Introduction
Smooth muscle is a major component of human tissues
and is essential for the normal function of many organs, including
the intestines, urinary tract and vascular system (1). Smooth muscle tissue defects or damage
caused by congenital or acquired abnormalities may result in severe
dysfunctions. Tissue engineering and regenerative medicine may be
used to repair these defects using seed cells and biomaterials,
including smooth muscle cells (SMCs) (2), bone marrow stem cells (3) and adipose-derived stem cells (ASCs)
(4). However, mature
differentiated SMCs demonstrate a limited ability to proliferate,
and usually lose their contractile phenotype and convert to a
synthetic phenotype during in vitro expansion (5). Therefore, further investigation into
alternative cell sources for blood vessel engineering is required,
as a large number of functional cell types are usually involved in
this process (6).
Adipose tissue consists of an abundance of
mesenchymal stem cells (MSCs, known as ASCs), with faster growth
and higher proliferative capacities when compared with MSCs from
other sources (7). In addition,
the multipotency of ASCs is independent of the age of the donor
(8,9). ASCs demonstrate the potential to
differentiate into osteocytes (10), neural cells (11) and muscular cells (12).
ASCs express smooth muscle-specific contractile
proteins when stimulated by transforming growth factor-β1 (TGF-β1)
and bone morphogenetic protein-4 (BMP4), and the differentiated
cells exhibit levels of contractility similar to that of SMCs
(13). TGF-β1 and BMP4 are potent
inducers of genes involved in contractility (14). Transcription of contractile genes
is positively regulated by a regulatory DNA element known as the
CArG box (15). The CArG box is
activated by binding of the serum response factor (SRF) together
with its coactivators myocardin (MYOCD) and myocardin-related
transcription factors (MRTFs) (16–18).
Krüppel-like factor 4 (KLF4) inhibits activation of the CArG box
(19). These observations suggest
that modulation of KLF4 may be a prerequisite for the induction of
contractile genes by TGF-b1 and BMP4 (20).
MicroRNAs (miRNAs) are a group of
post-transcriptional regulators that serve a major role in a number
of diverse functions, including cell proliferation, apoptosis and
organogenesis (21). In addition,
miRNAs are regulators of the differentiation, self-renewal and
division of stem cells (22,23).
Previous studies have identified an important role of miR-145 in
mechanisms of smooth muscle differentiation and function, including
the direct and indirect effects of miRNA-145 on MYOCD expression
(24,25), angiotensin signaling (26) and actin polymerization (27). Cheng et al (25) demonstrated that overexpression of
miR-145 increased the expression of vascular smooth muscle cell
(VSMC) differentiation marker genes, including smooth muscle
α-actin, calponin and smooth muscle-myosin heavy chain (SM-MHC).
The levels of these marker genes were decreased in cultured VSMCs
following treatment with a miR-145 inhibitor. In addition to
regulating VSMC differentiation markers, miR-145 alone was able to
maintain the differentiated spindle-like shape and inhibit VSMC
proliferation.
The present study investigated human ASCs treated
with TGF-β1 and BMP4 following transfection with miR-145 mimics or
antisense oligonucleotides. Expression alterations in smooth muscle
contractile proteins and their associated genes were examined to
investigate the effects of miR-145. Short-interfering RNA (si-RNA)
targeting KLF4 was used to mimic the differentiation of ASCs to
SMCs by TGF-β1 and BMP4. This aimed to further clarify the role of
miR-145 in the differentiation process.
Material and methods
Isolation and culture of human ASCs
(hASCs)
Fresh human lipoaspirate fractions were obtained
from 12 donors who had received abdominal liposuction (4 male, 8
female; age, 25–52 years; weight, 82–97 kg). The donors were
admitted by the Research Ethical Committee of The First Affiliated
Hospital of Xinjiang Medical University (Urumqi, China) and were
admitted to the same hospital in 2013. All donors had provided
written informed consent for the use of their samples in the
present study. The study was approved by the Research Ethical
Committee of The First Affiliated Hospital of Xinjiang Medical
University (Urumqi, China). Fresh lipoaspirate fractions were
washed with phosphate-buffered saline (PBS) and treated with 0.075%
type I collagenase (Sigma-Aldrich; Merck Millipore, Darmstadt,
Germany) under shaking (120 rpm) at 37°C for 60 min. The enzyme
activity was neutralized with low-glucose Dulbecco's Modified
Eagle's Medium (LG-DMEM; Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) containing 10% fetal bovine serum (FBS; Hylone;
GE Healthcare Life Sciences, Logan, UT, USA). The digested
lipoaspirate samples were then centrifuged at 1,200 × g for 10 min
at 20°C to obtain the high density stromal vesicular fraction,
which was filtered through a 50 µm nylon mesh to remove undigested
tissue, and subsequently centrifuged at 1,000 × g for 10 min at
20°C. The supernatant was discarded and the pellet was resuspended
in LG-DMEM supplemented with 10% FBS, 100 U/ml penicillin
(Sigma-Aldrich; Merck Millipore) and 100 mg/ml
penicillin/streptomycin (Sigma-Aldrich; Merck Millipore). The cells
were seeded in 100-mm culture dishes at a density of
4×104 cells/cm2 and the medium was refreshed
twice each week. When cells reached 70–80% confluence, they were
passaged and cells from passage 3 to 5 were used for the purposes
of this study. The ability of hASCs to differentiate into
osteogenic, adipogenic and chondrogenic lineages was examined as
previously reported (8).
Induction of SMC differentiation
The hASCs were induced to differentiate into SMCs
using TGF-β1 and BMP4 (R&D Systems, Inc., Minneapolis, MN, USA)
following serum starvation. The differentiation medium consisted of
5 ng/ml TGF-β1, 2.5 ng/ml BMP4 and 1% FBS. The media was refreshed
every 2 days. Cell characterization and functional evaluation was
performed following 7 days of culture.
miRNA mimic and antisense
oligonucleotide transfection
The miRNA mimics, antisense oligonucleotides and
negative control were synthesized by Guangzhou RiboBio Co., Ltd.
(Guangzhou, China). The miR-145 and anti-miR-145 oligonucleotides
were designed according to the miRBase sequence database
(mirbase.org) (28).
The sequence of the miR-145 mimic was
5′-GUCCAGUUUUCCCAGGAAUCCCU-3′; and the anti-miR-145 sequence was
5′-AGGGAUUCCUGGGAAAACUGGAC-3′. A random sequence was used as the
negative control: 5′-CGGCGGTTGAGATGAAGCACTG-3′.
