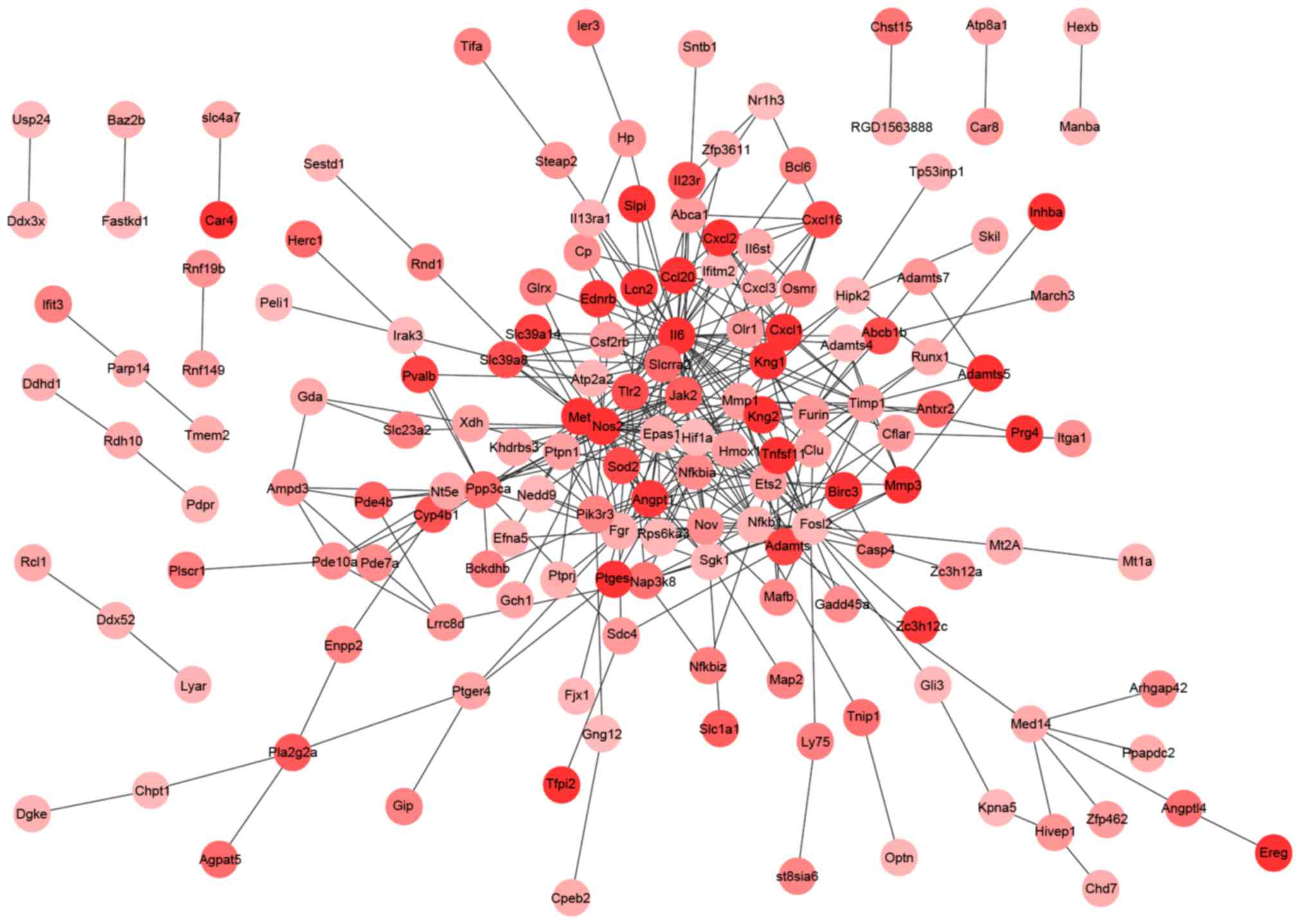
