of tumor cell migration and invasion (15). The tight regulation of MALAT-1
transcription by Sp1 suggests a critical role for Sp1 in regulating
tumor metastasis.
Notably, Sp1 interacts directly with estrogen
receptor (ER) (16). The binding
activity of Sp1 protein to GC-rich oligonucleotides has been
observed to be enhanced by the addition of ER, indicating
functional synergy between Sp1 and ER. Although the regulatory
mechanism of MALAT-1 transcription by the ER/Sp1 complex remains
unclear, one possible mechanism is through 17β-estradiol (E2)
stimulation. The regulatory role of E2 on MALAT-1 expression levels
has been reported to be dose-dependent (17), however, the detailed mechanisms
remain to be fully elucidated.
In the current study, the effects of E2 treatment on
the osteosarcoma cell line U2OS were detected. It was identified
that treatment with E2 at a concentration of 10–100 pM
significantly upregulated MALAT-1 expression. Silencing of either
Sp1 or ERα was observed to abolish the regulatory effects of E2 on
MALAT-1 expression. Electrophoretic mobility shift assay (EMSA) and
the chromatin immunoprecipitation (ChIP) assay confirmed the effect
of E2 treatment on DNA binding activity of Sp1 by translocating ER
into nuclei.
Materials and methods
Plasmid construction
For Sp1 knockdown, the Sp1 shRNA plasmid (h) was
purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA).
For ERα knockdown, the short hairpin RNA (shRNA)-expressing
lentiviral vector (pLKO.1-shERα) was constructed by targeting to
5′-GTACCAATGACAAGGGAAGT-3′; scrambled control sequence,
5′-AGCCAGGCAGTATAGAGAAT-3′. For constructing the ERα expressing
vector, an approximately 1788 base pairs (bp) coding sequence was
polymerase chain reaction (PCR)-amplified from cDNA reverse
transcribed using 1 µg MCF-7 total RNA (MCF-7 cells from the
American Type Culture Collection, Manassas, VA, USA) with the First
Strand Synthesis kit (Guangzhou RiboBio Co., Ltd., Guangzhou,
China) with a poly(dT) oligonucleotide. MCF-7 RNA was used due to
high levels of ERα mRNA expression. For the following synthesis of
the coding sequence of ERα, 0.5 µl cDNA template was amplified
using PrimeSTAR® Max DNA Polymerase (Takara Bio, Inc.,
Shiga, Japan). For PCR amplification, the following primers were
used: ERα, forward 5′-GCCGGCGCTAGCATGCCATGACCCTCCACACCA−3′ and
reverse 5′-CCTTAACTTAAGCAGACCGTGGCAGGGAAACCC−3′. Subsequent to 5
min initial denaturation at 98°C, the mixture was amplified for a
total 30 cycles with a two-step cycle process that began with the
denaturation at 98°C for 15 sec and annealing and extension at 65°C
for 2 min. Subsequent to digestion with Nhe I and Hind III, the
fragment was inserted into pcDNA3.1. For construction of the
different expression vectors, the same procedure was followed
described above with the following primer pairs: Hemagglutinin
(HA)-ERα, forward
5′-GCCGGCGCTAGCTACCCATACGACGTCCCAGACTACGCTATGCCATGACCCTCCACACCA−3′
and reverse 5′-CCTTAACTTAAGCAGACCGTGGCAGGGAAACCC−3′; Flag-Sp1
forward
5′-GCCGGCGCTAGCGATTACAAGGATGACGACGATAAGATGGATGAAATGACAGCTGTGGTGA
and reverse 5′-CCTTAACTTAAGTCAGAAGCCATTGCCACTGATA.
Cell line and transfection
The human osteosarcoma cell line U2OS was purchased
from the American Type Culture Collection. The cells were cultured
in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal
bovine serum (FBS; Life technologies; Thermo Fisher Scientific,
Inc.) and 100X antibiotic-antimycotic mixed stock solution (Life
technologies; Thermo Fisher Scientific, Inc.).
For transfection, Lipofectamine 2000 (Life
technologies; Thermo Fisher Scientific, Inc.) was employed
following the manufacturer's instructions. These methods were used
to establish U2OS-shER, U2OS-shSp1, U2OS-ERα, U2OS-Sp1 and
U2OS-shScrambled.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA of the target cells were isolated using
TRIzol (Life technologies; Thermo Fisher Scientific, Inc.).
First-strand cDNA was synthesized from 1 µg total RNA using the
First Strand Synthesis kit (Guangzhou RiboBio Co., Ltd.). qPCR was
performed with a SoEva Green Super Mix (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA), and the Applied Biosystems 7500 Real-time
system (ABI 7500HT instrument; Applied Biosystems; ThermoFisher
Scientific, Inc.) was used for measurement. The reaction mixture
contained 0.2 µl cDNA, 10 µl SoEva Green Super Mix (Bio-Rad
Laboratories, Inc., Hercules, CA, USA), 0.5 µl of each primer and
8.8 µl deionized water. The primers used in the current study were
as follows: MALAT-1, forward 5′-GACTTCAGGTCTGTCTGTTCT−3′ and
reverse 5′-CAACAATCACTACTCCAAGC-3′; NEAT1, forward
5′-GCTCTGGGACCTTCGTGACTCT-3′ and reverse
5′-CTGCCTTGGCTTGGAAATGTAA-3′; HN1, forward
5′-CACAGCAAGACGAGAAGACCCTATGGAGC-3′ and reverse
5′-GTCAAGTTATTGGATCAA−3′; HOTAIR forward 5′-GACAGGGTCTGGGACAGAAG-3′
and reverse 5′-GAGTCAGAGTTCCCCACTGC-3; GAPDH, forward
5′-CTTTGGTATCGTGGAAGGACTC-3′ and reverse
5′-GTAGAGGCAGGGATGATGTTCT-3′.