The ASCs were seeded onto a 24-well culture plate at
a density of 1×105 cells/cm2 at 24 h prior to
transfection. Cells were transfected when cells reached 80%
confluency. The miR-145 mimics, miR-145 inhibitor and negative
controls were diluted using 30 µl 1X riboFECT™ CP Buffer
(Guangzhou RiboBio Co., Ltd.) and incubated at room temperature for
5 min. A total of 3 µl riboFECT™ CP Reagent (Guangzhou
RiboBio Co., Ltd.) was added and incubated at room temperature for
15 min. riboFECT™ CP mixture (Guangzhou RiboBio Co.,
Ltd.) was added to 465.75 µl serum-free, cell culture medium
without penicillin/streptomycin. All cells were cultured for 6 h
with above conditions. Culture medium was then changed to growth
medium (DMEM with 10% FBS without penicillin/streptomycin). Cells
were harvested 48 h following transfection and were then
analyzed.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
For RT-qPCR analysis, RNA was extracted using TRIzol
Reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's instructions, and cDNA was synthesized using
PrimeScript™ RT Master Mix (Takara Bio, Inc., Otsu, Japan). A total
of 1 µl total RNA was reverse transcribed into cDNA. The 5X
PrimerScript master mix (Takara Bio, Inc.) was used to reverse
transcribe RNA. The reactions were performed and monitored in a
Biometra T3 thermocycler (Biometra GmbH, Göttingen, Germany). qPCR
was performed using a 7500 Fast Real Time PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.) with thermocycling
parameters consisting of 94°C for 3 min, followed by 40 cycles of
94°C for 15 sec and 60°C for 40 sec, according to the
manufacturer's protocol. SYBR Premix Ex Taq (Takara Bio, Inc.) was
used in each reaction. The PCR primers were as follows: a-smooth
muscle actin (α-SMA), forward, 5′-GGTGATGGTGGGAATGGG-3′ and
reverse, 5′-GCAGGGTGGGATGCTCTT-3′; smooth muscle protein-22α
(SM22α), forward, 5′-AACAGCCTGTACCCTGATGG-3′ and reverse,
5′-CGGTAGTGCCCATCATTCTT-3′; Calponin, forward,
5′-ATGTCCTCTGCTCACTTCA-3′ and reverse, 5′-TTTCCGCTCCTGCTTCTCT-3′;
SM-MHC, forward, 5′-TGCTTTCGCTCGTCTTCC-3′ and reverse,
5′-CGGCAACTCGTGTCCAAC-3′; KLF4, forward, 5′-CCCAATTACCCATCCTTCCT-3′
and reverse, 5′-CGTCCCAGTCACAGTGGTAA-3′; miR-145, forward,
5′-CGGCGGTGTCCAGTTTTCCCAGGA-3′ and reverse,
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGGGAT-3′. β-actin
forward, 5′-ATCATGTTTGAGACCTTCAA-3′ and reverse,
5′-CATCTCTTGCTCGAAGTCCA-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and
reverse, 5′-AACGCTTCACGAATTTGCGT-3′. All experiments were performed
in triplicate. The relative expression of target mRNA was
normalized to the expression of β-actin. The expression level of
miR-145 was normalized to U6 and the fold-change was calculated
using the 2−∆∆Cq method of relative quantification
(29).
Western blotting
Cells were harvested and lysed in
radioimmunoprecipitation buffer with added protease and phosphatase
inhibitors (Roche Diagnostics, Indianapolis, IN, USA). The protein
concentration was determined using the bicinchoninic acid method
(Bio-Rad Laboratories, Inc., Hercules, CA, USA). Total protein
(~20–50 µg) was separated using a 10% SDS-PAGE gel, before it was
transferred to polyvinylidene difluoride membranes. Membranes were
blocked in Tris-buffered saline containing 0.05% Tween-20
(Sigma-Aldrich; Merck Millipore) and 5% skim milk, and then
incubated with the following primary antibodies from Abcam
(Cambridge, UK): Rabbit polyclonal to SMA (cat. no, ab5694;
dilution, 1:1,000), rabbit polyclonal anti-SM22α (cat. no, ab14106;
dilution, 1:1,000), rabbit monoclonal anti-calponin (cat. no,
ab46794; dilution, 1:20,000) rabbit polyclonal to SM-MHC (cat. no,
ab53219; dilution, 1:1,000), rabbit monoclonal anti-KLF4 (cat. no,
ab151733; dilution, 1:1,000) and GAPDH (cat. no, ab181603;
dilution, 1:2,000). Membranes were then washed with Tris-HCl with
Tween-20 (TBST; Sigma Aldrich; Merck-Millipore), incubated with the
secondary antibody (horseradish peroxidase-conjugated goat
anti-rabbit antibody, dilution, 1:2,000; AP307P; EMD Millipore,
Billerica, MA, USA) at room temperature for 2 h and detected using
enhanced chemiluminescence (Bio-Rad Laboratories, Inc.). GAPDH was
used as an internal loading control.
RNA interference
As a negative control, a non-targeting scrambled
siRNA (Shanghai Usen Biotechnology, Shanghai, China) was used. The
sequence of the KLF4 siRNA was 5′-GGACGGCUGUGGAUGGAAATT-3′
(Shanghai Usen Biotechnology). The hASCs were seeded into 24-well
plates at density of 1×105 cells/cm2 and
transfected with 100 pmol/well siRNA using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's instructions. The medium was refreshed following 6 h
transfection, and the cells were harvested 48 h following
transfection for total RNA and protein isolation.
Immunofluorescence staining
Immunofluorescence was performed on methanol-fixed
cells (density, 1×105 cells/cm2; 4% methanol)
using the following primary antibodies purchased from Abcam: Rabbit
polyclonal anti-α-SMA (dilution, 1:100; cat. no. ab5694), rabbit
polyclonal anti-SM22α (dilution, 1:250; cat. no. ab14106), rabbit
monoclonal anti-calponin (dilution, 1:150; cat. no. ab46794) and
rabbit polyclonal anti-SM-MHC (dilution, 1:50; cat. no. ab53219).
Following incubation with primary antibodies for 60 min at room
temperature, the cells were washed with PBS three times. The Alexa
Fluor 594-conjugated donkey anti-rabbit secondary antibody
(dilution, 1:5,000; cat. no. R37119; Thermo Fisher Scientific,
Inc.) was used to detect the localization of anti-α-SMA,
anti-SM22α, anti-calponin and anti-SM-MHC antibodies. Cell nuclei
were stained with DAPI. The images were visualized using a confocal
laser scanning platform microscope (TCS SP8; Leica Microsystems
GmbH, Wetzlar, Germany).
