Introduction
Mesenchymal stem cells (MSCs) were discovered in
bone marrow and are multipotent stromal cells that have the ability
to differentiate into numerous cell types (1). Therefore, MSCs have become one of the
most promising seed cells for the treatment of osteoblastic or
cardiovascular diseases (2,3).
Osteoblastic or adipocytic differentiation occurs when MSCs are
cultured in three conditions in vitro: i) Three-dimensional
culture format; ii) serum-free nutrient medium; and iii) with the
addition of inducible factors, such as bone morphogenetic protein 6
(BMP-6) and dexamethasone (4).
BMP-6 or dexamethasone may induce osteoblastic or adipocytic
differentiation of MSCs, respectively, via crosstalk with signaling
pathways, including mitogen-activated protein kinase, Wnt, Notch
and nuclear factor-kappa B (5–7).
Osteoblast- and adipocyte-associated diseases, such
as bone disorders, cartilage injury, bone damage, angiopathy and
liposarcoma, put tremendous burden on patients worldwide (8–10).
Previous studies have demonstrated that although certain treatments
have been successful, overall treatment outcomes remain
unsatisfactory (11,12). Various studies have examined the
differential expression of genes or microRNAs (miRNAs) during MSC
differentiation using bioinformatics. For example, Chen et
al (13) demonstrated that
MSCs secreted microparticles enriched in pre-miRNAs. De Luca et
al (14) revealed that
epidermal growth factor receptor signaling affected the secretome
of MSCs in breast cancer. Mrugala et al (15) performed large-scale expression
profiling using DNA microarrays on MSCs at different time points
during chondrogenic differentiation. Although a number of studies
have screened for differentially expressed genes (DEGs) or
differentially expressed miRNAs during MSC differentiation into
osteoblasts or adipocytes, an association between these two
differentiation pathways has rarely been examined.
The present study used RNA sequencing (RNA-seq)
analysis to screen for DEGs and differentially expressed miRNAs
during the differentiation of MSCs into osteoblasts or adipocytes.
Comprehensive bioinformatics methods were used to analyze the
functions of DEGs and to investigate the interaction between DEGs
and differentially expressed miRNAs. The present study aimed to
screen and identify target genes and miRNAs that are different
between osteoblastic and adipocytic differentiation of MSCs, which
may provide a theoretical basis for targeted prevention in MSC
directional differentiation.
Materials and methods
MSC differentiation induction
Human MSCs were obtained from Biomedical Sciences
Cell Center of Fudan University (Shanghai, China) and cultured in
Dulbecco's modified Eagle's medium-low glucose (DMEM-LG; Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10%
fetal bovine serum (Hyclone; GE Healthcare, Logan, UT, USA), 100
U/ml penicillin and 100 U/ml streptomycin (Gibco; Thermo Fisher
Scientific, Inc.) in an incubator at 37°C with 5% CO2.
Medium was changed every other day; non-adherent cells were removed
and MSCs were cultured to passage 3.
Passage 3 MSCs (5×104 per well) were
seeded on 6-well plates and cultured in DMEM-LG and induced towards
osteoblastic differentiation by adding 1×10−9 mol/l
dexamethasone (Sigma-Aldrich; Merck Millipore, Darmstadt, Germany),
10 mmol/l β-glycerophosphoric acid, disodium salt pentahydrate
(Meryer Chemical Technology Co., Ltd., Shanghai, China) and 0.2
mmol/l sodium ascorbate (Merck Millipore). Adipocytic
differentiation was induced by culturing MSCs in DMEM-high glucose
medium with 1×10−7 mol/l dexamethasone, 0.5 mmol/l
3-isobutyl-1-methylxanthine and 0.05 mmol/l indomethacin. Prior to
induction, 50 ng/ml BMP-6 (Prospec-Tany TechnoGene Ltd., East
Brunswick, NJ, USA) was added to each well and cells were cultured
for 24 h. The induced samples were separated into 3 groups based on
the time point (n=3/group): i) Osteoblast (OB)/at day (AD) 7, the
BMP-6 induced sample at 7 days; ii) OB/AD14, the BMP-6 induced
sample at 14 days; and iii) OB/AD21, the BMP-6 induced sample at 21
days. Induced MSCs without BMP-6 at 0 day were used as the control
(n=3).