Western blotting
Samples were separated by 10% SDS-PAGE and
transferred to PVDF membranes (EMD Millipore, Billerica, MA, USA)
pre-blocked in 5% powdered milk + 5% bovine serum albumin
(Sigma-Aldrich; Merck Millipore, Darmstadt, Germany) in PBS
containing 0.3% Tween 20 for 30 min at room temperature.
Subsequently, the blot-transferred membrane was incubated at 4°C
overnight with the following primary antibodies from Abcam
(Cambridge, UK): Anti-SP1 (ab13370; 1:1,000), anti-ERα (ab32063;
1:1,000), anti-GAPDH (ab8245; 1:1,000) and anti-TIM antibody
(ab66062; 1:1,000). After washing three times with PBS containing
0.3% Tween 20, the membrane was incubated with goat anti-rabbit
horseradish peroxidase (HRP)-labeled IgG secondary antibodies
(ab6721; 1:5,000; Abcam) for 1 h at room temperature. Then membrane
was then developed by using ECL detection systems (Thermo Fisher
Scientific, Inc.).
ChIP
For the ChIP assay, 5×106 cells were
cross-linked with ice-cold 1% formaldehyde in 4°C for 15 min and
washed three times with ice-cold PBS. Cells were collected with
lysis buffer. DNA was then sheared to an approximate length of
200–500 bp. Sheared DNA (100 µl) was incubated with 10–15 µg
primary antibody or rabbit IgG (negative control) followed by IP
with 50 µl protein A agarose beads (Life Technologies; Thermo
Fisher Scientific, Inc.) during an overnight incubation at 4°C with
rotation. Enriched DNA was extracted from the DNA/antibody/protein
A bead complexes by proteinase K digestion, reverse crosslinking
process at 65°C for 4 h and purification via centrifugation at
1,000 × g for 10 min at 4°C. The IP product was amplified
using the MALAT-1 promoter region: Forward GGAAGTTGGGCAGCAGCTCCACG
and reverse CCACTGGTTCTAACCGGCTCTAG. Dihydrofolate reductase
5′untranslated region was included as a negative control; forward
ACCTGGTCGGCTGCACCT and reverse TTGCCCTGCCATGTCTCG.
Co-IP
Protein A agarose bead slurry (50 µl) was incubated
with 10 µg anti-Flag or anti-HA antibody overnight at 4°C with
rotation. The beads were then centrifuged at 1,000 × g for
10 min at 4°C and washed three times with PBS. The protein
A/antibody complex was then incubated with 500 µg lysate protein
overnight with rotation, followed by three washes with PBS. The
complex was then heated at 100°C for 10 min, and 15 µl eluted
sample was loaded for the following semiquantitative western blot
assay.
Isolation of nuclei
Intact nuclei were isolated from target cells using
a nuclei isolaton kit (cat. no. NUC201; Sigma-Aldrich; Merck
Millipore) following the manufacturer's instructions.
EMSA
The Sp1-binding fragment of the MALAT-1 promoter
region was 5′-biotinylated and annealed. The sequence was as
follows: 5′-CAGGCGTTAGGGCGGGGCGCGCGTGC-3′. Either 10 or 30 ng
target protein was incubated with 10 pM biotinylated DNA fragment
for 30 min at 4°C. The complexes were fractionated using 4% PAGE
gel in 0.5X TBE and transferred onto Hybond-N+ membranes (Life
Technologies; Thermo Fisher Scientific, Inc. Subsequently, the
assay was conducted and detected using HRP-conjugated streptavidin
(LightShift™ Chemiluminescent EMSA kit; Thermo Fisher Scientific,
Inc.) according to the manufacturer's instructions.
Cell counting kit-8 (CCK-8) assay for
proliferation
Target cells were seeded into a 96-well plate at a
concentration of 5,000 cells/well. After 24 h, E2 was added if
required. At 1, 2, 3 and 4 days after transfection, the cell
proliferation assay was conducted by the addition of 10 µl CCK-8
solution [Sangon Biotech (Shanghai) Co., Ltd., Shanghai, China] to
each well, followed by incubation at 37°C for 2 h. Absorbance was
measured at a wavelength of 450–620 nm using a Multiskan spectrum
microplate reader (Thermo Fisher Scientific, Inc.).
Invasion and migration analyses
Cell migration was analyzed using a scratch assay.
The cell layer that reached confluence was scratched by a 200 µl
pipette tip and incubated at 37°C for 24 h. The average extent of
wound closure was imaged. For analyzing invasion, the under surface
of the membrane was coated with Matrigel (0.01%) at 37°C for 2 h.
The lower chamber was filled with 1 ml DMEM supplemented with 10%
FBS. A total of 1×106 cells in a volume of 0.2 ml were
added to the upper chamber. Following incubation at 37°C for 24 h,
the cells on the upper surface of the transwell membrane were
removed. The cells attached to the lower surface of membrane were
stained with 1% crystal violet and imaged.
Statistical analysis and presentation
of data
The results were expressed as the means ± standard
deviation. The statistical analysis involving two groups was
performed using the means of Student's t-test. All data were
processed using SPSS software, version 19. P<0.05 was considered
to indicate a statistically significant difference.
Results
E2 treatment upregulates MALAT-1 RNA
levels in the U2OS osteosarcoma cell line
Due to the fact that ERα is strongly expressed in
U2OS cells (data not shown), the effects of E2 treatment on the
expression of lncRNAs was analyzed in U2OS cells. MALAT-1, NEAT1
and HOX transcript antisense RNA (HOTAIR), which are all associated
with malignancy and upregulated in osteosarcoma, were detected
quantitatively. As presented in Fig.