Luciferase reporter assay
The wild-type 3′-untranslated (UTR) regions of KLF-4
(nt=549-1988) and corresponding mutations were amplified using PCR,
followed by cloning into a pLVX report vector (Thermo Fisher
Scientific, Inc.). 293T cells (Shanghai Institutes for Biological
Sciences, Chinese Academy of Sciences, Shanghai, China) were seeded
onto 24-well plates at a density of
1×105/cm2. When the cell confluency reached
70–80%, the reporter gene plasmid and miR-145 were co-transfected
into 293T cells by using Lipofectamine 2000 (Thermo Fisher
Scientific, Inc.). Following 30 h after transfection, cells were
collected and luciferase activity was detected. The experiment was
repeated in triplicate.
Statistical analysis
Data are expressed as mean ± standard deviation from
at least three independent experiments. Statistical analysis was
performed using Student's t-test using SPSS software (version,
17.0; SPSS Inc., Chicago, IL, USA). P<0.05 was considered to
indicate a statistically significant difference.
Results
hASCs differentiate into SMCs when
treated with TGF-β1 and BMP4
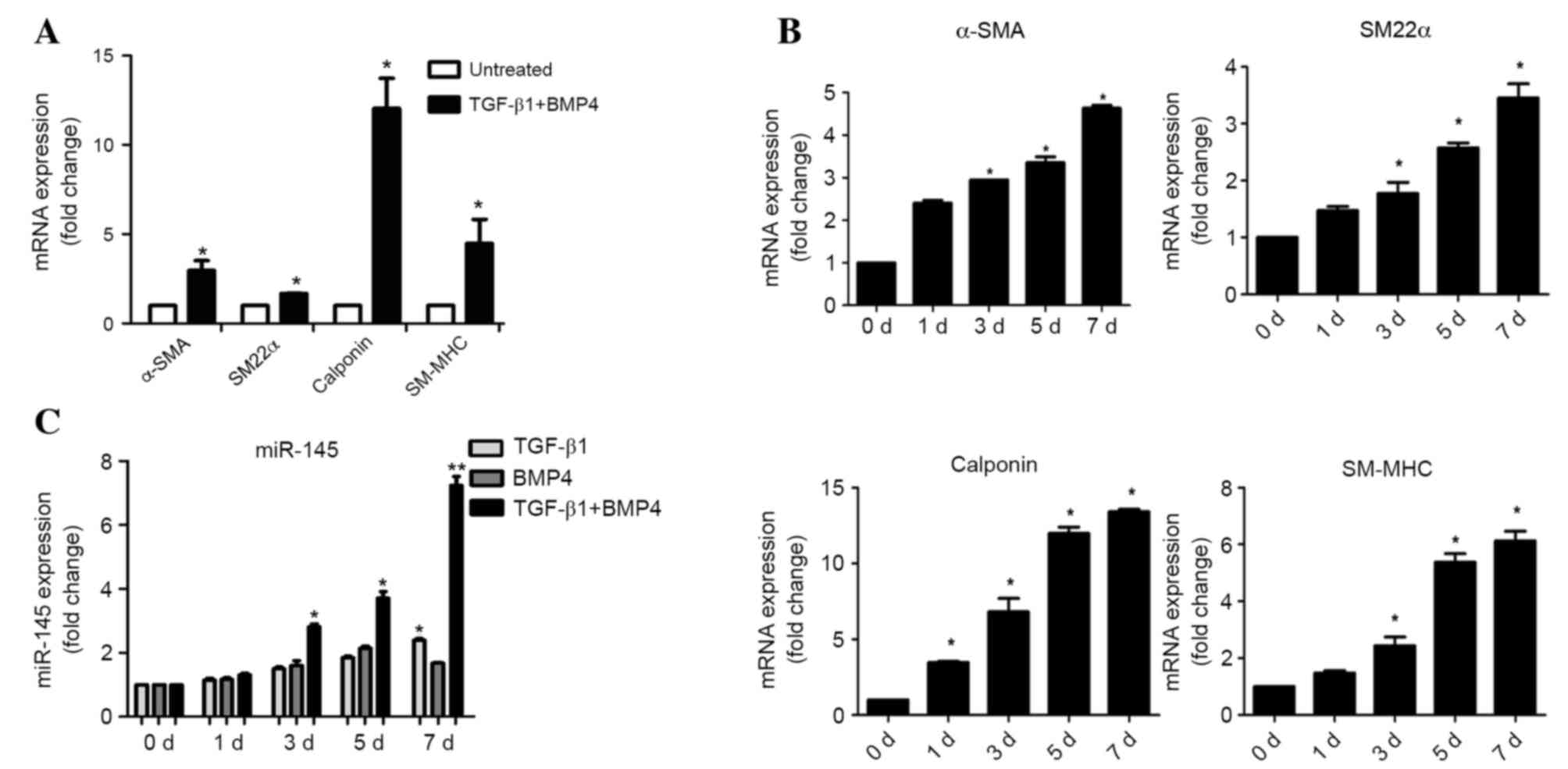
In preliminary experiments, hASCs were successfully
induced into SMCs. The hASCs exhibited a fine, elongated,
fibroblast-like morphology when subcultured in normal medium. Cells
at passages 3–5 stimulated by TGF-β1 and BMP4 for 7 days
demonstrated a spindle-like morphology and proliferated at
fluctuating rates (data not shown). To confirm whether hASCs
differentiated into SMCs when treated with TGF-β1 and BMP4, mRNA
levels of smooth muscle-specific contractile proteins α-SMA, SM22α,
calponin and SM-MHC were detected by RT-qPCR analysis. The mRNA
levels of these markers were significantly increased 7 days
following differentiation (α-SMA, P=0.032; SM22α, P=0.041;
calponin, P=0.018; SM-MHC, P=0.027) (Fig. 1A and B). To investigate the role of
miR-145 in SMC differentiation, miR-145 expression levels were
detected by RT-qPCR on day 0, 1, 3, 5 and 7 following
differentiation. The expression of miR-145 significantly increased
at day 3, 5 and 7 following induction of differentiation with
TGF-β1 and BMP4 (3 d, P=0.019; 5 d, P=0.011; 7 d, P=0.002; Fig. 1C).