Data preprocessing
Total RNAs was extracted using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) and the quality of
RNAs were evaluated by modified formaldehyde agarose gel
electrophoresis and OD260/OD280 absorbance
ratio detection using a Genova Nano spectrophotometer (Bibby
Scientific; Cole-Palmer, Stone, UK). RNA-seq data from the
osteoblastic or adipocytic differentiation induced MSCs were
obtained at 7, 14 or 21 days using SMARTer Universal Low Input RNA
kit for sequencing (Clontech Laboratories, Inc., Mountainview, CA,
USA), according to the manufacturer's protocol. The Fast-QC
software (Babraham Bioinformatics, Cambridge, UK; http://www.bioinformatics.babraham.ac.uk/projects/fastqc)
was used to assess the data, including the quality value
distribution of bases. RNA sequences were subsequently mapped to
human genome sequences using TopHat software 1.3 release (Center
for Computational Biology, John Hopkins University, Baltimore, MD,
USA; https://ccb.jhu.edu/software/tophat/index.shtml)
(16). Expression values of mRNAs
were calculated based on the fragments per kilobase of exon, per
million of fragments mapped and fragment length.
Screening differentially expressed
mRNA and miRNA
DEGs in each group at each of the three time points
were screened and compared with the control (cells at day 0 that
had not been exposed to BMP-6) using the t-test function in
Bioconductor 3.4 (https://bioconductor.org) with a false discovery rate
<0.05 and |log2FC| >2, where FC is fold change. In
addition, differentially expressed miRNAs at each time point were
screened compared with the control using |log2FC|
>2.
Series cluster analysis
The DEGs and differentially expressed miRNAs
screened from four time point samples (0, 7, 14 and 21 days) were
used for series cluster analysis. The correlation between time and
expressed value of differentially expressed genes/miRNAs was
calculated using the agglomerative hierarchical clustering method
using SPSS 19.0 (IBM SPSS, Armonk, NY, USA); the correlation index
was 0.85 for series cluster analysis.
Gene ontology (GO) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathway enrichment
analysis of DEGs
To further explore the functions of selected DEGs,
biological processes and pathways were analyzed using the GO
database (http://www.geneontology.org/page/go-database) and the
KEGG database (http://www.genome.jp/kegg), respectively (17,18).
Enrichment analysis was performed by Fisher's exact test using the
Database for Annotation, Visualization and Integrated Discovery
(https://david.ncifcrf.gov)
bioinformatics resource based on a hypergeometric distribution
(19); P<0.01 was used as the
threshold.
Target gene prediction for
differentially expressed miRNAs
All screened differentially expressed miRNAs were
imported into the TargetScanHuman database version 7.1 (http://www.targetscan.org/vert_71) to predict
miRNA target genes (20).
Construction of miRNA-mRNA interaction
network
Since osteoblastic and adipocytic differentiation
were both induced from the MSCs, the DEGs with opposite expression
characteristics were used to indicate the differentiation status of
the two types of cells. To investigate the negative correlations
between the DEGs and differentially expressed miRNAs, a miRNA-mRNA
interaction network was constructed with the protein-protein
interaction network using Cytoscape 3.4.0 software (http://www.cytoscape.org) and the Reactome Pathway
Database (http://www.reactome.org); miRNAs and
mRNAs with a plus tendency value of 25 were chosen for interaction
pairs.
Results
Screening differentially expressed
mRNAs and miRNAs
DEGs that were identified in the
BMP-6/dexamethasone-induced samples at three time points (7, 14 and
21 days) were screened and compared with the control samples (day
0) that were not treated with BMP-6. Among the cells undergoing
osteoblastic differentiation, 148 (58 upregulated and 90
downregulated) DEGs were selected at the 7-day time point, 293 (181
upregulated and 112 downregulated) at day 14 and 417 (253
upregulated and 164 downregulated) at day 21; whereas 45 (33
upregulated and 12 downregulated), 28 (11 upregulated and 17
downregulated) and 203 (21 upregulated and 182 downregulated)
differentially expressed miRNAs were selected at the respective
time points. Among the adipocyte differentiating groups, 572 (340
upregulated and 232 downregulated) DEGs were selected at the 7-day
time point, 753 (419 upregulated and 334 downregulated) at day 14
and 829 (466 upregulated and 363 downregulated) at day 21. A total
of 48 (20 upregulated and 28 downregulated), 48 (22 upregulated and
26 downregulated) and 66 (26 upregulated and 40 downregulated)
differentially expressed miRNAs were selected at the three
respective time points (data not shown).
