Introduction
Gastric cancer (GC) is the fifth most common cancer
worldwide, and is currently the third leading cause of
cancer-associated mortality. GC is particularly prevalent in Asia
(1). Unfortunately, the majority
of patients with GC present with late stage cancer, and therefore
require palliative chemotherapy (2). Treatment of GC continues to present a
challenge, particularly with regards to the high
mortality-to-incidence ratio, despite significant progress being
made in early detection and treatment (3). Genetic alterations are thought to be
important factors in the vast majority of solid tumors. Recently,
gene therapy has been considered an attractive therapeutic option.
In addition, autophagy-related genes have attracted attention as
novel potential targets in cancer treatment (4,5).
Autophagy is a dynamic process, during which cells
recycle and remove damaged organelles and proteins, in order to
ensure cell survival in response to cellular stress (6). Furthermore, autophagy can induce cell
death under certain conditions. Therefore, the two opposing effects
of autophagy serve an important role in cellular differentiation,
developmental processes and human disease (7). Several proteins are involved in the
detection of autophagic activity, including microtubule-associated
protein 1A/1B-light chain 3 (LC3) and the LC3 binding protein,
sequestome 1/p62 (8).
The significance of autophagy in cell cycle
progression and cell death has previously been reported. A previous
study demonstrated that activated autophagy contributes to
matrine-induced cell death of the SGC-7901 GC cell line (9). Conversely, autophagy-mediated high
mobility group box 1 release promotes GC cell survival via receptor
for advanced glycation endproducts activation of extracellular
signal-regulated kinases 1/2 (10). However, the function of autophagy
in GC remains to be elucidated.
Previous studies indicated the potential roles of
zinc finger proteins in gastric cancer progression and the
association with autophagy (11).
The importance of zinc finger protein like 1 (ZFPL1) in autophagy
and human GC resulted in the aim of the present study; to explore
the potential antitumor effects of ZFPL1 knockdown, using short
hairpin (sh)RNA against ZFPL1 (shZFPL1). The present study explored
whether knockdown of ZFPL1 would increase autophagy, inhibit
protein glycosylation and promote autophagy-related cell death. In
addition, the present study evaluated whether the effects of ZFPL1
on autophagy correlated with the interaction between ZFPL1 and
GM130.
Materials and methods
Reagents and antibodies
The MTT Cell Proliferation and Cytotoxicity Assay
kit was purchased from Beyotime Institute of Biotechnology (Haimen,
China). Immunoblotting was performed using anti-ZFPL1 (cat. no.
sc-515393; Santa Cruz Biotechnology, Inc., Dallas, TX, USA),
anti-β-actin (cat. no. sc-47778; Santa Cruz Biotechnology, Inc.),
anti-LC3 (cat. no. L8918; Sigma-Aldrich; Merck Millipore,
Darmstadt, Germany), anti-p62 (cat. no. N1163; Sigma-Aldrich; Merck
Millipore), cleaved caspase antibody sampler kit (cat. no. 2855;
Cell Signaling Technology, Inc., Danvers, MA, USA), anti-RL2 (cat.
no. SAB1304907; igma-Aldrich; Merck Millipore), anti-p53 (cat. no.
ab1431; Abcam, Cambridge, MA, USA), anti-BH3 interacting domain
death agonist (Bid; cat. no. ab32060; Abcam), anti-B-cell lymphoma
2 (Bcl-2)-associated X protein (Bax; cat. no. 554104; BD
Pharmingen, San Diego, CA, USA), anti-Bcl-x (cat. no. 551269; BD
Pharmingen) and anti-Bcl-2 (cat. no. 610539; BD Biosciences, San
Jose, CA, USA). Horseradish peroxidase (HRP)-conjugated secondary
antibodies (cat. no. 58203) were purchased from Cell Signaling
Technology, Inc. Brefeldin A (BFA) and bafilomycin A1 were
purchased from Sigma-Aldrich; TRIzol reagent was obtained from
Invitrogen (Thermo Fisher Scientific, Inc., Waltham, MA, USA);
RevertAid First Strand cDNA Synthesis kit was purchased from Thermo
Fisher Scientific, Inc.; and TransStart Green qPCR SuperMix was
from Beijing Transgen Biotech Co., Ltd. (Beijing, China). All other
reagents were of analytical grade.
Cell culture and lentiviral
infection
The NCI-N87 and BGC-823 human GC cell lines were
purchased from the American Type Culture Collection (Manassas, VA,
USA). The NCI-N87 and BGC-823 cells were cultured in RPMI-1640
medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with
10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.), 100
µg/ml streptomycin and 100 U/ml penicillin at 37°C in a humidified
atmosphere containing 5% CO2. Autophagy was induced by
culturing cells in medium supplemented with Hanks' balanced salt
solution (HBSS; ph 7.2–7.3) or BFA (1 µg/ml) overnight (3%
CO2, 37°C) prior to lentiviral infection. The
recombinant plasmids were constructed according to our previous
study (12). Transient
transfections were performed using FUGENE 6 (Roche Diagnostics,
Basel, Switzerland) according to the manufacturer's instructions.
The ZFPL1 overexpression was performed using ZFPL1 mimics (cat. no.
HMI0070; Sigma-Aldrich; Merck Millipore). RNA interference was
carried out using shRNA. A shRNA sequence targeting Lamin A
(5′-AACTGGACTTCCAGAAGAACA-3′; Gibco; Thermo Fisher Scientific,
Inc.) was used as a negative control. ZFPL1 was targeted with two
oligonucleotides (obtained from Gibco; Thermo Fisher Scientific,
Inc.): shZFPL1#1 (5′-CGACCCGCCTTGTCTGCTA-3′) and shZFPL1#2
(5′-GCTCCAAGATAGCGACTAC-3′). All shRNA sequences were cloned into
pSUPER.retro.puro vectors (Shanghai Ling Feng Chemical Reagent Co.,
Ltd., Shanghai, China) for further use. The results presented in
the current study were obtained using shZFPL1#1; shZFPL1#2 worked
similarly. For lentiviral transduction, human embryonic kidney
(HEK)293T cells (Shanghai Ling Feng Chemical Reagent Co., Ltd.)
were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific,
Inc.) supplemented with 10% fetal bovine serum (Gibco; Thermo
Fisher Scientific, Inc.), 100 µg/ml streptomycin and 100 U/ml
penicillin at 37°C in a humidified atmosphere containing 5%
CO2. When 75–80% confluent, the HEK293T cells were
transduced (incubated at 3% CO2, 37°C, 25 min) with the
third-generation packaging plasmids (Cell Signaling Technology,
Inc.) pMD2.VSVG, pRSV-REV and pMDLg/pRRE, alongside the transfer
constructs. The fresh supernatant was filtered using a 0.45 µm
filter for further use. The first harvest pool was placed into 50
ml tubes and stored at 4°C and then centrifuged for 10 min at 1,153
× g to produce a pellet consisting of cells and debris. The
cell-free supernatant was subjected to ultra-centrifugation for 120
min (16°C) at 49,460 × g, after which the supernatant was discarded
and the pellet was resuspended in HBS. The vector stock solutions
were pooled to produce a homogenous vector stock solution and
stored at −80°C. Freezing and thawing of the lentiviral stock were
avoided as much as possible. Infected NCI-N87 and BGC-823 cells
were rinsed with PBS, and were allowed to recover for 12 h prior to
further experimentation.