1A, 10–100 pM E2 treatment specifically and significantly
upregulated the MALAT-1 RNA levels, however did not affect NEAT1 or
HOTAIR. To determine whether the E2 treatment is mediated by the
stimulation of ERα, ERα was efficiently knockdown by a plasmid that
encodes a shERα sequence (Fig. 1B;
knockdown efficiency ~65±4.5%). As expected, knockdown of ERα
desensitized U2OS-shERα to E2, indicating the necessity of ERα. As
mentioned previously, the direct interaction between ERα and Sp1,
and the transcriptional regulatory role of Sp1 on MALAT-1 suggests
a role of Sp1 in E2-induced MALAT-1 upregulation. Similar to the
results of ERα knockdown, Sp1 knockdown also desensitized
U2OS-shSp1 to E2. In order to establish whether ERα may regulate
MALAT-1 expression without E2 stimulation, ERα was overexpressed in
U2OS. It was identified that, compared with the U2OS-vector,
MALAT-1 RNA levels were not significantly different without E2
treatment (Fig. 1E, left panel),
however were significantly upregulated with E2 treatment (Fig. 1E, right panel).
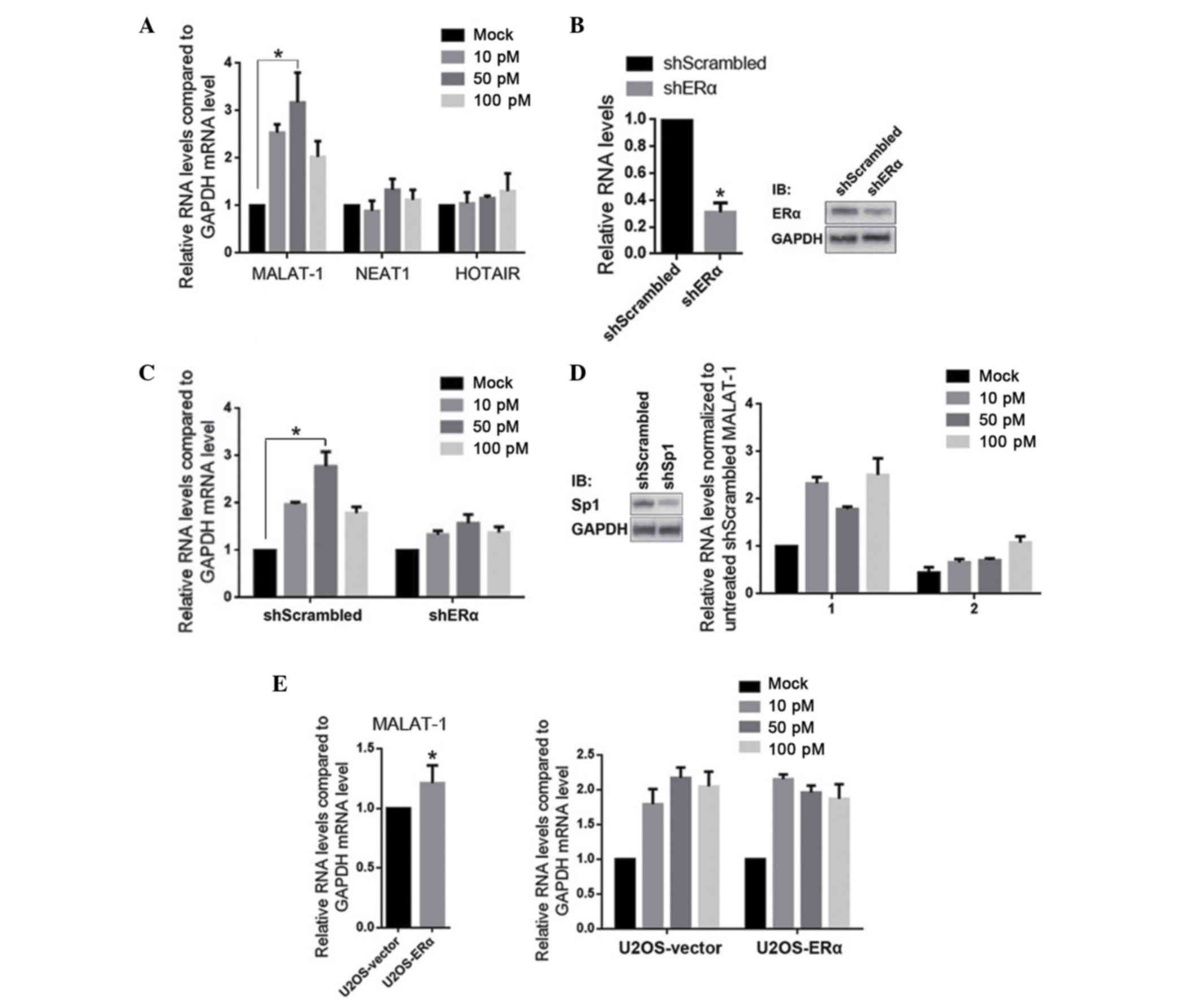 | Figure 1.E2 treatment upregulates MALAT-1,
however not NEAT1 or HOTAIR, in ERα- and Sp1-dependent manners. (A)
U2OS cells were treated with 10, 50 and 100 pM E2. After 24 h,
MALAT-1, NEAT1 and HOTAIR were detected by RT-qPCR. (B) ERα
expression was knocked down by transfection of shERα, then the ERα
mRNA (left panel) and protein (right panel) were detected. (C)
Subsequent to ERα knockdown, U2OS-shERα was exposed to 10, 50 and
100 pM E2 respectively, then the MALAT-1 levels were measured by
RT-qPCR. (D) Subsequent to the efficient knockdown of Sp1 (left
panel), the effects of E2 treatment on MALAT-1 were measured (right
panel). (E) The effect of overexpressed ERα on MALAT-1 level was
measured with (right panel) or without (left panel) E2 treatment.