GO and KEGG pathway enrichment
analysis of DEGs
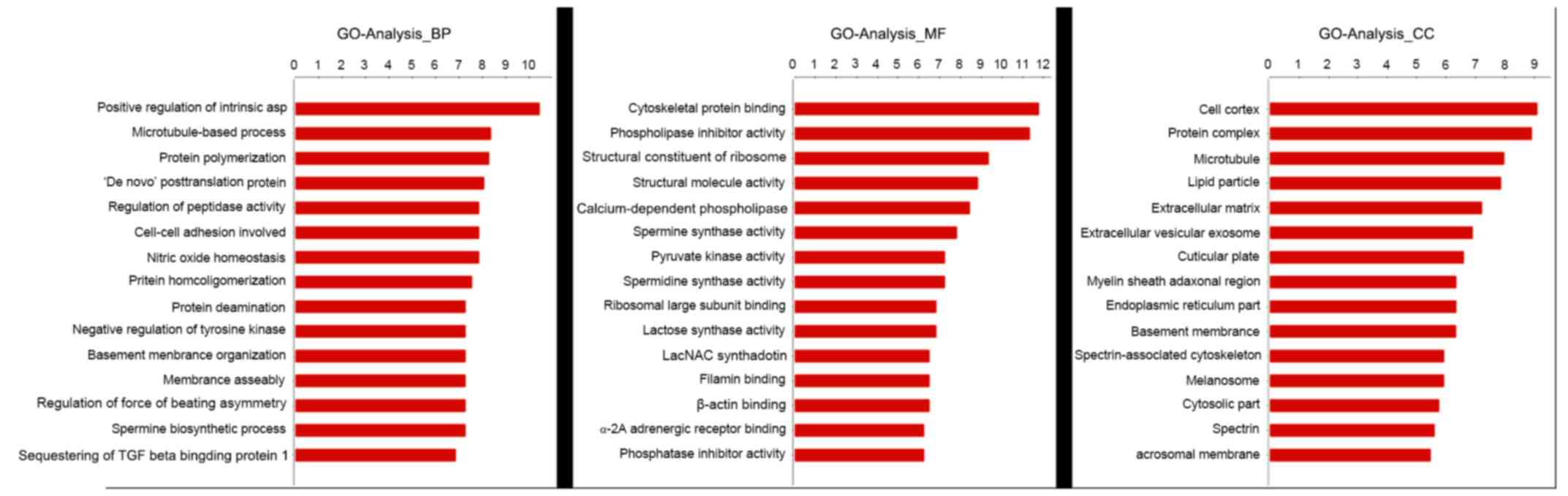
The results of data analysis suggested that GO terms
analyzed for MSCs included biological processes (such as the
positive regulation of integrin, microtubule-based process and
protein polymerization), molecular functions (such as cytoskeletal
protein binding and phospholipase inhibitor activity) and cellular
components (such as the cell cortex, protein complex and
microtubule) as presented in Fig.
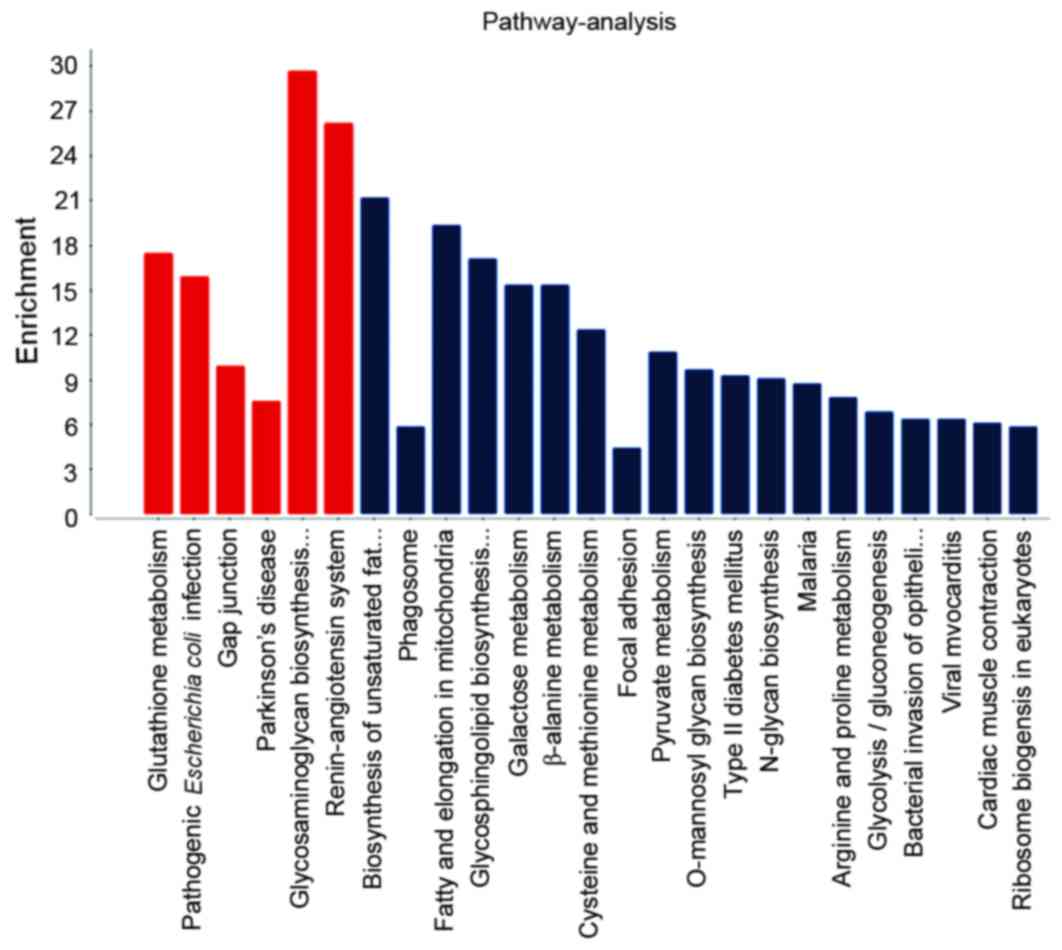
1. KEGG pathway analysis for MSC differentiation-related DEGs
revealed enrichment in glutathione metabolism, pathogenic
Escherichia coli infection and Parkinson's disease (Fig. 2).