Cell growth inhibition assay
Cell growth inhibition was detected using the MTT
Cell Proliferation and Cytotoxicity Assay kit (Siemens AG, Munich,
Germany). The present study consisted of the following four groups:
Control shRNA group, shZFPL1 group, control shRNA + bafilomycin A1
group and shZFPL1 + bafilomycin A1 group. Bafilomycin A1 group
cells were incubated with 3 mg/ml bafilomycin A1 at room
temperature for 3 h. Briefly, cells in the logarithmic growth phase
were collected and reseeded into 96-well plates at 10,000
cells/well. Following a 24 h incubation, the medium was gently
removed from all wells and fresh medium containing 10 µl MTT (5
mg/ml) was added to each well. Following a further 4 h incubation
at 37°C, 150 µl dimethyl sulfoxide (Sigma-Aldrich; Merck Millipore)
was added, and the plates were slowly agitated for 10 min at room
temperature. Subsequently, Sorensen's glycine buffer (0.1 M
glycine, 0.1 M NaCl; pH 10.5) was added, and the absorbance of each
well was measured at 490 nm in 10 min. The cell growth inhibition
rate was calculated as follows: (1 - experimental group
absorbance/control group absorbance) ×100. A graph was plotted
using GraphPad Prism 5 software (GraphPad Software, Inc., La Jolla,
CA, USA). All experiments were carried out in triplicate.
Western blotting
The cells were harvested in PBS, and were
homogenized in lysis buffer [25 mM Tris-HCl (pH 7.4), 5 mM
MgCl2, 200 mM NaCl, 1 mM EDTA, 2 mM
Na3VO4, 0.1 mM EGTA, 1% NP-40, 0.1% SDS, 10%
glycerol, 1 mM phenylmethylsulfonyl fluoride (cat. no. 8553; Cell
Signaling Technology, Inc.) and Protease Inhibitor Cocktail (cat.
no. 5871; Cell Signaling Technology, Inc.)] on ice for 30 min.
Lysates were then centrifuged at 28,000 × g for 30 min at
4°C. The supernatants were carefully transferred into fresh
Eppendorf® tubes and the protein concentration was
detected using the Bradford method. Subsequently, the protein
samples were denatured at 100°C for 10 min and were cooled on ice.
Protein samples (35 µg) were separated by 10–15% SDS-PAGE (150 V,
70 min) and were transferred to a polyvinylidene difluoride
membrane (90 V, 60 min). After blocking with 5% nonfat milk in
TBS-0.1% Tween 20 for 1 h at room temperature, the PVDF membranes
were blotted with primary antibodies (1:500 dilution) overnight at
4°C. Subsequently, the membranes were incubated with HRP-conjugated
secondary antibodies (1:1,000 dilution) at room temperature for 1
h. After washing, the band intensity was analyzed using HYBOND-ECL
(Sangon Biotech Co., Ltd., Shanghai, China) and the LAS-3000
luminescent image analyzer system (Fujifilm, Tokyo, Japan).
Co-immunoprecipitation
Co-immunoprecipitation experiments were performed as
previously described (12).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the cells using
TRIzol® reagent, according to the manufacture's
protocol. RNA (4 µg) then underwent RT-qPCR. The reaction mixture
contained 4 µl RNA, 4 µl 5*PrimeScript Buffer, 1 µl PrimerScript RT
Enzyme Mix I (Applied Biosystems; Thermo Fisher Scientific, Inc.),
1 µl oligo dT primer and 1 µl random hexamers in a total of 20 µl.
The primers used were as follow: GAPDH, forward
5′-AAGCTCATTTCCTGGTATGACAACG-3′, reverse
5′-TCTTCCTCTTGTGCTCTTGCTGG-3′, which served as an internal
reference gene (126 bp); and ZFPL1, forward
5′-GCCGATCAGTAAACACAGA-3′ and reverse 5′-ACTGGACGATGCACTTG-3′ (149
bp). The amplification conditions were as follows: Pre-denaturation
at 95°C for 5 min, followed by 40 cycles of denaturation at 95°C
for 30 sec, annealing at 58°C for 30 sec and extension at 72°C for
30 sec, and a final extension at 72°C for 5 min. Changes in ZFPL1
expression levels were calculated using the 2−ΔΔCq
method (13). Each sample was
tested in triplicate.
Microscopy
Cells stably expressing red fluorescent protein
(RFP)-LC3 plasmid (Addgene, Inc., Cambridge, MA, USA) were cultured
in a 25-cm2 culture bottle. A fluorescence microscope
(Nikon Corporation, Tokyo, Japan) was used to observe cells
expressing RFP. Images were obtained using a charge-coupled device
camera (Coolsnap; Nikon Corporation) with MetaMorph 7.8 software
(Molecular Devices, LLC, Sunnyvale, CA, USA). At least 200 cells
from 10 different fields were counted for statistical analysis.