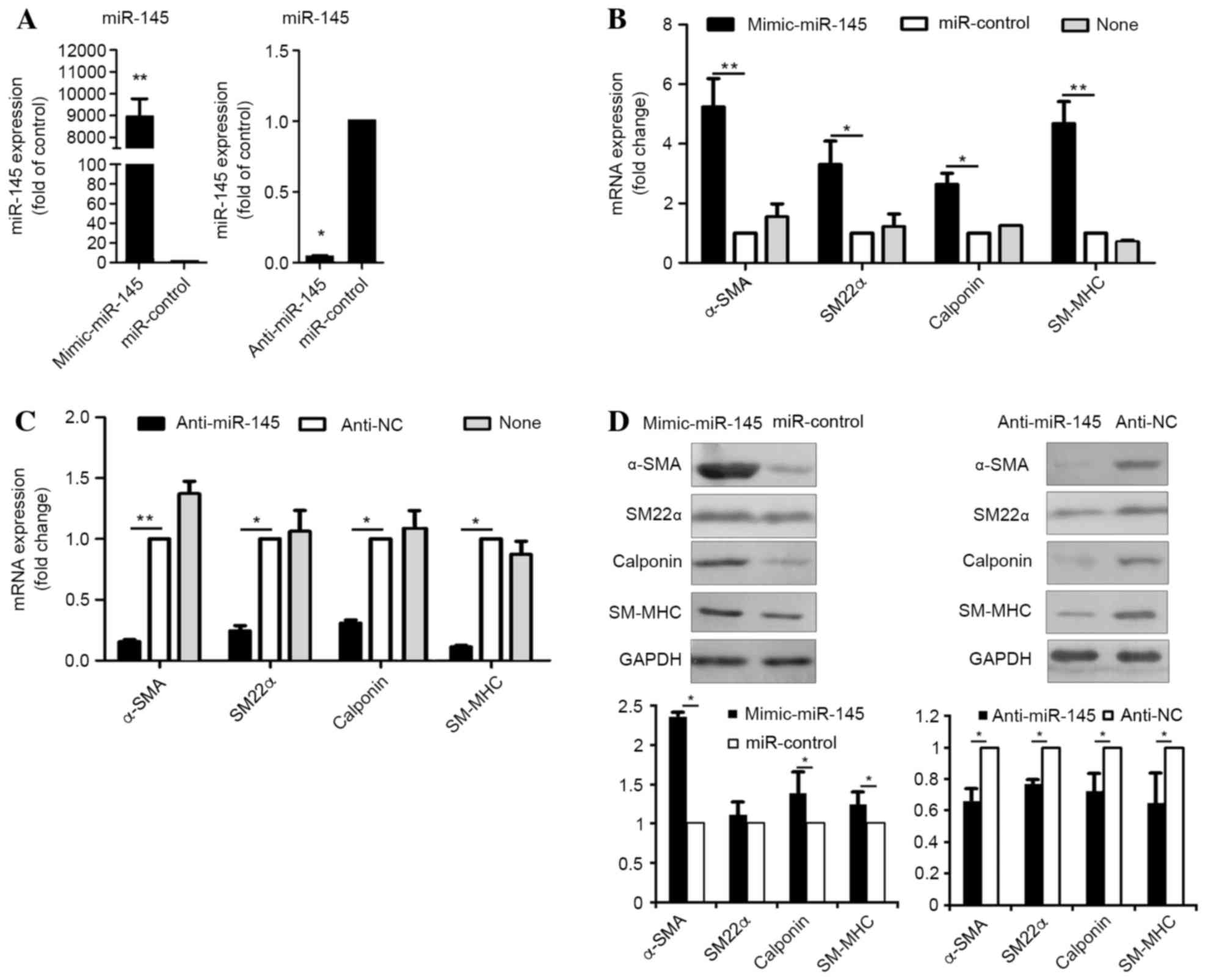
Overexpression of miR-145 levels in
hASCs undergoing smooth muscle cell differentiation
For functional evaluation of miR-145, hASCs were
transfected with miR-145 mimics, their counterpart inhibitor
(miR-control), anti-miR-145 or a negative control (anti-NC).
Quantification of miR-145 expression levels by RT-qPCR analysis
demonstrated that intracellular miR-145 levels in hASCs transfected
with miR-145 mimics was ~9,000-fold higher when compared with cells
transfected with the negative control (P=0.0001) (Fig. 2A). In addition, hASCs transfected
with anti-miR-145 exhibited a ~10-fold lower level of miR-145
expression when compared with cells transfected with the control
(P=0.029) (Fig. 2A). Following
transfection, hASCs were cultured in medium supplemented with
TGF-β1 and BMP4 for seven days following starvation. The markers of
SMCs were analyzed by RT-qPCR analysis. The mRNA levels of these
markers were significantly increased following transfection with
miR-145 mimic (α-SMA, P=0.0072; SM22α, P=0.031; calponin, P=0.038;
SM-MHC, P=0.0007; Fig. 2B),
whereas they were significantly decreased by anti-miR-145 treatment
(α-SMA, P=0.0081; SM22α, P=0.036; calponin, P=0.015; SM-MHC,
P=0.035; Fig. 2C), when compared
with negative controls. In addition, the results demonstrated that
smooth muscle-specific protein expression levels were significantly
increased following transfection of miR-145 mimics (α-SMA, P=0.024;
SM22α, P=0.345; calponin, P=0.029; SM-MHC, P=0.022), but were
significantly decreased by anti-miR-145 (α-SMA, P=0.039; SM22α,
P=0.040; calponin, P=0.026; SM-MHC, P=0.011; Fig. 2D), when compared with the negative
controls. The results suggest that miR-145 may function as a
positive regulator of smooth muscle differentiation in hASCs.