miRNA-mRNA interaction network
The miRNA might negatively regulate mRNA expression
via degradation and translation processes. Therefore, to further
investigate interaction correlations between the screened DEGs and
the differentially expressed miRNAs, a miRNA-mRNA interaction
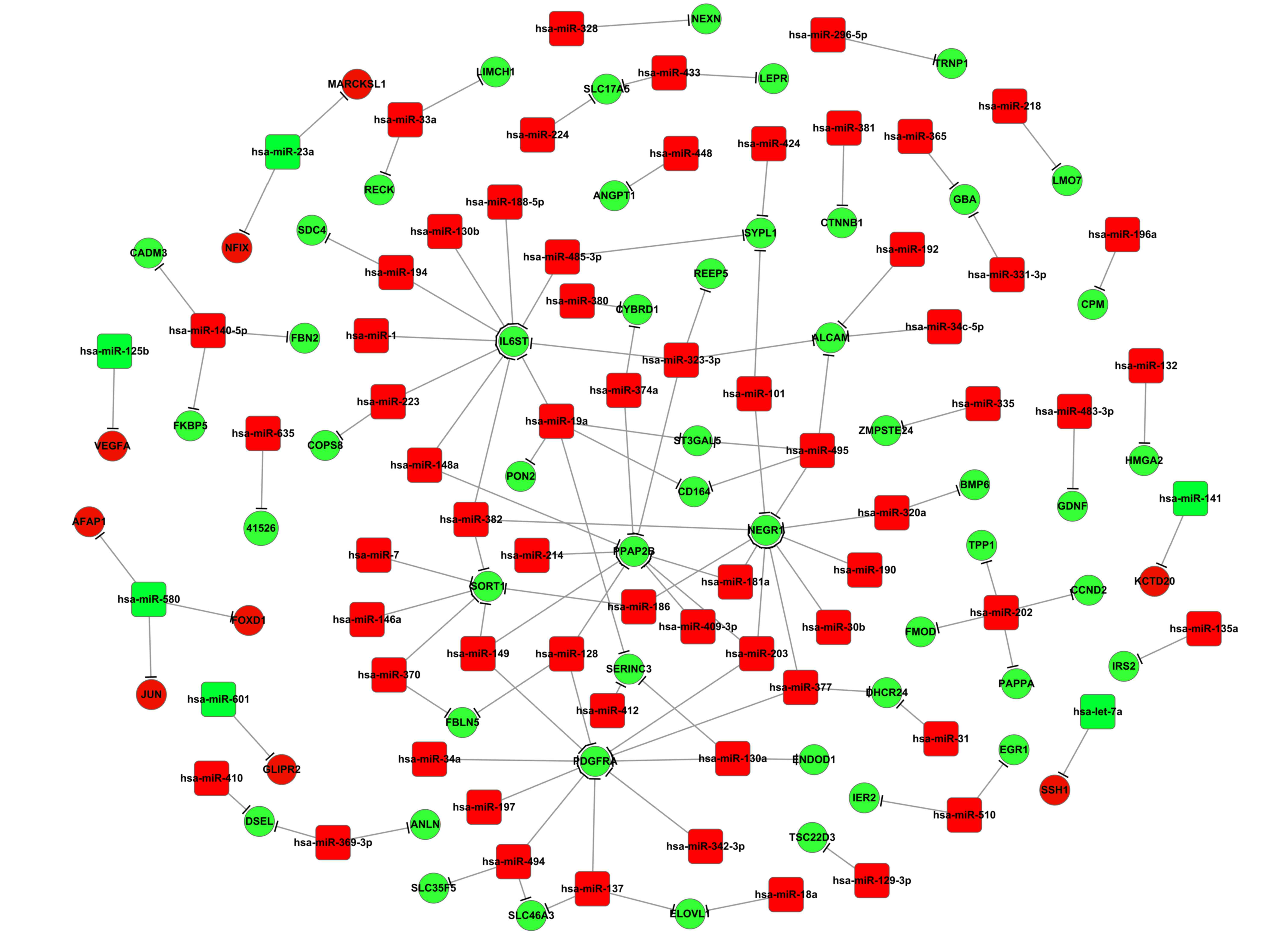
network was constructed. Results from the present study
demonstrated that during osteoblastic differentiation, one target
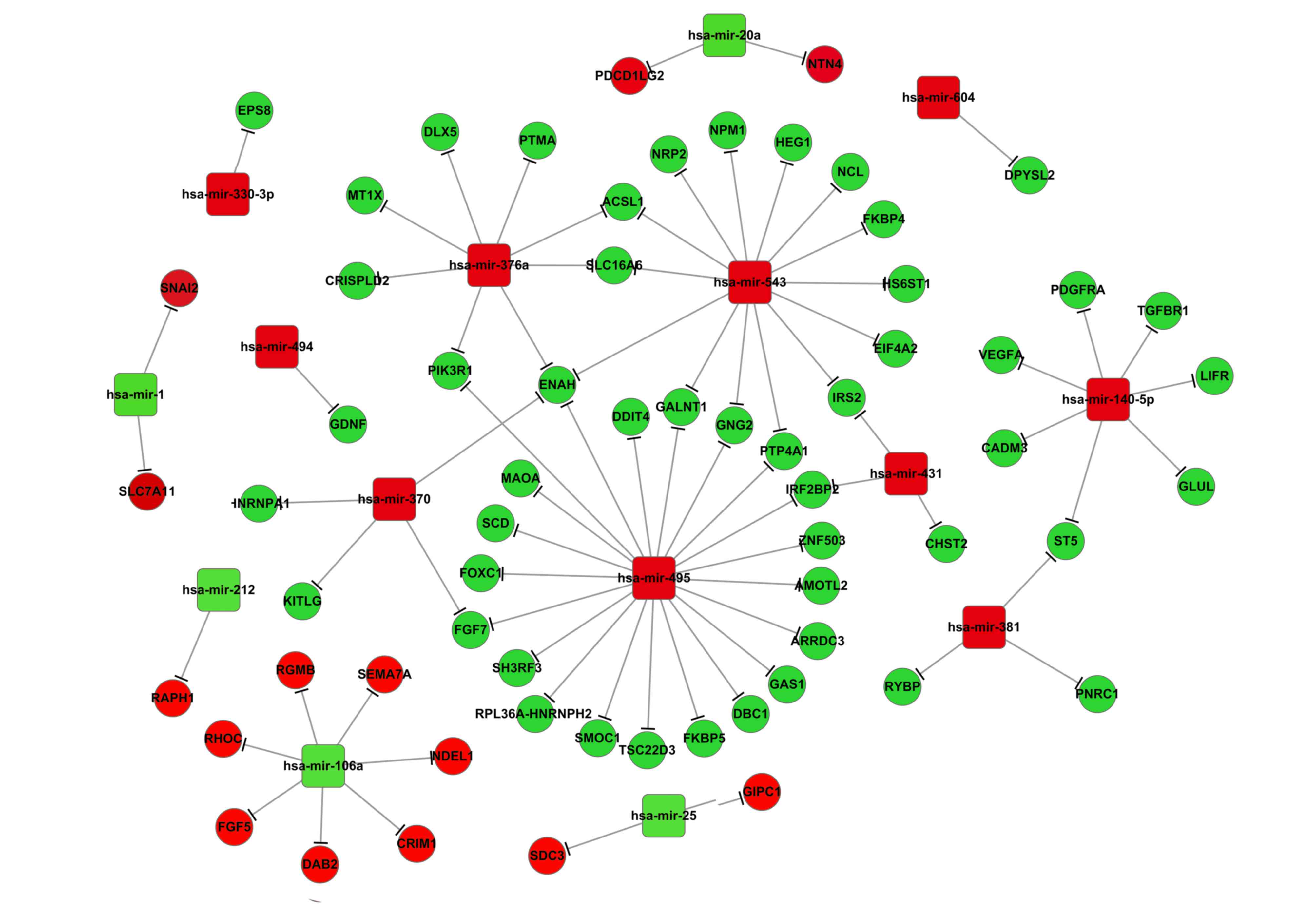
gene may be regulated by several miRNAs (Fig. 3). This was also observed in
adipocytes (Fig. 4). For example,
upregulated DEGs, including neuronal growth regulator 1 (NEGR1),
interleukin 6 signal transducer (IL6ST), platelet-derived growth
factor receptor (PDGFR), enhanced homolog (ENAH), and activated
leukocyte cell adhesion molecule (ALCAM), may be targeted by >1
differentially expressed miRNA (Fig.
3). During adipocytic differentiation, the downregulated
miRNAs, including miR-495, miR-543, miR-376a and miR-140-5p, and
the upregulated miR-106a, had more target genes compared with the
other miRNAs examined in adipocytic differentiation (Fig. 4). It was also demonstrated that
miR-145 was downregulated in both adipocytes and osteoblasts, and
miR-140-5p was associated with the expression of CADM3 in the two
differentiation pathways (Fig.