Statistical analysis
All statistical analyses were conducted using
GraphPad Prism 5.0 software. Data were analyzed using one-way
analysis of variance and least significant difference test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
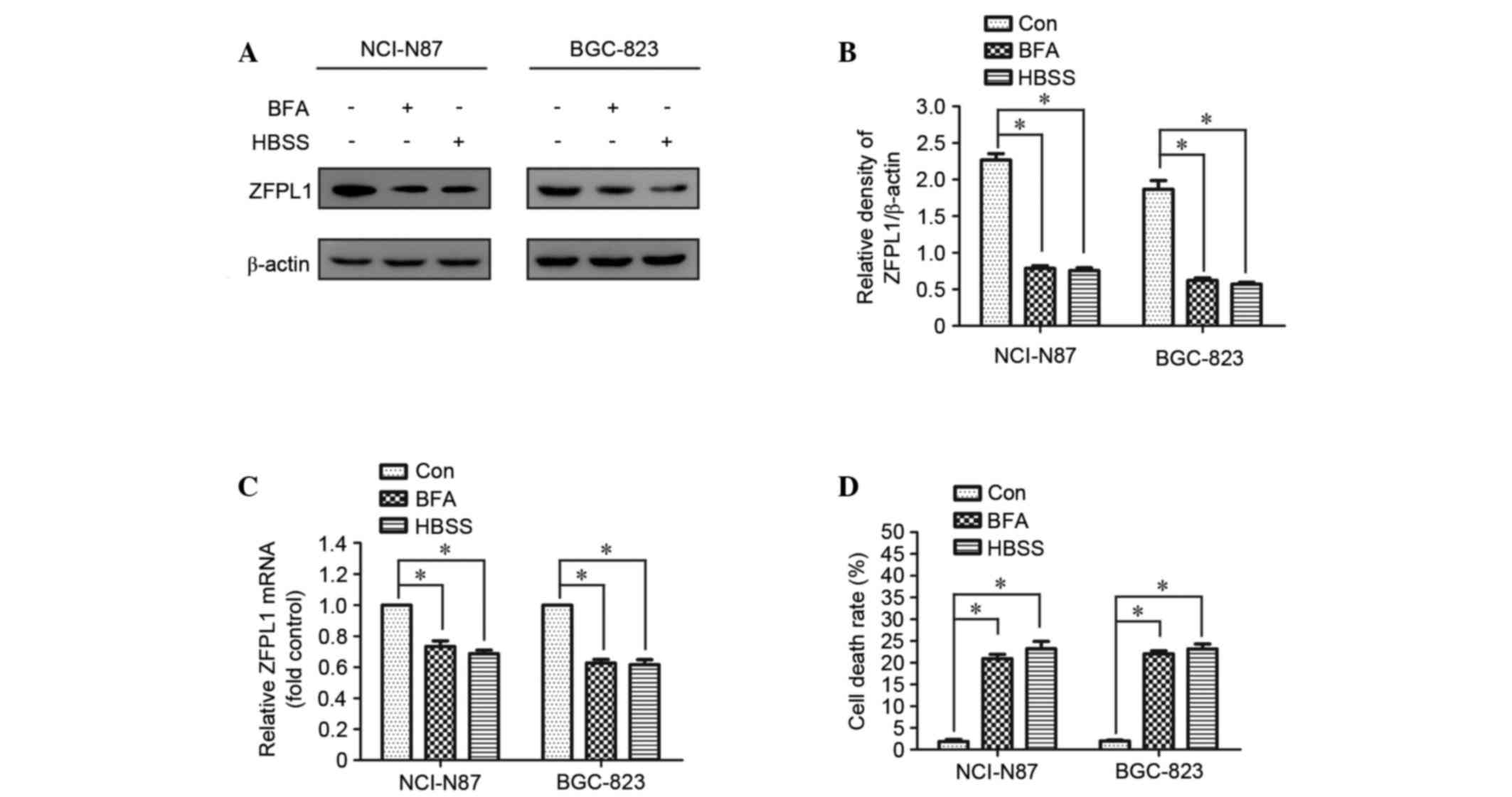
ZFPL1 expression is decreased and cell
death is increased in autophagy-activated cells
It is well known that BFA and nutrient deprivation
induce autophagy in several cell lines. In the present study, ZFPL1
expression was decreased in NCI-N87 cells following treatment with
HBSS or BFA (1 µg/ml); similar results were observed in another
human GC cell line, BGC-823 (Fig.
1A-C). In addition, the cell death rate was markedly increased
in HBSS- or BFA-treated cells compared with in the control group
(Fig. 1D).
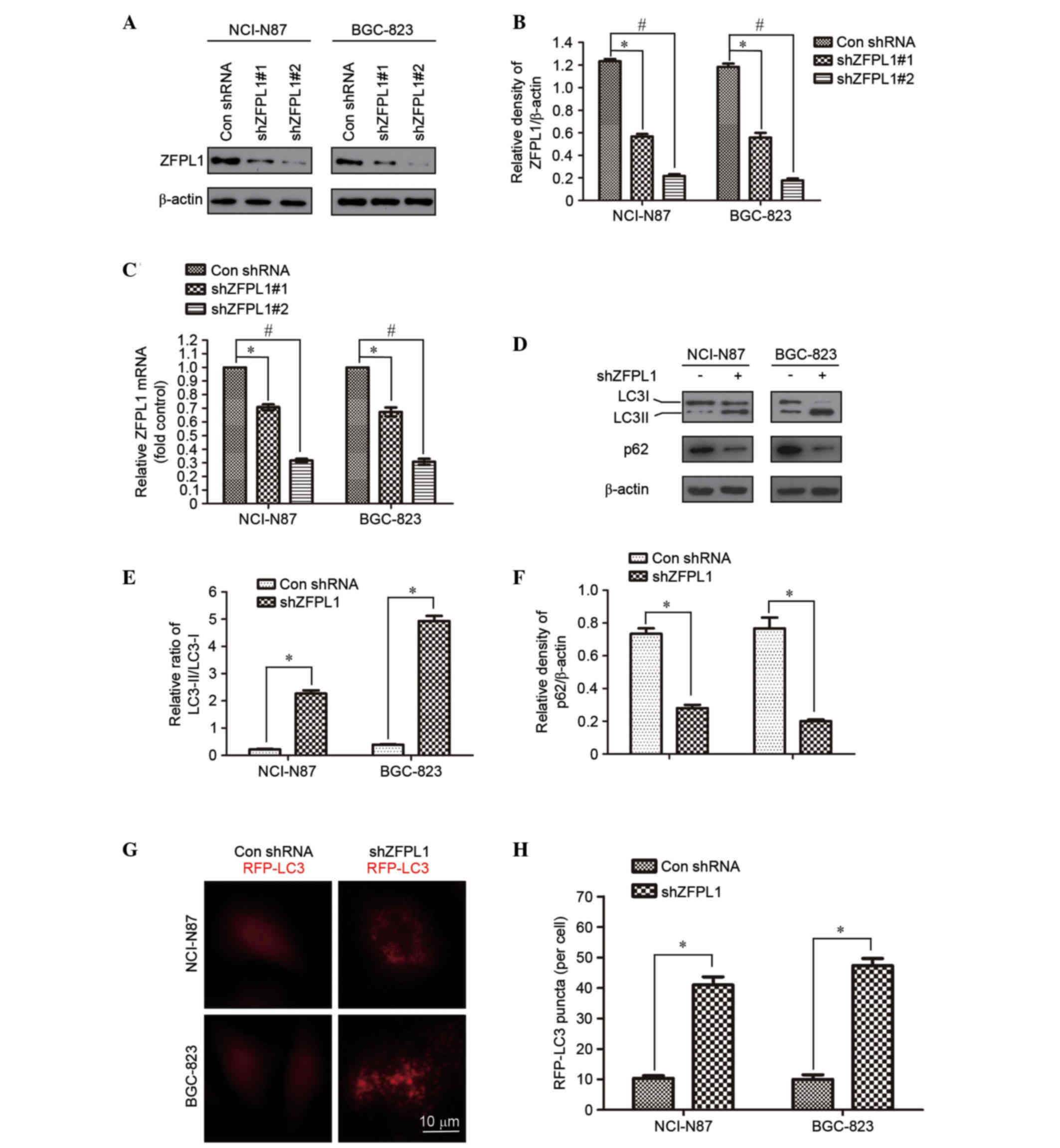
Knockdown of ZFPL1 induces autophagy
in NCI-N87 and BGC-823 cells
To determine the effects of ZFPL1 on autophagy
regulation, a ZFPL1 knockdown system was generated using lentiviral
infection. To investigate the silencing efficiency of two shZFPL1
shRNA molecules, western blotting was performed in NCI-N87 and
BGC-823 cells. The results demonstrated that shZFPL1 significantly
decreased ZFPL1 protein levels compared with the control group, as
detected by western blotting and densitometric analysis (Fig. 2A and B). The marked decrease in
ZFPL1 mRNA expression was further confirmed by qPCR (Fig. 2C). It has previously been reported
that ZFPL1 directly interacts with the cis-Golgi matrix protein
golgin A2/GM130 (14), which is a
key factor in stacking of Golgi cisternae and Golgi structural
maintenance (15). Previous
studies have indicated that Golgi and autophagy-related membrane
trafficking are functionally interdependent (16). In addition, autophagosome formation
has been detected in GM130-downregulated cells (17). To determine whether ZFPL1 was
associated with autophagy, LC3-I to LC3-II conversion and p62
degradation were detected by western blotting and densitometric
analysis in ZFPL1 knockdown cells. LC3-I and LC3-II are cellular
forms of the LC3 protein; LC3-I is the cytoplasmic form, whereas
LC3-II is the autophagosome membrane-bound form (18,19).