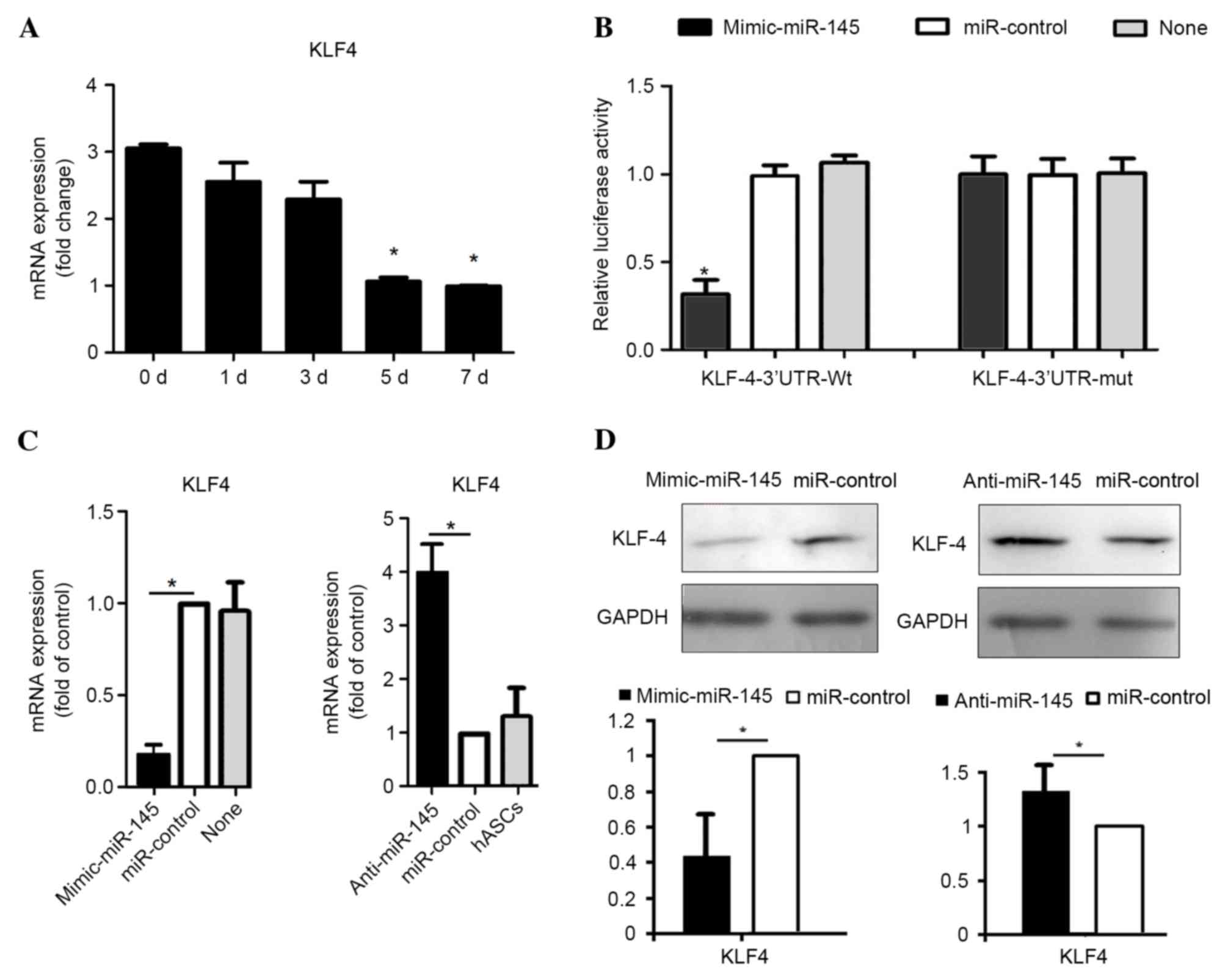
miR-145 targets KLF4
In order to understand the molecular mechanisms
underlying miR-145-mediated regulation of SMC differentiation,
potential targets of miR-145 that are implicated in SMC
differentiation were identified, using miRNA target prediction
algorithms, TargetScan and PicTar (30). The results demonstrated that MYOCD
KlF-5, fascin, calcium/calmodulin kinase II-δ and adducin-3 are the
predicted targets of miR-145. Among the predicted targets, KLF4 was
identified as serving an established role in smooth muscle cell
differentiation (20). Therefore,
the expression level of KLF4 in SMC differentiation was examined in
the present study. The results demonstrated that the mRNA level of
KLF4 significantly decreased during treatment with TGF-β1 and BMP4
at 5 (P=0.028) and 7 (P=0.016) days (Fig. 3A), which contrasted that of miR-145
expression (Fig. 1C). This
suggests that miR-145 may suppress KLF4 expression during SMC
differentiation. To verify that KLF4 is a functional target of
miR-145, the wild-type 3′-UTR of KLF-4 (KLF4-3′-UTR-Wt) was cloned
into a reporter plasmid. Co-transfection of the miR-145 mimic and
KLF4-3′-UTR-Wt strongly decreased luciferase activity (P=0.033),
whereas co-transfection of miR-control with KLF4-3′-UTR-Wt did not
alter the luciferase activity (Fig.
3B). These results suggest that miR-145 inhibited the 3′-UTR of
KLF4. A KLF4 luciferase reporter construct containing a predicted
miR-145 binding site was mutated (KLF4-3′-UTR-Mut) to determine
whether miR-145 targets KLF4 directly through the predicted binding
site. This binding site is located at position 278–284 of the KLF4
3′ UTR (14) Co-transfection of
the miR-145 mimic and KLF4-3′-UTR-Mut did not alter the luciferase
activity (Fig. 3B).
To investigate whether miR-145 may be able to
downregulate KLF4 expression, the mRNA and protein levels were
detected after hASCs were transfected with miR-145 mimics and
inhibitors using RT-qPCR and western blotting analyses,
respectively. The results indicated that the mRNA level of KLF4 was
significantly decreased by miR-145 overexpression (P=0.029),
whereas it was significantly increased by anti-miR-145 transfection
(P=0.044), when compared with cells transfected with negative
controls (Fig. 3C). In addition,
the protein expression level of KLF4 was significantly higher in
cells transfected with anti-miR-145 (P=0.017) and significantly
lower in cells overexpressing miR-145 compared with the controls
(P=0.031) (Fig. 3D). These results
demonstrate that miR-145 may downregulate KLF4 in hASCs treated
with TGF-β1 and BMP4.
KLF4 demonstrates negative effect on
the smooth muscle differentiation of hASCs
To examine the role of KLF4 in the control of
contractile gene expression, siRNA was used to knockdown KLF4
expression, thus mimicking the effect of miR-145 expression during
induction by TGF-β1 and BMP4. KLF4 protein expression was
downregulated by 90% and mRNA expression was downregulated by 40%,
following transfection with KLF4 siRNA (si-KLF4) in hASCs (Fig. 4A and B). Under these conditions,
the levels of contractile proteins, α-SMA, SM22α, calponin and
SM-MHC were elevated, as determined by western blot analysis
(α-SMA, P=0.039; SM22α, P=0.152; calponin, P=0.018; SM-MHC,
P=0.035; Fig. 4C) and
immunofluorescence staining (Fig.
4D), indicating that endogenous KLF4 constitutively represses
the expression of contractile genes. These results provide further
evidence of the inhibitory role of KLF4 in the induction of
contractile genes by TGF-β1 and BMP4.