*P<0.05. E2, 17β-estradiol; MALAT-1, metastasis-associated lung
adenocarcinoma transcript 1; NEAT1, nuclear-enriched abundant
transcript 1; HOTAIR, HOX transcript antisense RNA; ERα, estrogen
receptor α; Sp1, specificity protein 1; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction. |
Following E2 treatment, ERα promotes
the nuclear translocation of Sp1 and results in the increase of
Sp1/MALAT-1 promoter binding
Due to the fact that Sp1 is reported to bind to
MALAT-1 promoter region, its binding activity in ERα-knockdown U2OS
cells was analyzed. ChIP was performed with an anti-Sp1 antibody.
IgG was employed as a negative control. As presented in Fig. 2A, the binding activity of Sp1 to
the MALAT-1 promoter region increased in U2OS-shScrambled, however
not in U2OS-shERα. For the quantitative assay, IP products were
included in the qPCR reaction. Consistently, the stimulation of
MALAT-1 promoter binding activity of Sp1 by E2 treatment was
abolished when ERα was knocked down (Fig. 2B). As it has been previously
demonstrated, E2 treatment will translocate ERα from the cell
membrane into the nucleus, it was hypothesized that the increase of
Sp1/DNA binding activity may be associated with the subcellular
location. The immunofluorescence assay indicated that, following E2
treatment, Sp1 accumulated in the nuclei instead of the cytoplasm
(Fig. 2C). For further
confirmation, nuclear fractions from E2-treated U2OS cells were
isolated. GAPDH (cytoplasm specific protein) and translocase of the
inner membrane (mitochondrial specific protein) were employed as
indicators of purity. Western blotting identified nuclear
fractions, indicating acceptable cytoplasm and mitochondria
contamination (Fig. 2D). Following
E2 treatment, ERα and Sp1, particularly Sp1, were significantly
accumulated in the nuclear fraction.
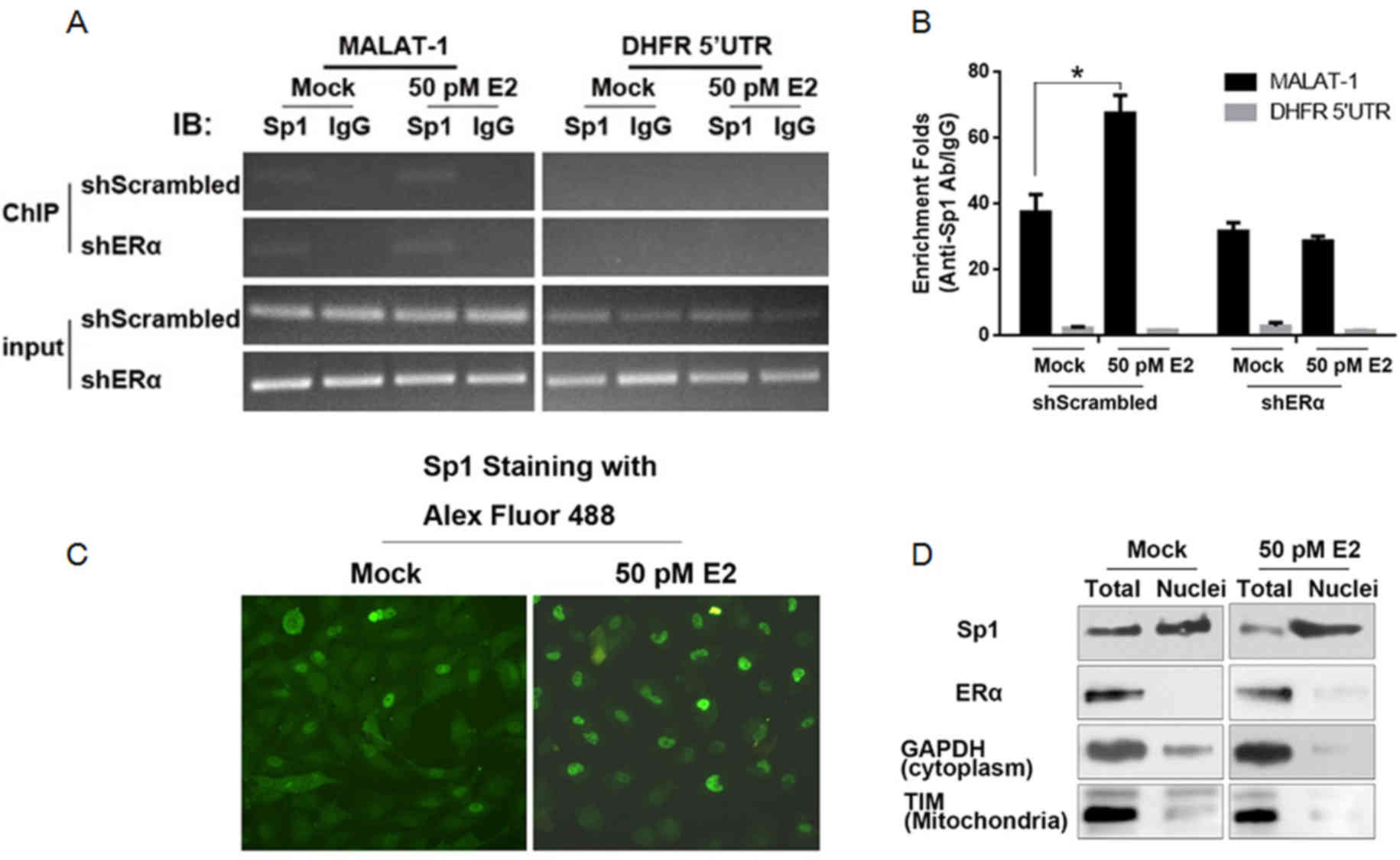 | Figure 2.E2 treatment promotes nuclei
translocation of Sp1 in the presence of ERα. (A) ChIP assay was
performed to detect the binding of Sp1 to MALAT-1 promoter region
following E2 stimulation. (B) ChIP products were detected by
semiquantitative PCR and quantitative PCR. (C) Following E2
treatment, Sp1 was stained with the Alex Fluor 488-labeled
antibody. (D) Following E2 treatment, nuclei of U2OS cells were
isolated. The mitochondria-specific TIM, cytoplasm-specific GAPDH,