Therefore, an increase in the ratio of LC3-II to LC3-I protein is
correlated with the extent of autophagosome formation. p62, which
is a ubiquitin-binding scaffold protein, is associated with
autophagy by directly binding to the autophagic effector proteins
gamma-aminobutyric acid receptor-associated protein and LC3 for
degradation (20). Therefore,
total cellular levels of p62 reflect autophagic activity. The
present study demonstrated that knockdown of ZFPL1 significantly
increased the ratio of LC3-II to LC3-I and p62 degradation, as
compared with the shRNA control (Fig.
2D and E). In the present study, RFP-LC3 was examined in
shZFPL1 NCI-N87 and BGC-823 cells, and autophagy induction was
evaluated using confocal microscopy. As shown in Fig. 2F, shZFPL1 cells exhibited an
increase in LC3 puncta compared with the control cells. These
results clearly indicate that downregulation of ZFPL1 induces
autophagy in NCI-N87 and BGC-823 cells.
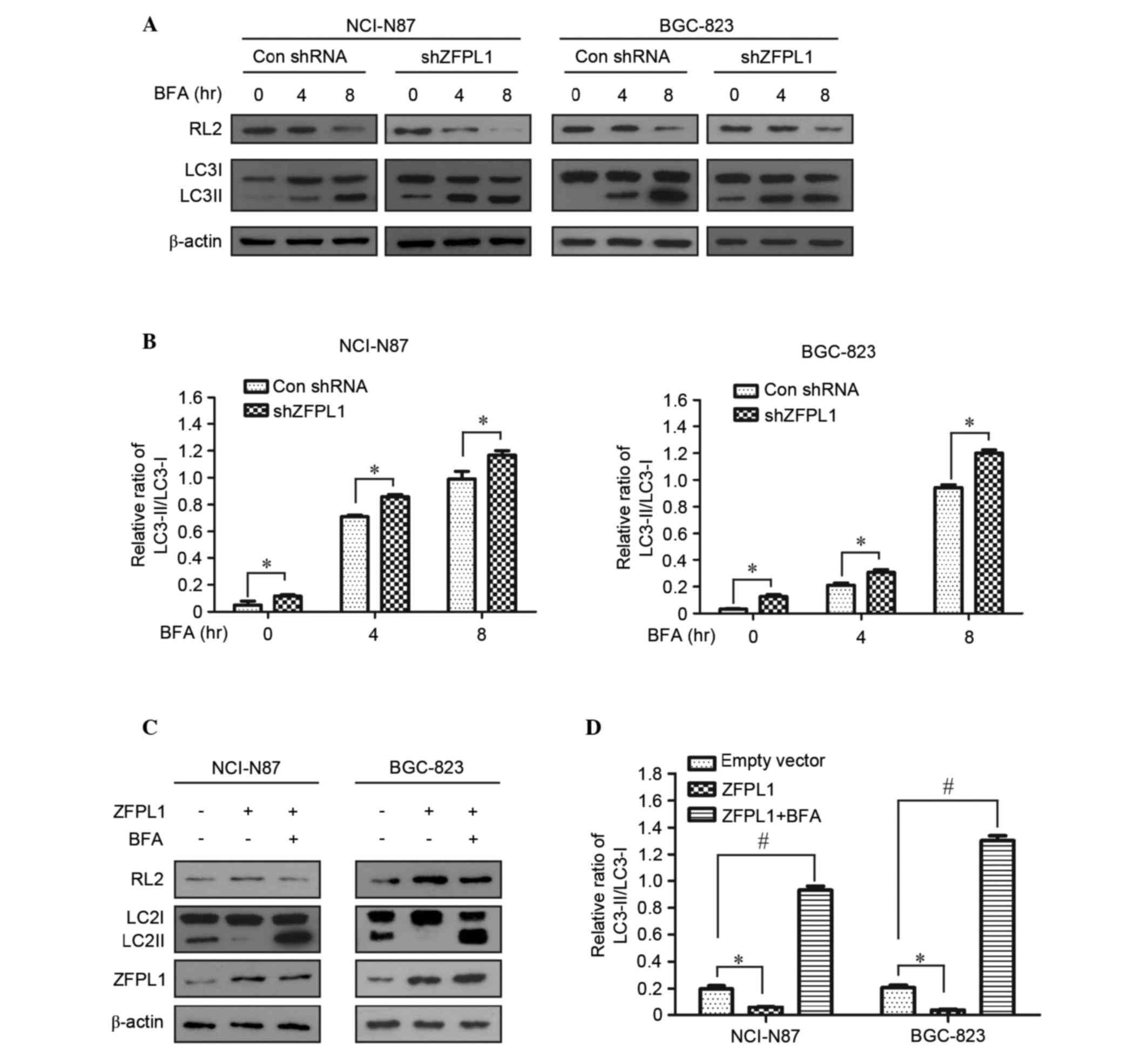
Effects of ZFPL1 knockdown on
autophagy and glycosylation in NCI-N87 and BGC-823 cells
The cis-Golgi matrix protein GM130, which directly
interacts with ZFPL1, serves an essential role in protein
glycosylation, and downregulation of GM130 decreases the level of
RL2 (17). To determine the
effects of ZFPL1 knockdown on protein glycosylation, the expression
levels of RL2 in ZFPL1 knockdown cells were detected by western
blotting. The expression of RL2 was markedly decreased in a
time-dependent manner, whereas the ratio of LC3II to LC3I was
increased in stably transfected cells treated with BFA (Fig. 3A and B). To further confirm the
function of ZFPL1 on protein glycosylation regulation, a ZFPL1
overexpression assay was conducted using a lentiviral system.