4).
Discussion
Previous studies have demonstrated that MSCs have
the ability to differentiate into multiple cell types (21), and have identified possible
applications of MSCs in cell replacement therapies in various
diseases (4,22). In the present study, DEGs and
differentially expressed miRNAs were screened during MSC
differentiation into either adipocytes or osteoblasts. miRNAs
including miR-382 and miR-203, and DEGs including NEGR1,
phosphatidic acid phosphatase 2B (PPAP2B), PDGFRA, IL6ST and
sortilin 1 (SORT1) were identified as having a potential
involvement in osteoblastic differentiation. In addition, the
downregulated miR-495, miR-376a and miR-543, the upregulated
miR-106a, the upregulated DEGs including ENAH,
N-acetylgalactosaminyltransferase 1 (GALNT1) and acyl-CoA
synthetase long-chain family member 1 (ACSL1), and the
downregulated repulsive guidance molecule family member B (RGMB)
and semaphorin 7A (SEMA7A) were identified as having a possible
role in adipocytic differentiation.
Results from the present study suggested that
miR-203 was a common regulator for certain DEGs, including NEGR1,
PPAP2B and PDGFRA, whereas miR-382 was a common regulator of IL6ST,
NEGR1 and SORT1 (Fig. 3),
suggesting their crucial roles in regulating osteoblastic
differentiation of MSCs. NEGR1 has been indicated to be involved in
the cell adhesion pathway (23).
Arai et al (24)
demonstrated that MSCs expressed ALCAM when involved in bone marrow
formation. PPAP2B, which is a member of the phosphatidic acid
phosphatase family, has been revealed to convert phosphatidic acid
to diacylglycerol (25); and a
previous study demonstrated that PPAP2B was involved in peroxisome
proliferator-activated receptor-γ-induced osteoblastic cell
differentiation in MSCs (26).
Alison and Islam (27) revealed
that miR-203 was involved in MSC differentiation by targeting
several genes. Therefore, the present study hypothesized that the
downregulated expression of miR-203 may promote MSC osteoblastic
differentiation, which may be associated with the upregulated
expression of DEGs, including NEGR1, PPAP2B and PDGFRA. In
addition, IL6ST and NEGR1 have been demonstrated to be associated
with MSC osteoblastic differentiation (26,28),
whereas SORT1 was indicated to participate in the regulation of
osteoblastic differentiation of human MSCs in vitro
(29). The decay of miR-382 has
been demonstrated to be important in osteoblastic differentiation
of human MSCs (10,30). The present study indicated that
miR-382 was likely to be involved in regulating osteoblastic
differentiation from human MSCs by targeting DEGs, such as IL6ST,
NEGR1 and SORT1.
Results from the present study indicated that ENAH
mRNA may be a common target of miRNAs, including miR-495, miR-376a,
miR-543 and miR-370, suggesting a role for this DEG in MSCs
differentiation into adipocytes (Fig.
4). ENAH is an actin-associated protein that is involved in
various cytoskeleton remodeling and cell polarity processes, such
as axon guidance and lamellipodia formation during cell migration
(31). Microarray analyses by
Charoenpanich et al (32)
demonstrated that ENAH was differentially expressed in human
adipose-derived adult stem cells undergoing osteoblastic
differentiation under uniaxial cyclic tensile strain. The
differentially expressed miRNAs identified in the present study,
including miR-495, miR-376a, miR-543 and miR-370, have been
examined in previous MSC differentiation studies. For example, Gao
et al (33) revealed that
miR-495 was overexpressed during MSC differentiation into
osteoblasts, whereas Brodie and Slavin (34) demonstrated that downregulation of
miR-495 expression was able to be used in the astrocytogenesis
induction. The present study revealed that miR-495 expression was
decreased during differentiation into adipocytes. Together, these
results indicated that the expression of one miRNA may be affected
by different conditions or different cell types, resulting in
different expression levels.
miR-543 has been demonstrated to serve a role in
human adult stem cell regeneration (35). Data from the present study led to
the hypothesis that the upregulated expression of ENAH may be
regulated by miR-495, miR376a, miR-543 and miR-370, and thus may
serve a contributing role in MSC differentiation into
adipocytes.
Notably, the present study demonstrated that the
expression of miR-495 was downregulated in MSCs undergoing either
osteoblastic or adipocytic differentiation; however, the target
DEGs of miR-495 were different (Figs.
3 and 4). For example, during
osteoblastic differentiation, ALCAM, NEGR1, CD164 and ST3
β-galactoside α-2,3-sialyltransferase 5 (ST3GAL5) were upregulated,
suggesting that they may be targets for miR-495. However, none of
these DEGs were identified as targets of miR-495 during MSC
adipocytic differentiation. These suggested that the regulatory
mechanism of the two types of MSC differentiation was
different.