ERα and Sp1 were measured in this fraction compared with total
protein. *P<0.05. E2, 17β-estradiol; Sp1, specificity protein 1;
ERα, estrogen receptor α; ChIP, chromatin immunoprecipitation;
MALAT-1, metastasis-associated lung adenocarcinoma transcript 1;
PCR, polymerase chain reaction; TIM, translocase of the inner
membrane. |
ERα binds directly to Sp1, however not
to the MALAT-1 promoter region in vitro
Plasmids containing the HA-tagged ERα or Flag-tagged
Sp1 coding sequences were established, and they were introduced
into HEK293 cells for transient expression. Cell lysates were
immunoprecipitated with anti-HA or anti-Flag antibodies
individually. Subsequently, the IP products were assayed by
immunoblotting using anti-HA or anti-Flag antibodies separately. As
presented in Fig. 3A, Flag-tagged
and HA-tagged proteins were detectable in the IP products,
suggesting the direct binding of Sp1/ERα. It was additionally
investigated whether ERα binds to the MALAT-1 promoter by itself.
In order to establish this, the EMSA assay was conducted. The
MALAT-1 promoter region DNA sequence was labeled with biotin was
incubated with purified HA-tagged ERα or Flag-tagged Sp1 separately
and fractionated by 4% 0.5X TBE PAGE gel. The binding bands were
observed only in Sp1 involved lanes. For further confirmation, E2
treated U2OS cells underwent a ChIP assay. Following E2 treatment,
anti-ERα and anti-Sp1 antibodies enriched the MALAT-1 promoter
region. However, without E2 treatment, the anti-ERα antibody failed
to enrich the MALAT-1 promoter region, while the anti-Sp1 antibody
was observed to exhibit no difference to the E2-treated sample
(Fig. 3C). Taken together, ERα
binds to the Sp1 protein, however not to the MALAT-1 promoter.
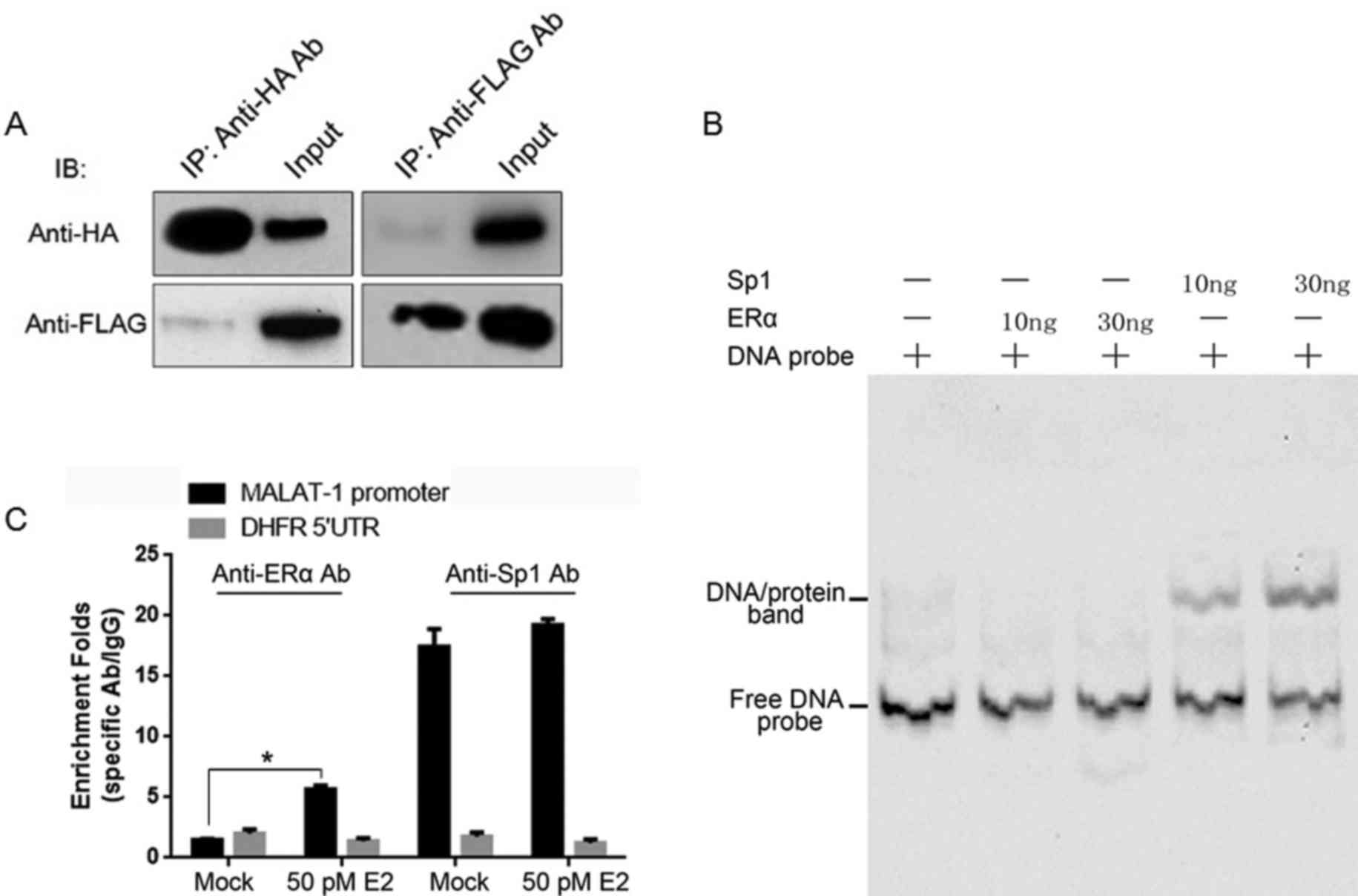 | Figure 3.ERα binds to Sp1, however not to the
MALAT-1 promoter region. (A) Plasmids expressing Flag-tagged ERα
and HA-tagged Sp1 were co-transfected into HEK293 cells. After 48
h, co-IPs were performed using anti-Flag or -HA antibodies,
respectively. (B) The binding of Sp1 or ERα to the MALAT-1 promoter
region was further confirmed in vitro by the electrophoretic
mobility shift assay. (C) Plasmids expressing Flag-tagged ERα and
HA-tagged Sp1 were co-transfected into U2OS cells, and after 48 h,
ChIPs were performed with anti-Flag and -HA antibodies or IgG.