Notably, the expression levels of RL2 and the ratio of LC3II to
LC3I were reversed in cells overexpressing ZFPL1 compared with
ZFPL1 knockdown cells (Fig. 3C and
D). These results suggest that knockdown of ZFPL1 may induce
autophagy and reduce the protein glycosylation process.
ZFPL1 knockdown induces
autophagy-associated cell death, rather than apoptosis, in NCI-N87
and BGC-823 cells
Autophagy is an important mechanism of cell survival
in response to stress conditions, including nutrient deprivation,
reactive oxygen species and ursulic acid; however, autophagic
dysfunction also leads to cell death, which is known as autophagic
cell death or type II programmed cell death (21,22).
The present study demonstrated that cell death was significantly
increased in ZFPL1 knockdown NCI-N87 and BGC-823 cells. However, in
ZFPL1 knockdown cells treated with bafilomycin A1 (10 µM), cell
death was markedly reduced compared with in the untreated cells
(Fig. 4A). Immunoblotting analysis
indicated that transfection with shZFPL1 or control shRNA did not
affect the expression of apoptosis-associated proteins, including
Bcl-2, Bcl-x, Bax, Bid, p53, and the classical caspase family
members, caspase-3, caspase-8 and caspase-9 (Fig. 4B and C). Conversely, the induction
of autophagy by ZFPL1 knockdown was confirmed using the autophagy
inhibitor bafilomycin A1, which blocks the fusion process of
autophagosomes and mature lysosomes (23). Western blotting indicated that the
expression levels of LC3II, the autophagy biomarker, increased
following knockdown of ZFPL1 (Fig. 2D
and E). As shown in Fig. 2E,
the ratio of LC3II to LC3I was increased following knockdown of
ZFPL1 compared with the control. The autophagy inhibitor
bafilomycin A1 was used to determine the effects of ZFPL1 knockdown
on autophagy. Cells stably transfected with shZFPL1 and treated
with bafilomycin A1 (10 µM) exhibited no difference in LC3
expression compared with the control cells (Fig. 4D and E). These results suggest that
ZFPL1 knockdown may induce autophagy-related cell death in NCI-N87
and BGC-823 cells.
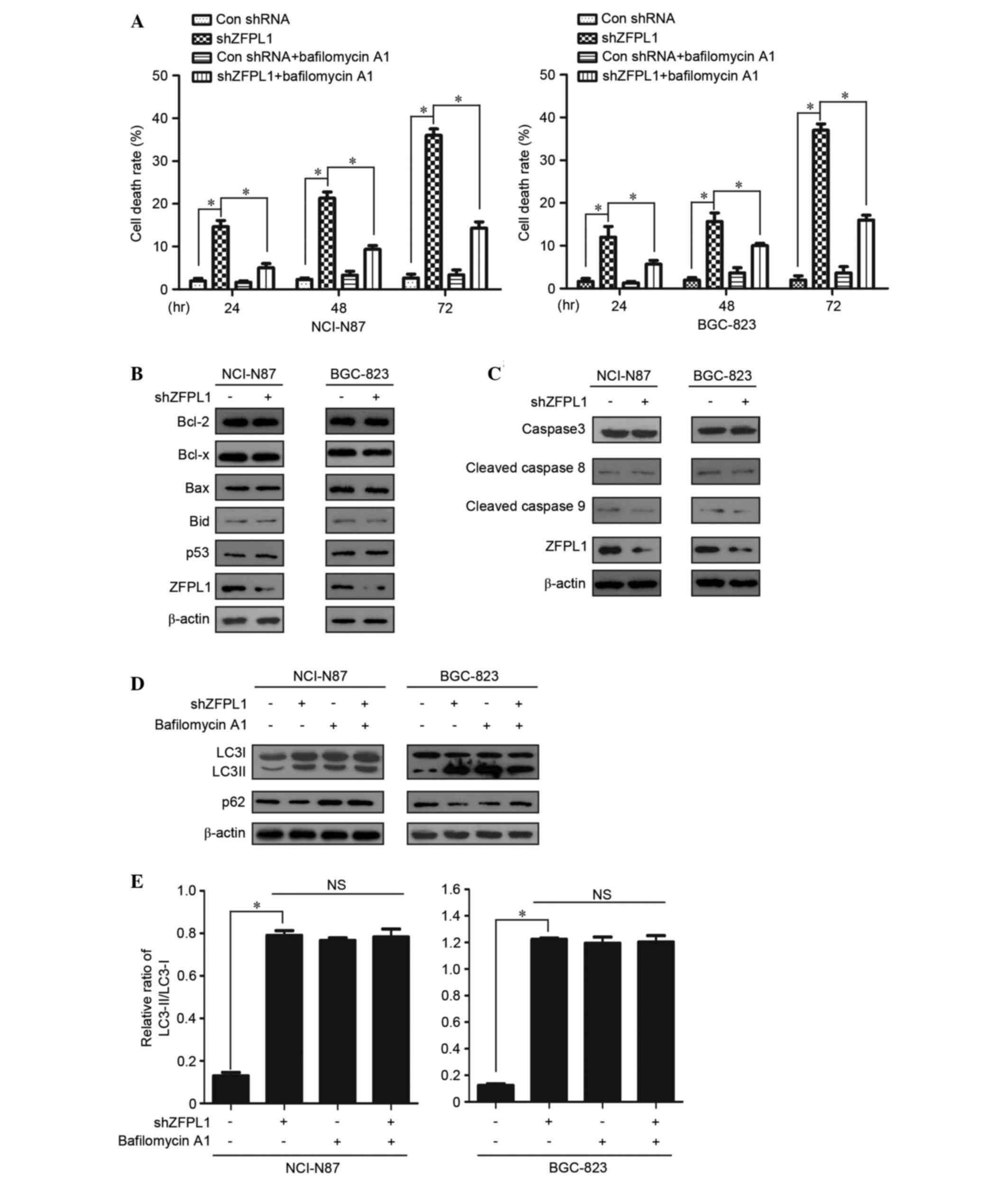 | Figure 4.Knockdown of ZFPL1 induces
autophagy-related cell death, rather than apoptosis, in NCI-N87 and
BGC-823 cells. (A) Cell death rate was measured using the MTT assay
(n=3). Knockdown and control cells were treated with bafilomycin A1
(10 µM) for 24, 48 or 72 h. (B and C) Cell lysates were collected
post-transfection with shZFPL1 or control shRNA for 48 h, and
protein expression was determined using western blotting, with
β-actin as an internal control. (D and E) Knockdown and control
cells were treated with bafilomycin A1 (10 µM) for 48 h. Western
blotting was used to detect LC3 and p62 expression. Data are
presented as the mean ± standard error of the mean of three
independent experiments. *P<0.05. NS, no significance; ZFPL1,
zinc finger protein like 1; shRNA, short hairpin RNA; Bcl-2, B-cell
lymphoma 2; Bax, Bcl-2-associated X protein; Bid, BH3 interacting
domain death agonist; LC3, microtubule-associated protein
1A/1B-light chain 3; Con, control. |
Effects of ZFPL1 on autophagy are
associated with the interaction between ZFPL1 and GM130
To determine whether the interaction between ZFPL1
and GM130 affects autophagy, a co-immunoprecipitation assay was
conducted. An endogenous ZFPL1-GM130 association was identified in
NCI-N87 cells (Fig. 5A); similar
results were observed in BGC-823 cells (data not shown). Chiu et
al (14) revealed that the
first zinc finger domain (ZFD; 1–43 amino acid) of ZFPL1 contained
the GM130-binding site. Therefore, the present study overexpressed
ZFD-FLAG to further analyze the effects of ZFPL1-GM130 interaction
on autophagy. As shown in Fig. 5B and
C, ZFD-FLAG competed with ZFPL1 for GM130 binding; however, no
effect on ZFPL1 expression was observed in the process. The results
of the present study indicated that ZFPL1 knockdown induced
autophagy in NCI-N87 and BGC-823 cells. Therefore, knockdown of
ZFPL1 should result in decreased endogenous ZFPL1-GM130
association. In addition, autophagy was induced after using
competitive binding reactions with ZFPL1 and ZFD-Flag (Fig. 5C and D). These results suggest that
ZFPL1 regulates autophagy via a GM130-interacting mechanism.