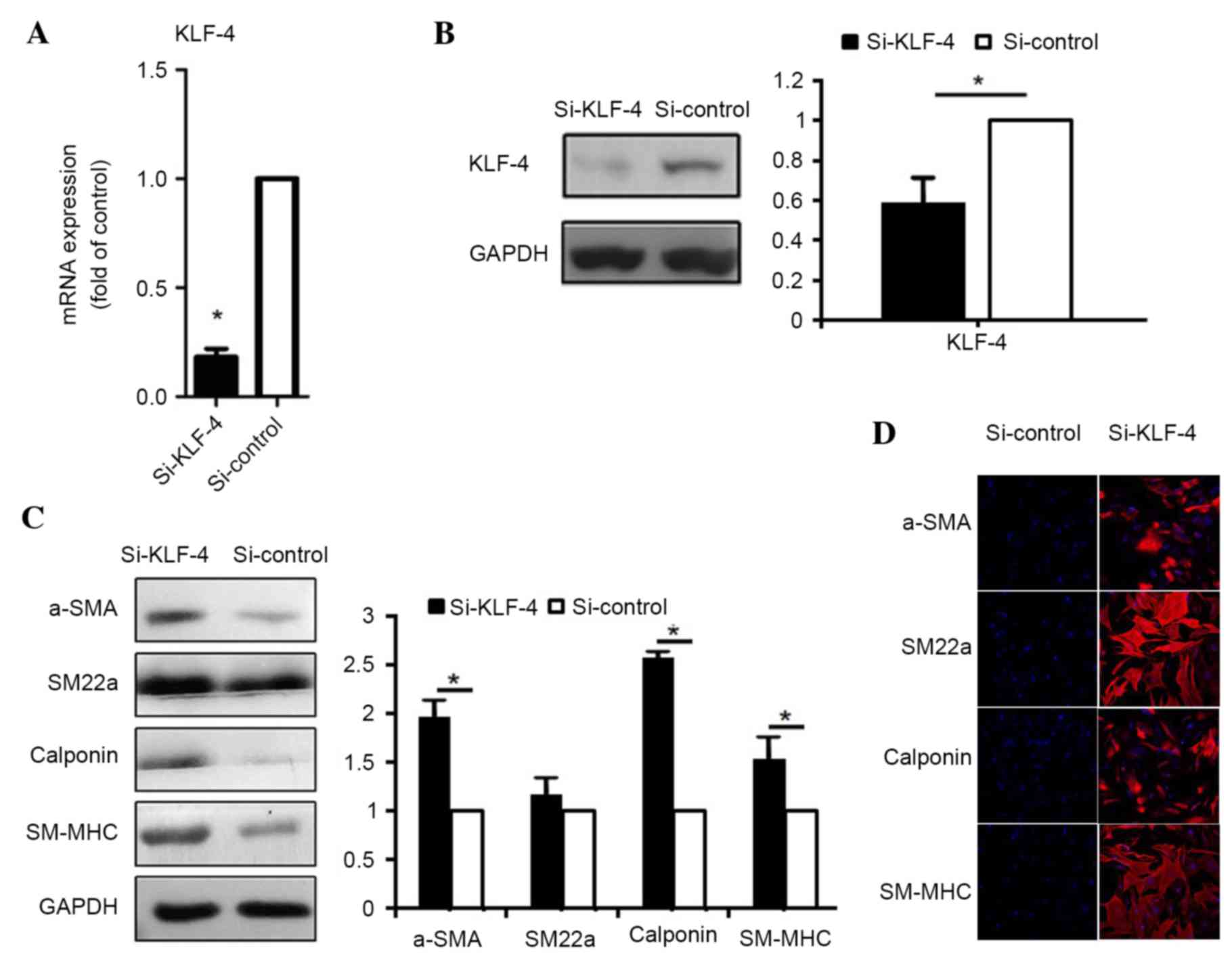 | Figure 4.KLF4 represses the expression of
smooth muscle contractile proteins and associated genes. The (A)
mRNA and (B) protein expression levels of KLF4 following
transfection with non-targeting siRNA (si-control) and siRNA
targeting KLF4 (si-KLF-4). (C) Smooth muscle contractile protein
expression following transfection with si-control and si-KLF4. (D)
Following transfection with si-control and si-KLF4, cells were
subject to immunofluorescence staining with antibodies against
α-SMA, SM22α, calponin and SM-MHC (red) and nuclear staining with
DAPI (blue), respectively (magnification, ×200). Data are presented
as the mean ± standard deviation (n=3). *P<0.05 vs. si-control.
KLF4, Krüppel-like factor 4; α-SMD, α-smooh muscle actin; siRNA,
small interfering RNA; SM22α, smooth muscle protein-22α; SM-MHC,
smooth muscle-myosin heavy chain. |
Discussion
SMCs exhibit a spectrum of phenotypes, ranging from
the more differentiated ‘contractile’ state, in which high levels
of SMC differentiation markers are expressed, to the less
differentiated ‘synthetic’ state, in which SMC markers are
downregulated (15,31,32).
Alterations in SMC differentiation states serve a major role in a
number of cardiovascular diseases such as atherosclerosis,
restenosis, hypertension and aneurysm (33). Therefore, an improved understanding
of the molecular mechanisms that control SMC differentiation of
ASCs is critical for enabling the development of novel strategies
to prevent and treat these diseases.
miRNAs are small noncoding RNAs that regulate gene
and protein expression by interacting with the 3′-UTR of the target
mRNA sequences. This interaction results in mRNA degradation and/or
inhibition or activation of protein translation (34). A number of miRNAs serve integral
roles in smooth muscle development and contractile differentiation.
The aim of the present study was to clarify the role of miRNAs in
the SMC differentiation of hASCs. The results demonstrated that
miR-145 promoted smooth muscle differentiation of hASCs. Firstly,
ASCs were induced to differentiate to SMCs using TGF-b1 and BMP4,
according to a previous study (13). The expression of smooth muscle
contractile proteins and genes were increased following this
induction. These results confirmed that ASCs successfully
differentiated into SMCs. In addition to smooth muscle contractile
proteins and genes, miR-145 expression was similarly upregulated
during this process. ASCs were transfected with miR-145 mimics,
antisense oligonucleotides and negative controls to investigate the
effect of miR-145 on SMC differentiation of ASCs. The results
indicated that miR-145 promotes smooth muscle contractile protein
expression, while ASCs transfected with anti-miR-145 demonstrated
the opposite effect. These findings are in accordance with earlier
reports (20,24,35).
To investigate the molecular mechanisms by which
miR-145 regulates the smooth muscle differentiation of ASCs,
potential target genes of miR-145 were investigated. One gene,
KLF4, was hypothesized to be a target of miR-145 using miRNA target
prediction software. miR-145 has a target site in the 3′-UTR of
KLF4 (20). Previous studies have
demonstrated that KLF4 is involved in smooth muscle differentiation
(36,37).
In addition, miR-145 overexpression downregulated
KLF4 at the protein and mRNA level, whereas functional inhibition
of miR-145 by anti-miR-145 led to increased expression of KLF4.