*P<0.05. E2, 17β-estradiol; Sp1, specificity protein 1; MALAT-1,
metastasis-associated lung adenocarcinoma transcript 1; ERα,
estrogen receptor α; HA, hemagglutinin; IP, immunoprecipitation;
ChIP, chromatin IP. |
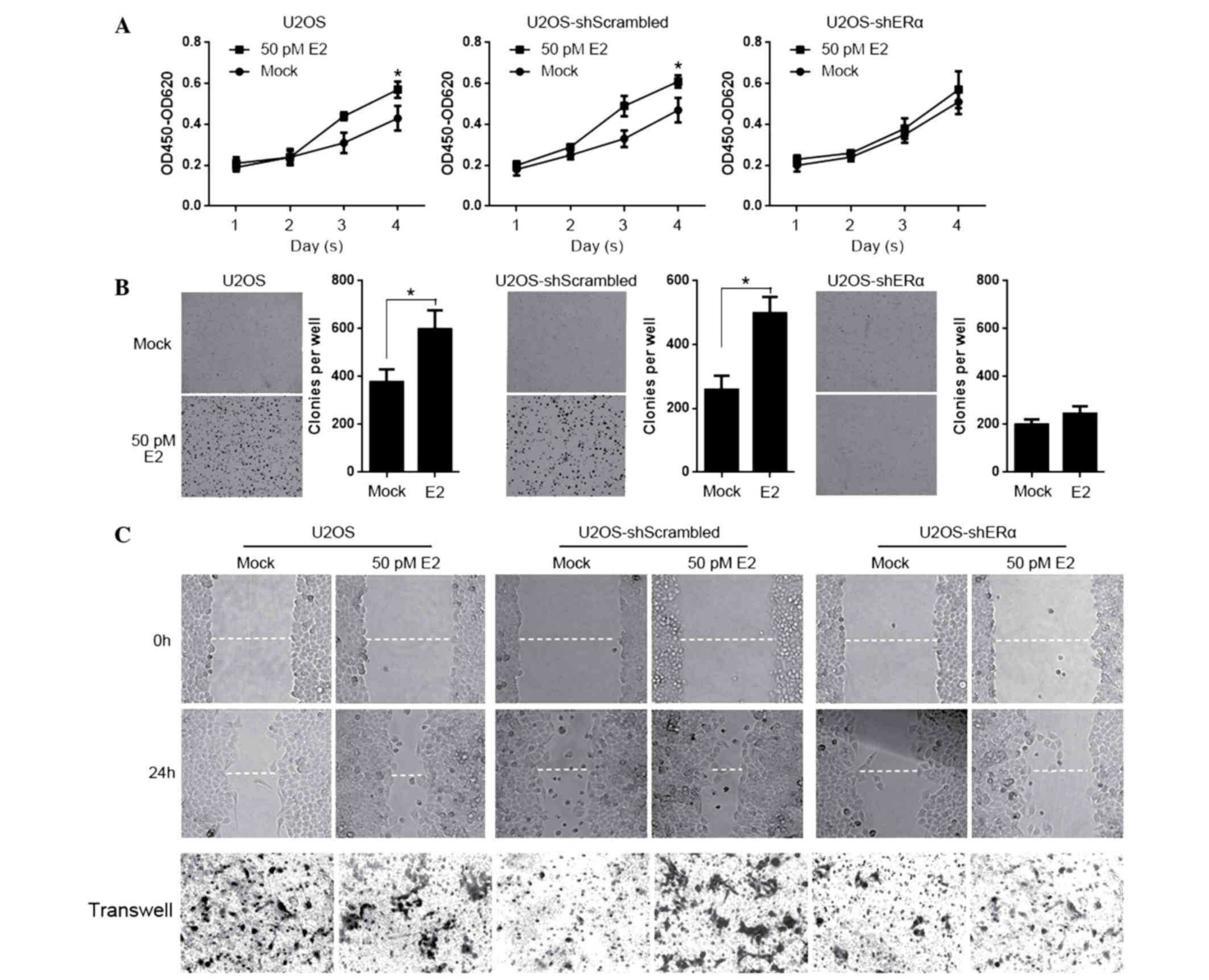
E2 treatment tightly regulates
physiological processes of U2OS cells in an ERα-dependent manner by
upregulating MALAT-1 RNA
MALAT-1 tightly regulates the physiological
processes in osteosarcoma, and aids in the determination of the
effects of E2 treatment on these processes in an ERα-dependent
manner. For testing proliferation of E2 treated cells, U2OS,
U2OS-shScrambled and U2OS-shERα were seeded into 12-wells and were
stained with CCK-8 on days 1, 2, 3 and 4. As presented in Fig. 4A, E2 treatment significantly
promoted proliferation, however not in U2OS-ERα. Subsequently,
cells were seeded in 0.3% soft agar for testing colony formation
for 14 days. Consistently, E2 promoted the colony formation,
however not in ERα-knockdown U2OS cells (Fig. 4B). In order to further determine
the effects of E2 treatment on migration and invasion, the scratch
assay and transwell assay were performed with the above mentioned
cell lines. It is observed that E2 treatment affected the migration
and invasion independent of the presence of ERα. (Fig. 4C).