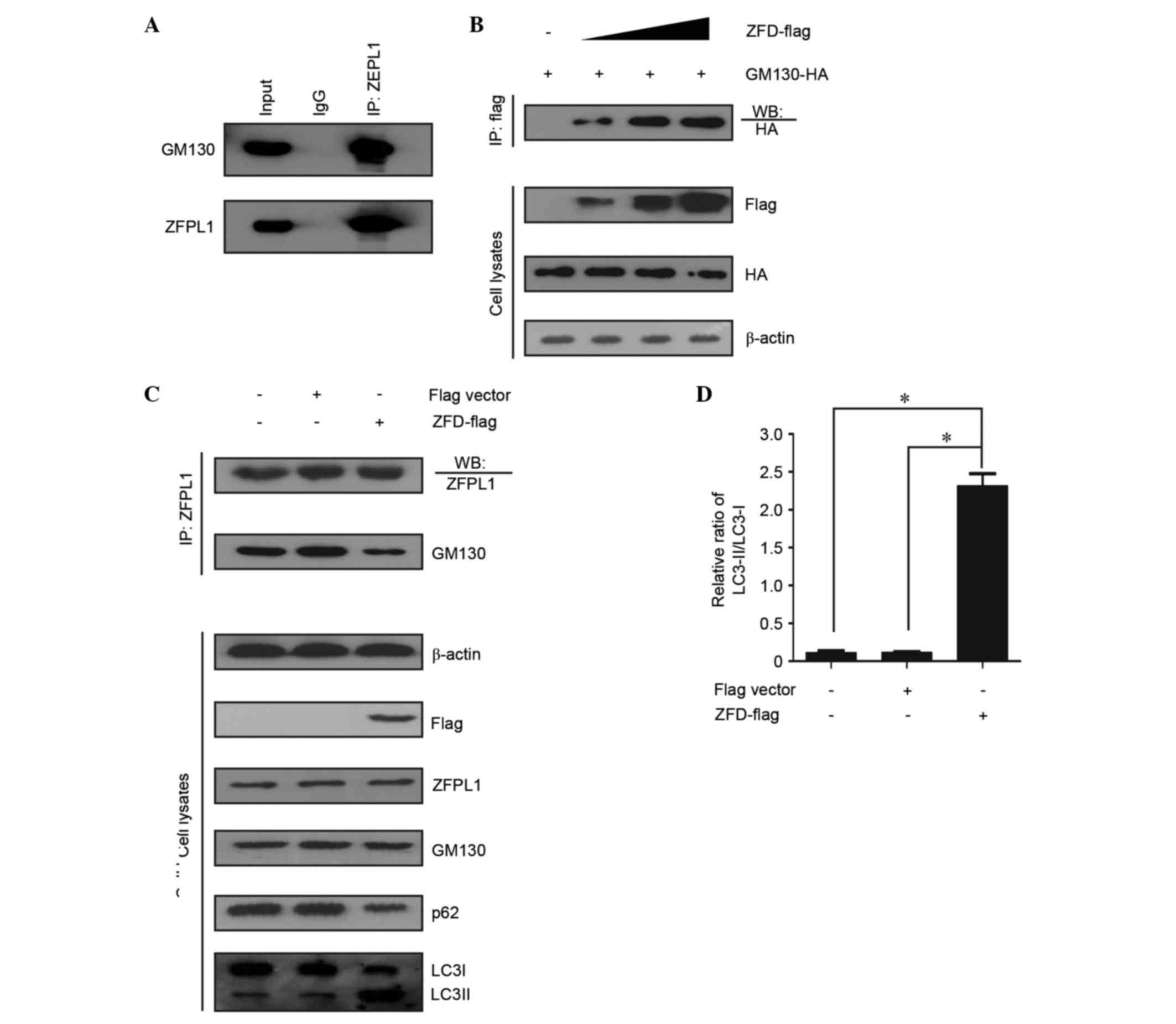 | Figure 5.Effects of ZFPL1 on autophagy are
associated with the interaction between ZFPL1 and GM130. (A)
Endogenous ZFPL1 coimmunoprecipitates with GM130 in NCI-N87 cells.
(B) ZFD-FLAG coimmunoprecipitates with HA-GM130. Human embryonic
kidney 293T cells were grown to 80% confluence, and were then
transiently co-transfected with pCMV HA-GM130 plasmid (6 µg) and
pcDNA3 ZFD-FLAG plasmid (0, 2, 4 and 6 µg). (C) ZFD-FLAG competed
with ZFPL1 for GM130 binding in NCI-N87 cells. NCI-N87 cells stably
expressed FLAG vector or ZFD-FLAG; two thirds of the cell lysate
was used for coimmunoprecipitation experiments, and the remaining
third was used to assess LC3 and p62 expression. (D) LC3II/LC3I
levels were measured using densitometric analysis. Data are
presented as the mean ± standard error of the mean of three
independent experiments. *P<0.05. ZFPL1, zinc finger protein
like 1; shRNA, short hairpin RNA;LC3, microtubule-associated
protein 1A/1B-light chain 3; IP, immunoprecipitation; HA,
hemagglutinin; IgG, immunoglobulin G; Con, control. |
Discussion
GC is the fifth most common cancer globally, and is
the third leading cause of cancer-associated mortality; >723,000
individuals succumb to GC annually, despite significant progress in
early detection and improvements to systemic cytotoxic chemotherapy
regimens (1). The high mortality
rate associated with GC is predominantly due to its silent nature,
advanced stage, and underlying biological and genetic
heterogeneity. A more detailed molecular understanding of the GC
pathogenesis is required to improve patient outcomes. Recent
research regarding biochemical pathways has guided researchers to
identify potential targets. In cancer therapy, tumor cell death is
a vital event in the clearance of cancer cells, and anticancer
therapies often involve caspase-dependent apoptosis (24–26).