In conclusion, the present study screened several
DEGs and differentially expressed miRNAs during osteoblastic and
adipocytic differentiation of MSCs. The results revealed that DEGs,
such as NEGR1, PPAP2B, PDGFRA, SORT1 and IL6ST, and differentially
expressed miRNAs, including miR-203 and miR-382, may be involved in
the MSCs differentiation into osteoblasts, whereas the
downregulated expression of miR-495, miR-376a, miR-543 and miR-370,
may be involved in regulating the expression of ENAH mRNA in MSCs
differentiating into adipocytes The present study may provide a
theoretical basis for further investigation into the differing
pathways of MSC differentiation into osteoblasts or adipocytes,
which may lead to the use of MSCs in the treatment of various
diseases. Further experimental studies are required to confirm the
roles of the selected DEGs or miRNAs in MSC differentiation.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 81160113) and the
Landed Science Project of Jiangxi Province Institution of Higher
Education (grant no. KJLD13094).
References
|
1
|
Cells MS: Mesenchymal stem cells.
Springer; Berlin: 2010
|
|
2
|
Wang T: Bone marrow mesenchymal stem cells
and cardiovascular diseases. Chinese Journal of Critical Care
Medicine. 2008.
|
|
3
|
Sekiya I, Muneta T, Morio T, Shimizu N and
Kuroiwa Y: Application of synovium-derived mesenchymal stem cells
(mscs) for cartilage or meniscus regeneration. Va Us. 2008.
|
|
4
|
Barry FP and Murphy JM: Mesenchymal stem
cells: Clinical applications and biological characterization. Int J
Biochem Cell Biol. 36:568–584. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hamidouche Z, Fromigué O, Nuber U, Vaudin
P, Pages JC, Ebert R, Jakob F, Miraoui H and Marie PJ: Autocrine
fibroblast growth factor 18 mediates dexamethasone-induced
osteogenic differentiation of murine mesenchymal stem cells. J Cell
Physiol. 224:509–515. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sammons J, Ahmed N, El-Sheemy M and Hassan
HT: The role of BMP-6, IL-6 and BMP-4 in mesenchymal stem
cell-dependent bone development: Effects on osteoblastic
differentiation induced by parathyroid hormone and vitamin D(3).
Stem Cells Dev. 13:273–280. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jing L and Jia Y, Lu J, Han R, Li J, Wang
S, Peng T and Jia Y: MicroRNA-9 promotes differentiation of mouse
bone mesenchymal stem cells into neurons by Notch signaling.
Neuroreport. 22:206–211. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Vam Pham P, Hong-Thien Bui K, Ngo D Quoc,
Tan Khuat L and Kim Phan N: Transplantation of nonexpanded adipose
stromal vascular fraction and platelet-rich plasma for articular
cartilage injury treatment in mice model. J Med Eng.
2013:8323962013.PubMed/NCBI
|
|
9
|
Wan Q, Guan SJ, LI XC, Sun CP, Zhou JX,
Xia B, Xia TT and Zeng QS: Imaging features of thoracic
well-differentiated and myxoid liposarcoma: Comparison with
pathology. Chin J Med Imag Technol. 30:78–81. 2014.
|
|
10
|
Arfat Y, Xiao WZ, Ahmad M, Zhao F, Li DJ,
Sun YL, Hu L, Zhihao C, Zhang G, Iftikhar S, et al: Role of
microRNAs in osteoblasts differentiation and bone disorders. Curr
Med Chem. 22:748–758. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Longhi A, Neri S, Speranza C, Alberghini
M, Ferrari C, Abate M, Cesari M, Ferrari S, Palmerini E and Mercuri
M: Liposarcoma treatment: Role of radiotherapy and chemotherapy. J
Clin Oncol. 26:431–436. 2008. View Article : Google Scholar
|
|
12
|
Mithoefer K, McAdams TR, Scopp JM and
Mandelbaum BR: Emerging options for treatment of articular
cartilage injury in the athlete. Clin Sports Med. 28:25–40. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen TS, Lai RC, Lee MM, Choo AB, Lee CN
and Lim SK: Mesenchymal stem cell secretes microparticles enriched
in pre-microRNAs. Nucleic Acids Res. 38:215–224. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Luca AD: RNA-seq analysis reveals
significant effects of EGFR signalling on the secretome of
mesenchymal stem cells. Oncotarget. 5:10518–10528. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mrugala D, Dossat N, Ringe J, Delorme B,
Coffy A, Bony C, Charbord P, Häupl T, Daures JP, Noël D and
Jorgensen C: Gene expression profile of multipotent mesenchymal
stromal cells: Identification of pathways common to TGF beta
3/BMP2-induced chondrogenesis. Cloning Stem Cells. 11:61–76. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Trapnell C, Pachter L and Salzberg SL:
TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics.