Discussion
Large number of lncRNAs have now been identified and
characterized in mammals (18).
Although the function of these novel lncRNAs remains unclear,
growing evidence has indicated their crucial roles in various
biological processes through various mechanisms, including
epigenetic regulation, transcriptional regulation and
post-transcriptional regulation. Their functional activities are
identified not only in normal developmental processes, however
additionally in disease, particularly in cancer (19,20).
Originally, MALAT-1 was identified to be tightly associated with
tumorigenesis, and it is also termed nuclear-enriched abundant
transcript 2 (21). Further
research on the molecular mechanisms of MALAT-1 identified its
various regulatory roles, including regulation of cell cycle
arrest, and epigenetic regulation of collagen triple helix repeat
containing 1, chaperonin containing TCP1 subunit 4, nuclear
receptor subfamily 4 group A member 1 and polypyrimidine tract
binding protein 3 (6,22).
Previous studies have indicated that MALAT-1 is
upregulated during tumorigenesis, in metastatic tumor tissues and
additionally in patients who have poor prognosis (10,23).
However, the mechanism of MALAT-1 regulation remains unclear. In a
breast cancer cell line, it was reported that high doses of E2
treatment (100 nM) markedly downregulate MALAT-1 RNA levels in an
ERα-independent manner (24). A
previous study additionally identified this mechanism in the MG63
osteosarcoma cell (17). The
effects of physiological doses (≤100 pM) of E2 on MALAT-1
expression remains unclear.
Sp1 contains highly conserved C2H2 zinc finger
motifs in its C-terminus and binds to GC-rich sites (11). For example, it transcriptionally
activates the fibroblast growth factor receptor 1 promoter by
binding to its GC-rich region in proliferating myoblasts (25). It has additionally been observed to
activate the promoter of the human MALAT-1 gene. In vitro,
EMSA assay have indicated the binding of Sp1 protein to the MALAT-1
promoter region. The ChIP assay further confirmed this tight
binding and thus upregulation of MALAT-1 expression levels
(15). Notably, Sp1 binds directly
to ERα and serves a synergistic role in transcriptional regulation
of their target genes (26).
Considering this, the molecular mechanisms by which E2-ER
positively regulates the gene expression of MALAT-1 were
investigated in the current study.
U2OS cells were treated with with 10–100 pM E2, and
it was identified that E2 treatment significantly downregulated
MALAT-1 expression. In ERα-knockdown U2OS cells, the effects of E2
treatment were abolished, suggesting that the necessity of ERα on
the effects caused by E2 treatment. Consequently, ChIP and
immunofluorescence assays observed that, following E2 treatment,
Sp1 translocated into the nuclei dependent on the presence of ERα.
This result indicates the direct binding of ERα to Sp1 after E2
forms the E2-ER complex.
E2-ER signaling stimulates target gene expression in
two different ways. Firstly, the E2-ER complex binds directly to a
palindromic ERE or half-ERE in the promoter region of a target gene
(17); secondly, instead of
binding directly to the target gene, the E2-ER complex interacts
with other transcription factors, such as Sp1 (27). In the current study, direct binding
of ERα to Sp1 was observed with Co-IP. The presence of a
supershifted band of the ERα/Sp1/DNA complex was predicted
following addition of ERα, however no band of ERα/Sp1/DNA was
observed in EMSA (data not shown); however, the ChIP assay
indicated that ERα bound to the MALAT-1 promoter region in an
E2-dependent manner. To explain this, two theories were proposed;
firstly that the ERα/Sp1/DNA complex is too fragile to be detected
as a supershifted band in EMSAs; secondly that the unstable dynamic
balance of ERα/Sp1/DNA leads to the dissociation of this complex.
As a result of E2-treatment-induced MALAT-1 upregulation,
consistent with the results of previous studies, the proliferation,
colony formation, migration and invasion of U2OS cells were
observed to be significantly promoted.
Taken together, the current study demonstrated that
E2 treatment of U2OS cells significantly promotes the nuclear
translocation of Sp1 potentially through the formation of the
ERα/Sp1 complex. The nuclei-accumulated Sp1 activates the
transcriptional activity of MALAT-1, then the upregulated MALAT-1
RNA causes promotion of proliferation, colony formation, migration
and invasion.
Acknowledgements
The present study was supported by a Sichuan
Provincial Scientific Grant (grant nos. 2016FZ0093 and 2016FZ0096).
The authors would like to thank Mrs Huimin Shi for English language
editing.
References
|
1
|
Ma O, Cai WW, Zender L, Davaram T, Shen J,
Herron AJ, Lowe SW, Man TK, Lau CC and Donehower LA: MMP13, Birc2
(cIAP1), and Birc3 (cIAP2), amplified on chromosome 9, collaborate
with p53 deficiency in mouse osteosarcoma progression. Cancer Res.