However, it has been indicated that cell death could be categorized
to a few types, including ‘accidental cell death’ (ACD) and
‘regulated cell death’ (RCD), obviously caspase-dependent apoptosis
is one subtype of RCD. In the context of antitumor therapy,
autophagic pathways are being considered as potential targets for
therapeutic intervention (27,28).
Three types of autophagy have been discussed in the field of
antitumor therapy, based on their effects on cells: Cytoprotective,
nonprotective and cytotoxic. The three forms can not be clearly
distinguished via molecular or biochemical characteristics
(29). Therefore, autophagy
regulation may provide antitumor therapeutic opportunities.
The present study aimed to explore potential
anticancer effects by silencing the expression of ZFPL1, and
uncovered the mechanisms underlying the effects of ZFPL1 on
autophagy regulation. The results demonstrated that ZFPL1 was
downregulated when autophagy was activated by HBSS or BFA in
NCI-N87 and BGC-823 human gastric cancer cell lines (Fig. 1A). Previous studies regarding
autophagy have indicated that functional autophagy serves a central
role in cellular recycling, homeostasis maintenance and stress
response (30). However, excessive
autophagy disturbs cellular energy supply and negatively affects
cell survival; therefore, autophagic cell death is considered
hyperstimulated self-eating (29,31,32).
The present study also demonstrated that increased cell death was
induced by HBSS or BFA (Fig. 1D).
Therefore, it may be hypothesized that cell death is regulated by
ZFPL1. This hypothesis is supported by a series of experiments.
The results of the present study demonstrated that
ZFPL1 knockdown induces autophagy (Fig. 2D and G), and basic autophagy is
mitigated by overexpression of ZFPL1. It has previously been
reported that ZFPL1 has the properties of a Golgi matrix protein,
and directly interacts with GM130 (14). It is also well known that
downregulation of GM130 results in impaired protein glycosylation
and active autophagy (17). The
majority of mammalian proteins synthesized in the endoplasmic
reticulum undergo glycosylation. Glycosylation acts as a modulator
of GC cellular behavior, including cell differentiation, cell-cell
and cell-matrix interactions, cell-pathogen interactions, invasion,
and metastasis (33). It has
previously been observed that glycosylation modifications may be
good targets for cancer therapy, such as synthesis of the
β1,6GlcNAc branched N-glycans, and these modifications appear to be
an attractive tool for future applications (34,35).
The present study demonstrated that ZFPL1 knockdown was associated
with impaired protein glycosylation (Fig. 3A), and overexpression of ZFPL1
could rescue glycosylation (Fig.
3C). Therefore, as a mediator of glycosylation and autophagy,
downregulation of ZFPL1 in cancer cells may impair cell
survival.
The present study also demonstrated that cell death
rate was increased following ZFPL1 knockdown, and treatment with
the autophagy inhibitor bafilomycin A1 was able to reduce cell
death rate (Fig. 4A). Therefore,
it may be hypothesized that ZFPL1 knockdown-induced cell death is
dependent on autophagy. A series of apoptotic markers were detected
using western blotting, and the expression levels of hardly any of
these markers were increased (Fig. 4B
and C). Autophagy induced by ZFPL1 knockdown was markedly
suppressed using bafilomycin A1 (Fig.
4D). These results indicated that ZFPL1 may have an important
role in autophagy-related cell death. A previous study observed
that GM130 directly binds ZFPL1 (14), and loss of GM130 induces autophagy
(17). Therefore, the interaction
between ZFPL1 and GM130 may be important in autophagy
regulation.
A co-immunoprecipitation analysis was conducted in
NCI-N87 cells, and the interaction between ZFPL1 and GM130 was
detected, in accordance with previous studies (Fig. 5A) (36). There are two ZFDs at the N-terminus
(37), and the first ZFD (1–43
amino acid) has been verified as the GM130-binding region (14). In the present study, ZFD-FLAG
competed with ZFPL1 for GM130 binding; however, wild type ZFPL1
hardly inhibits the binding (Fig. 5B
and C). Notably, autophagy was activated after ZFPL1 was
replaced for GM130 binding by overexpressed ZFD-FLAG. These data
suggest that it is possible to induce autophagy-related cell death
using drugs that interfere with the association between ZFPL1 and
GM130.
In conclusion, the present study demonstrated that
ZFPL1 serves as a regulator of autophagy-related cell death, which
may explain how ZFPL1 in human GC cells underlies the onset of
autophagy. Due to the relevant role of ZFPL1, the reducing the
function of ZFPL1 may provide a potential intervention strategy for
the treatment of GC.
Acknowledgements
The present study was supported by a grant from the
Science and Technology Develop Project in Kaifeng (project number:
1403005).
References
|
1
|
Tan P and Yeoh KG: Genetics and molecular
pathogenesis of gastric adenocarcinoma. Gastroenterology.
149:1153–1162.e3. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wesolowski R, Lee C and Kim R: Is there a
role for second-line chemotherapy in advanced gastric cancer?
Lancet Oncol. 10:903–912. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Takahashi T, Saikawa Y and Kitagawa Y:
Gastric cancer: Current status of diagnosis and treatment. Cancers.
5:48–63. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bhutia SK, Das SK, Azab B, Dash R, Su ZZ,
Lee SG, Dent P, Curiel DT, Sarkar D and Fisher PB: Autophagy
switches to apoptosis in prostate cancer cells infected with
melanoma differentiation associated gene-7/interleukin-24
(mda-7/IL-24). Autophagy. 7:1076–1077. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Janku F, McConkey DJ, Hong DS and Kurzrock
R: Autophagy as a target for anticancer therapy. Nat Rev Clin
Oncol. 8:528–539. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Klionsky DJ and Emr SD: Autophagy as a
regulated pathway of cellular degradation. Science. 290:1717–1721.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Levine B and Kroemer G: Autophagy in the
pathogenesis of disease. Cell. 132:27–42. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Klionsky DJ, Codogno P, Cuervo AM, Deretic
V, Elazar Z, Fueyo-Margareto J, Gewirtz DA, Kroemer G, Levine B,
Mizushima N, et al: A comprehensive glossary of autophagy-related
molecules and processes. Autophagy. 6:438–448. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang J, Li Y, Chen X, Liu T, Chen Y, He
W, Zhang Q and Liu S: Autophagy is involved in anticancer effects
of matrine on SGC-7901 human gastric cancer cells. Oncol Rep.