This suggested that miR-145 regulates KLF4 during smooth muscle
differentiation. To examine the role of KLF4 in the control of
contractile gene expression, siRNA was used to knockdown KLF4
expression, thus mimicking the effect of miR-145. Under these
conditions, the expression levels of contractile proteins were
dramatically elevated. This result was in accordance with previous
reports (38–40) and indicated a negative effect of
KLF4 on smooth muscle differentiation. KLF4 is a potent negative
regulator of contractile genes, which functions through different
mechanisms, including suppressing the transcriptional activation of
the CArG box by SRF, MYOCD or MRTFs (19). In the current study, the functional
significance of KLF4 downregulation by miR-145 during TGF-β1 and
BMP4-mediated induction of smooth muscle differentiation was
investigated. The results established that basal expression of
contractile genes increased following the siRNA-mediated knockdown
of KLF4. By contrast, it was previously reported that, in rat
aortic SMCs (rAoSMCs) (41) and
mouse mesenchymal C3H10T1/2 cells (8), TGF-β1 and BMP4 induced KLF4
expression at the same time points. Primary rAoSMCs treated with
TGF-β1 or BMP4 did not upregulate miR-145, and no repression of
KLF4 was observed. Thus, the induction of miR-145 by TGF-β1 and
BMP4, and the subsequent suppression of KLF4, may be specific to
human SMCs (20).
The present study aimed to predict additional
targets of miR-145 alongside KLF4. The analysis demonstrated that
miR-145 targets a wide range of factors, including MYOCD KlF-5,
fascin, calcium/calmodulin kinase II-δ and adducin-3. It could
therefore be suggested that attenuation of the expression of these
proteins may promote differentiation and repress the proliferation
of SMCs. These targets are involved in a number of smooth muscle
differentiation processes (24),
and provide an example of how one miRNA may regulate several
related or unrelated cellular processes. In addition to miR-145,
miR-21, miR-221, miR-222 and miR-143 have established roles in VSMC
differentiation. miR-21 promotes VSMC differentiation by negatively
regulating the expression of programmed cell death protein 4
(42), whereas miR-221 and miR-222
promote VSMC proliferation by targeting the negative regulators of
the cell cycle, p27 and p57 (43,44).
miR-143 and miR-145-encoding genes are highly conserved, and are
positioned in close proximity on mouse chromosome 18 and human
chromosome 5 (24). Recently,
miR-143 and miR-145, which are encoded as a gene cluster, were
observed to target KLF4 and serve a critical role in regulation of
the VSMC phenotype (24,25,27).
miR-143 or miR-145 VSMC gene knockout mice exhibited abnormal
vascular tone and reduced contractile gene expression (24).
In conclusion, the current study demonstrated that
miR-145 is a promoter of smooth muscle differentiation of hASCs
induced by TGF-β1 and BMP4. miR-145 targets the KLF4 gene, whose
protein may function as a potent negative regulator of contractile
gene expression. Overexpression of miR-145 suppressed KLF4 and
promoted smooth muscle contractility induced by stem cells. There
are additional mechanisms of smooth muscle differentiation, which
will require further investigation. Further understanding of the
molecular mechanisms would facilitate the treatment or prevention
of smooth muscle-associated diseases.
Acknowledgements
The present study was financially supported by the
National Natural Science Foundation of China (grant no.
81201204).
References
|
1
|
Rodríguez LV, Alfonso Z, Zhang R, Leung J,
Wu B and Ignarro LJ: Clonogenic multipotent stem cells in human
adipose tissue differentiate into functional smooth muscle cells.
In: Proc Natl Acad Sci USA. 103. pp. 12167–12172. 2006; View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lai JY, Yoon CY, Yoo JJ, Wulf T and Atala
A: Phenotypic and functional characterization of in vivo tissue
engineered smooth muscle from normal and pathological bladders. J
Urol. 168:1853–1857. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Silva GV, Litovsky S, Assad JA, Sousa AL,
Martin BJ, Vela D, Coulter SC, Lin J, Ober J, Vaughn WK, et al:
Mesenchymal stem cells differentiate into an endothelial phenotype,
enhance vascular density, and improve heart function in a canine
chronic ischemia model. Circulation. 111:150–156. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bacou F, el Andalousi RB, Daussin PA,
Micallef JP, Levin JM, Chammas M, Casteilla L, Reyne Y and Nougues
J: Transplantation of adipose tissue-derived stromal cells
increases mass and functional capacity of damaged skeletal muscle.
Cell Transplant. 13:103–111. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xu ZC, Zhang WJ, Li H, Cui L, Cen L, Zhou
GD, Liu W and Cao Y: Engineering of an elastic large muscular
vessel wall with pulsatile stimulation in bioreactor. Biomaterials.
29:1464–1472. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang C, Cen L, Yin S, Liu Q, Liu W, Cao Y
and Cui L: A small diameter elastic blood vessel wall prepared
under pulsatile conditions from polyglycolic acid mesh and smooth
muscle cells differentiated from adipose-derived stem cells.
Biomaterials. 31:621–630. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kern S, Eichler H, Stoeve J, Klüter H and
Bieback K: Comparative analysis of mesenchymal stem cells from bone
marrow, umbilical cord blood, or adipose tissue. Stem Cells.
24:1294–1301. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cartwright MJ, Tchkonia T and Kirkland JL:
Aging in adipocytes: Potential impact of inherent, depot-specific
mechanisms. Exp Gerontol. 42:463–471. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Weinzierl K, Hemprich A and Frerich B:
Bone engineering with adipose tissue derived stromal cells. J
Craniomaxillofac Surg. 34:466–471. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zuk PA, Zhu M, Ashjian P, De Ugarte DA,
Huang JI, Mizuno H, Alfonso ZC, Fraser JK, Benhaim P and Hedrick
MH: Human adipose tissue is a source of multipotent stem cells. Mol
Biol Cell. 13:4279–4295. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ashjian PA, Elbarbary AS, Edmonds B,
DeUgarte D, Zhu M, Zuk PA, Lorenz HP, Benhaim P and Hedrick HK: In
vitro differentiation of human processed lipoaspirate cells into
early neural progenitors. Plast Reconstr Surg. 111:1922–1931. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Deslex S, Negrel R, Vannier C, Etienne J
and Ailhaud G: Differentiation of human adipocyte precursors in a
chemically defined serum-free medium. Int J Obes. 11:19–27.
1987.PubMed/NCBI
|
|
13
|
Wang C, Yin S, Cen L, Liu Q, Liu W, Cao Y
and Cui L: Differentiation of adipose-derived stem cells into
contractile smooth muscle cells induced by transforming growth
factor-beta1 and bone morphogenetic protein-4. Tissue Eng Part A.
16:1201–1213. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lagna G, Ku MM, Nguyen PH, Neuman NA,
Davis BN and Hata A: Control of phenotypic plasticity of smooth
muscle cells by bone morphogenetic protein signaling through the
myocardin-related transcription factors. J Biol Chem.