69:2559–2567. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Guise TA, O'keefe R, Randall RL and Terek
RM: Molecular biology and therapeutics in musculoskeletal oncology.
J Bone Joint Surg Am. 91:724–732. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fellenberg J, Bernd L, Delling G, Witte D
and Zahlten-Hinguranage A: Prognostic significance of
drug-regulated genes in high-grade osteosarcoma. Mod Pathol.
20:1085–1094. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hutchinson JN, Ensminger AW, Clemson CM,
Lynch CR, Lawrence JB and Chess A: A screen for nuclear transcripts
identifies two linked noncoding RNAs associated with SC35 splicing
domains. BMC Genomics. 8:392007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ji P, Diederichs S, Wang W, Böing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel noncoding RNA, and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lin R, Maeda S, Liu C, Karin M and
Edgington TS: A large noncoding RNA is a marker for murine
hepatocellular carcinomas and a spectrum of human carcinomas.
Oncogene. 26:851–858. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Perez DS, Hoage TR, Pritchett JR,
Ducharme-Smith AL, Halling ML, Ganapathiraju SC, Streng PS and
Smith DI: Long, abundantly expressed non-coding transcripts are
altered in cancer. Hum Mol Genet. 17:642–655. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yamada K, Kano J, Tsunoda H, Yoshikawa H,
Okubo C, Ishiyama T and Noguchi M: Phenotypic characterization of
endometrial stromal sarcoma of the uterus. Cancer Sci. 97:106–112.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tseng JJ, Hsieh YT, Hsu SL and Chou MM:
Metastasis associated lung adenocarcinoma transcript 1 is
up-regulated in placenta previa increta/percreta and strongly
associated with trophoblast-like cell invasion in vitro. Mol Hum
Reprod. 15:725–731. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gutschner T, Hämmerle M, Eißmann M, Hsu J,
Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Groß M, et al: The
noncoding RNA MALAT1 is a critical regulator of the metastasis
phenotype of lung cancer cells. Cancer Res. 73:1180–1189. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li L and Davie JR: The role of Sp1 and Sp3
in normal and cancer cell biology. Ann Anat. 192:275–283. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Safe S and Abdelrahim M: Sp transcription
factor family and its role in cancer. Eur J Cancer. 41:2438–2448.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Deacon K, Onion D, Kumari R, Watson SA and
Knox AJ: Elevated SP-1 transcription factor expression and activity
drives basal and hypoxia-induced vascular endothelial growth factor
(VEGF) expression in non-small cell lung cancer. J Biol Chem.
287:39967–39981. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang L, Wei D, Huang S, Peng Z, Le X, Wu
TT, Yao J, Ajani J and Xie K: Transcription factor Sp1 expression
is a significant predictor of survival in human gastric cancer.
Clin Cancer Res. 9:6371–6380. 2003.PubMed/NCBI
|
|
15
|
Li S, Wang Q, Qiang Q, Shan H, Shi M, Chen
B, Zhao S and Yuan L: Sp1-mediated transcriptional regulation of
MALAT1 plays a critical role in tumor. J Cancer Res Clin Oncol.
141:1909–1920. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Porter W, Saville B, Hoivik D and Safe S:
Functional synergy between the transcription factor Sp1 and the
estrogen receptor. Mol Endocrinol. 11:1569–1580. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fang D, Yang H, Lin J, Teng Y, Jiang Y,
Chen J and Li Y: 17β-estradiol regulates cell proliferation, colony
formation, migration, invasion and promotes apoptosis by
upregulating miR-9 and thus degrades MALAT-1 in Osteosarcoma cell
MG-63 in an Estrogen receptor-independent manner. Biochem Biophys
Res Commun. 457:500–506. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nielsen MM, Tehler D, Vang S, Sudzina F,
Hedegaard J, Nordentoft I, Orntoft TF, Lund AH and Pedersen JS:
Identification of expressed and conserved human noncoding RNAs.
RNA. 20:236–251. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Batista PJ and Chang HY: Long noncoding
RNAs: Cellular address codes in development and disease. Cell.
152:1298–1307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wapinski O and Chang HY: Long noncoding
RNAs and human disease. Trends Cell Biol. 21:354–361. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tano K, Mizuno R, Okada T, Rakwal R,
Shibato J, Masuo Y, Ijiri K and Akimitsu N: MALAT-1 enhances cell
motility of lung adenocarcinoma cells by influencing the expression
of motility-related genes. FEBS Lett. 584:4575–4580. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yang F, Yi F, Han X, Du Q and Liang Z:
MALAT-1 interacts with hnRNP C in cell cycle regulation. FEBS Lett.
587:3175–3181. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Okugawa Y, Toiyama Y, Hur K, Toden S,
Saigusa S, Tanaka K, Inoue Y, Mohri Y, Kusunoki M, Boland CR and
Goel A: Metastasis-associated long non-coding RNA drives gastric
cancer development and promotes peritoneal metastasis.
Carcinogenesis. 35:2731–2739. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhao Z, Chen C, Liu Y and Wu C:
17β-Estradiol treatment inhibits breast cell proliferation,
migration and invasion by decreasing MALAT-1 RNA level. Biochem
Biophys Res Commun. 445:388–393. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Parakati R and DiMario JX: Repression of
myoblast proliferation and fibroblast growth factor receptor 1
promoter activity by KLF10 protein. J Biol Chem. 288:13876–13884.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gruber CJ, Tschugguel W, Schneeberger C
and Huber JC: Production and actions of estrogens. N Engl J Med.
346:340–352. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Safe S and Kim K: Non-classical genomic
estrogen receptor (ER)/specificity protein and ER/activating
protein-1 signaling pathways. J Mol Endocrinol. 41:263–275. 2008.
View Article : Google Scholar : PubMed/NCBI
|