26:115–124. 2011.PubMed/NCBI
|
|
10
|
Zhang QY, Wu LQ, Zhang T, Han YF and Lin
X: Autophagy-mediated HMGB1 release promotes gastric cancer cell
survival via RAGE activation of extracellular signal-regulated
kinases 1/2. Oncol Rep. 33:1630–1638. 2015.PubMed/NCBI
|
|
11
|
Thedieck K, Holzwarth B, Prentzell MT,
Boehlke C, Kläsener K, Ruf S, Sonntag AG, Maerz L, Grellscheid SN,
Kremmer E, et al: Inhibition of mTORC1 by astrin and stress
granules prevents apoptosis in cancer cells. Cell. 154:859–874.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sun Y, Liu JH, Jin L, Sui YX, Han LL and
Huang Y: Effect of autophagy-related beclin1 on sensitivity of
cisplatin-resistant ovarian cancer cells to chemotherapeutic
agents. Asian Pac J Cancer Prev. 16:2785–2791. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chiu CF, Ghanekar Y, Frost L, Diao A,
Morrison D, McKenzie E and Lowe M: ZFPL1, a novel ring finger
protein required for cis-Golgi integrity and efficient ER-to-Golgi
transport. EMBO J. 27:934–947. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nakamura N, Rabouille C, Watson R, Nilsson
T, Hui N, Slusarewicz P, Kreis TE and Warren G: Characterization of
a cis-Golgi matrix protein, GM130. J Cell Biol. 131:1715–1726.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Keith SA, Maddux SK, Zhong Y, Chinchankar
MN, Ferguson AA, Ghazi A and Fisher AL: Graded proteasome
dysfunction in Caenorhabditis elegans activates an adaptive
response involving the conserved SKN-1 and ELT-2 transcription
factors and the autophagy-lysosome pathway. PLoS Genet.
12:e10058232016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chang SH, Hong SH, Jiang HL, Minai-Tehrani
A, Yu KN, Lee JH, Kim JE, Shin JY, Kang B, Park S, et al:
GOLGA2/GM130, cis-Golgi matrix protein, is a novel target of
anticancer gene therapy. Mol Ther. 20:2052–2063. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Klionsky DJ, Abdalla FC, Abeliovich H,
Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M,
Agostinis P, Aguirre-Ghiso JA, et al: Guidelines for the use and
interpretation of assays for monitoring autophagy. Autophagy.
8:445–544. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Levine B: Cell biology: Autophagy and
cancer. Nature. 446:745–747. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pankiv S, Clausen TH, Lamark T, Brech A,
Bruun JA, Outzen H, Øvervatn A, Bjørkøy G and Johansen T:
p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of
ubiquitinated protein aggregates by autophagy. J Biol Chem.
282:24131–24145. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kepp O, Galluzzi L, Lipinski M, Yuan J and
Kroemer G: Cell death assays for drug discovery. Nat Rev Drug
Discov. 10:221–237. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang J, Whiteman MW, Lian H, Wang G, Singh
A, Huang D and Denmark T: A non-canonical MEK/ERK signaling pathway
regulates autophagy via regulating Beclin 1. J Biol Chem.
284:21412–21424. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wu YC, Wu WK, Li Y, Yu L, Li ZJ, Wong CC,
Li HT, Sung JJ and Cho CH: Inhibition of macroautophagy by
bafilomycin A1 lowers proliferation and induces apoptosis in colon
cancer cells. Biochem Biophys Res Commun. 382:451–456. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fulda S: Targeting extrinsic apoptosis in
cancer: Challenges and opportunities. Semin Cell Dev Biol.
39:20–25. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Reed JC: Apoptosis-targeted therapies for
cancer. Cancer Cell. 3:17–22. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Galluzzi L, Pedro Bravo-San JM, Vitale I,
Aaronson SA, Abrams JM, Adam D, Alnemri ES, Altucci L, Andrews D,
Annicchiarico-Petruzzelli M, et al: Essential versus accessory
aspects of cell death: Recommendations of the NCCD 2015. Cell Death
Differ. 22:58–73. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gewirtz DA: Cytoprotective and
nonprotective autophagy in cancer therapy. Autophagy. 9:1263–1265.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gewirtz DA: The four faces of autophagy:
Implications for cancer therapy. Cancer Res. 74:647–651. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fulda S and Kögel D: Cell death by
autophagy: Emerging molecular mechanisms and implications for
cancer therapy. Oncogene. 34:5105–5113. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li Z, Wang J and Yang X: Functions of
autophagy in pathological cardiac hypertrophy. Int J Biol Sci.
11:672–678. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Galluzzi L, Pietrocola F, Levine B and
Kroemer G: Metabolic control of autophagy. Cell. 159:1263–1276.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pupyshev AB: Reparative autophagy and
autophagy death of cells. Functional and regulatory aspects.
Tsitologiia. 56:179–196. 2014.(In Russian). PubMed/NCBI
|
|
33
|
Pinho SS, Carvalho S, Marcos-Pinto R,
Magalhães A, Oliveira C, Gu J, Dinis-Ribeiro M, Carneiro F, Seruca
R and Reis CA: Gastric cancer: Adding glycosylation to the
equation. Trends Mol Med. 19:664–676. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zavareh Beheshti R, Lau KS, Hurren R,
Datti A, Ashline DJ, Gronda M, Cheung P, Simpson CD, Liu W,
Wasylishen AR, et al: Inhibition of the sodium/potassium ATPase
impairs N-glycan expression and function. Cancer Res. 68:6688–6697.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Contessa JN, Bhojani MS, Freeze HH, Ross
BD, Rehemtulla A and Lawrence TS: Molecular imaging of N-linked
glycosylation suggests glycan biosynthesis is a novel target for
cancer therapy. Clin Cancer Res. 16:3205–3214. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Vitale I, Manic G, Dandrea V and De Maria
R: Role of autophagy in the maintenance and function of cancer stem
cells. Int J Dev Biol. 59:95–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hoppener JW, De Wit MJ, Simarro-Doorten
AY, Roijers JF, van Herrewaarden HM, Lips CJ, Parente F, Quincey D,
Gaudray P, Khodaei S, et al: A putative human zinc-finger gene
(ZFPL1) on 11q13, highly conserved in the mouse and expressed in
exocrine pancreas. The European Consortium on MEN 1. Genomics.
50:251–259. 1998. View Article : Google Scholar : PubMed/NCBI
|