25:1105–1111. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Harris MA, Clark J, Ireland A, Lomax J,
Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C,
et al: The gene ontology (GO) database and informatics resource.
Nucleic Acids Res. 32(Database Issue): D258–D261. 2004.PubMed/NCBI
|
|
18
|
Altermann E and Klaenhammer TR:
PathwayVoyager: Pathway mapping using the Kyoto encyclopedia of
genes and genomes (KEGG) database. BMC Genomics. 6:602005.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for annotation,
visualization and integrated discovery. Genome Biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Varambally S, Cao Q, Mani RS, Shankar S,
Wang X, Ateeq B, Laxman B, Cao X, Jing X, Ramnarayanan K, et al:
Genomic loss of microRNA-101 leads to overexpression of histone
methyltransferase EZH2 in cancer. Science. 322:1695–1699. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nardi N Beyer and da Silva Meirelles L:
Mesenchymal stem cells: Isolation, in vitro expansion and
characterization. Handb Exp Pharmacol. 249–282. 2006. View Article : Google Scholar
|
|
22
|
Schiavi-Tritz J, Reppel L, Magdalou J,
Stoltz JF and Huselstein C: 229 improving hBM-MSCs differentiation
DM a stratified scaffold for osteoarticular repairing. Osteoarthrit
Cartil. S1122011. View Article : Google Scholar
|
|
23
|
Funatsu N, Miyata S, Kumanogoh H, Shigeta
M, Hamada K, Endo Y, Sokawa Y and Maekawa S: Characterization of a
novel rat brain glycosylphosphatidylinositol-anchored protein
(Kilon), a member of the IgLON cell adhesion molecule family. J
Biol Chem. 274:8224–8230. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Arai F, Ohneda O, Miyamoto T, Zhang XQ and
Suda T: Mesenchymal stem cells in perichondrium express activated
leukocyte cell adhesion molecule and participate in bone marrow
formation. J Exp Med. 195:1549–1563. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sánchez-Sánchez R, Morales-Lázaro SL,
Baizabal JM, Sunkara M, Morris AJ and Escalante-Alcalde D: Lack of
lipid phosphate phosphatase-3 in embryonic stem cells compromises
neuronal differentiation and neurite outgrowth. Dev Dyn.
241:953–964. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shockley KR, Lazarenko OP, Czernik PJ,
Rosen CJ, Churchill GA and Lecka-Czernik B: PPARgamma2 nuclear
receptor controls multiple regulatory pathways of osteoblast
differentiation from marrow mesenchymal stem cells. J Cell Biochem.
106:232–246. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Alison MR and Islam S: Attributes of adult
stem cells. J Pathol. 217:144–160. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shockley KR, Rosen CJ, Churchill GA and
Lecka-Czernik B: PPARgamma2 regulates a molecular signature of
marrow mesenchymal stem cells. PPAR Res. 2007:812192007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kulterer B, Friedl G, Jandrositz A,
Sanchez-Cabo F, Prokesch A, Paar C, Scheideler M, Windhager R,
Preisegger KH and Trajanoski Z: Gene expression profiling of human
mesenchymal stem cells derived from bone marrow during expansion
and osteoblast differentiation. BMC Genomics. 8:702007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kapinas K and Delany AM: MicroRNA
biogenesis and regulation of bone remodeling. Arthritis Res Ther.
13:2202011. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gurzu S, Ciortea D, Ember I and Jung I:
The possible role of mena protein and its splicing-derived variants
in embryogenesis, carcinogenesis, and tumor invasion: A systematic
review of the literature. Biomed Res Int. 2013:3651922013.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Charoenpanich A, Wall ME, Tucker CJ,
Andrews DM, Lalush DS and Loboa EG: Microarray analysis of human
adipose-derived stem cells in three-dimensional collagen culture:
Osteogenesis inhibits bone morphogenic protein and Wnt signaling
pathways, and cyclic tensile strain causes upregulation of
proinflammatory cytokine regulators and angiogenic factors. Tissue
Eng Part A. 17:2615–2627. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gao J, Yang T, Han J, Yan K, Qiu X, Zhou
Y, Fan Q and Ma B: MicroRNA expression during osteogenic
differentiation of human multipotent mesenchymal stromal cells from
bone marrow. J Cell Biochem. 112:1844–1856. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Brodie C and Slavin S: MicroRNAs for the
generation of astrocytes. WO. 2014.
|
|
35
|
Ng TK, Carballosa CM, Pelaez D, Wong HK,
Choy KW, Pang CP and Cheung HS: Nicotine alters MicroRNA expression
and hinders human adult stem cell regenerative potential. Stem
Cells Dev. 22:781–790. 2013. View Article : Google Scholar : PubMed/NCBI
|