282:37244–37255. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Owens GK, Kumar MS and Wamhoff BR:
Molecular regulation of vascular smooth muscle cell differentiation
in development and disease. Physiol Rev. 84:767–801. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yoshida T, Sinha S, Dandré F, Wamhoff BR,
Hoofnagle MH, Kremer BE, Wang DZ, Olson EN and Owens GK: Myocardin
is a key regulator of CArG-dependent transcription of multiple
smooth muscle marker genes. Circ Res. 92:856–864. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang Z, Wang DZ, Hockemeyer D, McAnally J,
Nordheim A and Olson EN: Myocardin and ternary complex factors
compete for SRF to control smooth muscle gene expression. Nature.
428:185–189. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Miano JM: Channeling to myocardin. Circ
Res. 95:340–342. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu Y, Sinha S, McDonald OG, Shang Y,
Hoofnagle MH and Owens GK: Kruppel-like factor 4 abrogates
myocardin-induced activation of smooth muscle gene expression. J
Biol Chem. 280:9719–9727. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Davis-Dusenbery BN, Chan MC, Reno KE,
Weisman AS, Layne MD, Lagna G and Hata A: Down-regulation of
Kruppel-like factor-4 (KLF4) by microRNA-143/145 is critical for
modulation of vascular smooth muscle cell phenotype by transforming
growth factor-beta and bone morphogenetic protein 4. J Biol Chem.
286:28097–29110. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Alvarez-Garcia I and Miska EA: MicroRNA
functions in animal development and human disease. Development.
132:4653–4662. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hatfield SD, Shcherbata HR, Fischer KA,
Nakahara K, Carthew RW and Ruohola-Baker H: Stem cell division is
regulated by the microRNA pathway. Nature. 435:974–978. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kanellopoulou C, Muljo SA, Kung AL,
Ganesan S, Drapkin R, Jenuwein T, Livingston DM and Rajewsky K:
Dicer deficient mouse embryonic stem cells are defective in
differentiation and centromeric silencing. Genes Dev. 19:489–501.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cordes KR, Sheehy NT, White MP, Berry EC,
Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN and Srivastava D:
miR-145 and miR-143 regulate smooth muscle cell fate and
plasticity. Nature. 460:705–710. 2009.PubMed/NCBI
|
|
25
|
Cheng Y, Liu X, Yang J, Lin Y, Xu DZ, Lu
Q, Deitch EA, Huo Y, Delphin ES and Zhang C: MicroRNA-145, a novel
smooth muscle cell phenotypic marker and modulator, controls
vascular neointimal lesion formation. Circ Res. 105:158–166. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Boettger T, Beetz N, Kostin S, Schneider
J, Krüger M, Hein L and Braun T: Acquisition of the contractile
phenotype by murine arterial smooth muscle cells depends on the
Mir143/145 gene cluster. J Clin Invest. 119:2634–2647. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xin M, Small EM, Sutherland LB, Qi X,
McAnally J, Plato CF, Richardson JA, Bassel-Duby R and Olson EN:
MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and
responsiveness of smooth muscle cells to injury. Genes Dev.
23:2166–2178. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kozomara A and Griffiths-Jones S: miRBase:
Annotating high confidence microRNAs using deep sequencing data.
Nucleic Acids Res. 42(Database Issue): D68–D73. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Reczko M, Maragkakis M, Alexiou P,
Papadopoulos GL and Hatzigeorgiou AG: Accurate microRNA target
prediction using detailed binding site accessibility and machine
learning on proteomics data. Front Genet. 2:1032012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sartore S, Chiavegato A, Faggin E, Franch
R, Puato M, Ausoni S and Pauletto P: Contribution of adventitial
fibroblasts to neointima formation and vascular remodeling: From
innocent bystander to active participant. Circ Res. 89:1111–1121.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Walsh K and Takahashi A: Transcriptional
regulation of vascular smooth muscle cell phenotype. Z Kardiol. 90
Suppl 3:S12–S16. 2001. View Article : Google Scholar
|
|
33
|
Hirschi KK, Lai L, Belaguli NS, Dean DA,
Schwartz RJ and Zimmer WE: Transforming growth factor-beta
induction of smooth muscle cell phenotype requires transcriptional
and post-transcriptional control of serum response factor. J Biol
Chem. 277:6287–6295. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ohnaka M, Marui A, Yamahara K, Minakata K,
Yamazaki K, Kumagai M, Masumoto H, Tanaka S, Ikeda T and Sakata R:
Effect of microRNA-145 to prevent vein graft disease in rabbits by
regulation of smooth muscle cell phenotype. J Thorac Cardiovasc
Surg. 148:676–682. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Salmon M, Gomez D, Greene E, Shankman L
and Owens GK: Cooperative binding of KLF4, pELK-1 and HDAC2 to a
G/C repressor element in the SM22α promoter mediates
transcriptional silencing during SMC phenotypic switching in vivo.
Circ Res. 111:685–696. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yoshida T, Kaestner KH and Owens GK:
Conditional deletion of Krüppel-like factor 4 delays downregulation
of smooth muscle cell differentiation markers but accelerates
neointimal formation following vascular injury. Circ Res.
102:1548–1557. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cherepanova OA, Pidkovka NA, Sarmento OF,
Yoshida T, Gan Q, Adiguzel E, Bendeck MP, Berliner J, Leitinger N
and Owens GK: Oxidized phospholipids induce type VIII collagen
expression and vascular smooth muscle cell migration. Circ Res.
104:609–618. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Pidkovka NA, Cherepanova OA, Yoshida T,
Alexander MR, Deaton RA, Thomas JA, Leitinger N and Owens GK:
Oxidized phospholipids induce phenotypic switching of vascular
smooth muscle cells in vitro and in vivo. Circ Res. 101:792–801.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu Y, Sinha S and Owens G: A transforming
growth factor-beta control element required for SM alpha-actin
expression in vivo also partially mediates GKLF-dependent
transcriptional repression. J Biol Chem. 278:48004–48011. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li HX, Han M, Bernier M, Zheng B, Sun SG,
Su M, Zhang R, Fu JR and Wen JK: Krüppel-like factor 4 promotes
differentiation by transforming growth factor-beta
receptor-mediated Smad and p38 MAPK signaling in vascular smooth
muscle cells. J Biol Chem. 285:17846–17856. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Davis BN, Hilyard AC, Lagna G and Hata A:
SMAD proteins control DROSHA-mediated microRNA maturation. Nature.
454:56–61. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Davis BN, Hilyard AC, Nguyen PH, Lagna G
and Hata A: Induction of microRNA-221 by platelet-derived growth
factor signaling is critical for modulation of vascular smooth
muscle phenotype. J Biol Chem. 284:3728–3738. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Liu X, Cheng Y, Zhang S, Lin Y, Yang J and
Zhang C: A necessary role of miR-221 and miR-222 in vascular smooth
muscle cell proliferation and neointimal hyperplasia. Circ Res.
104:476–487. 2009. View Article : Google Scholar : PubMed/NCBI
|