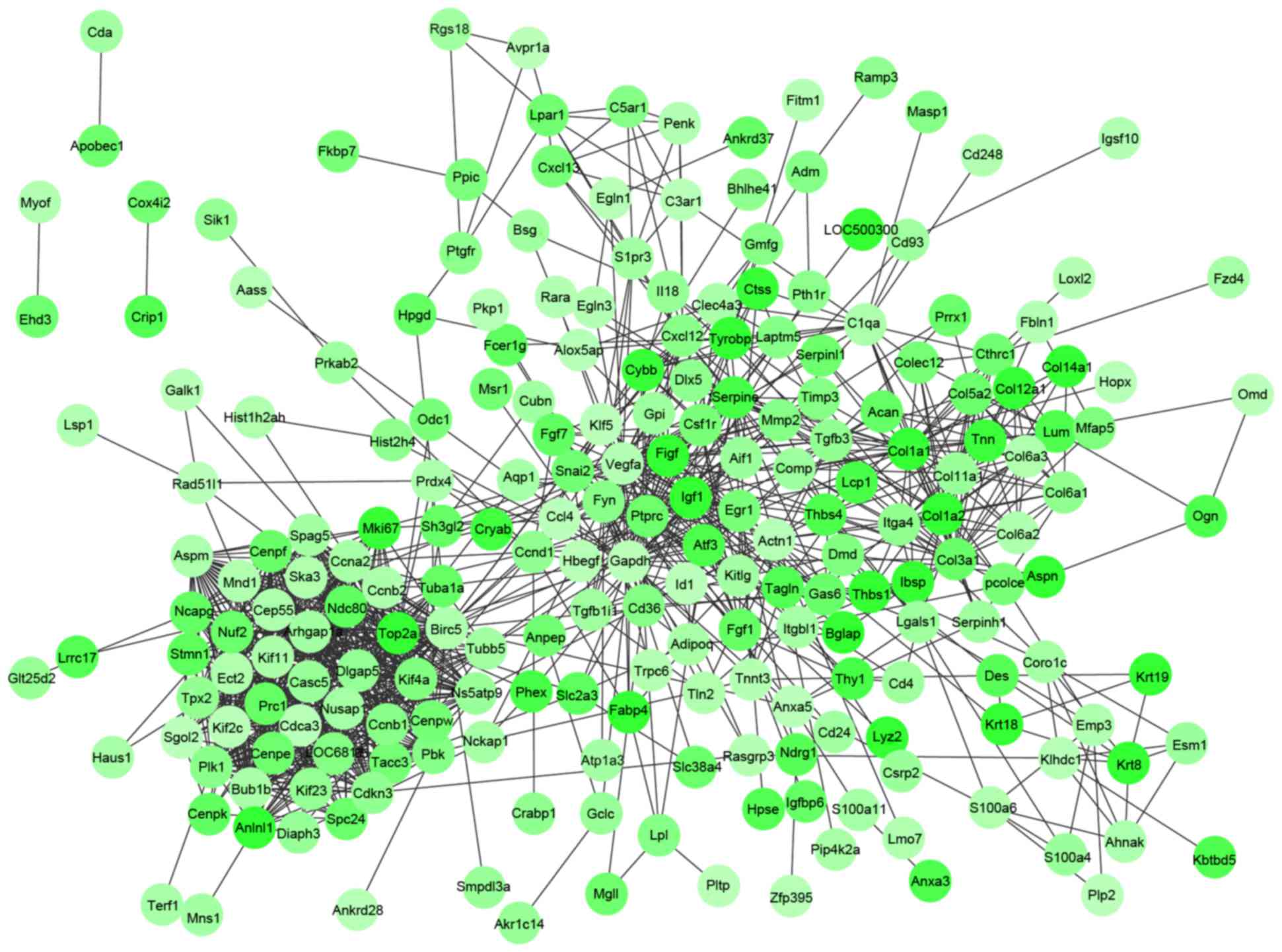
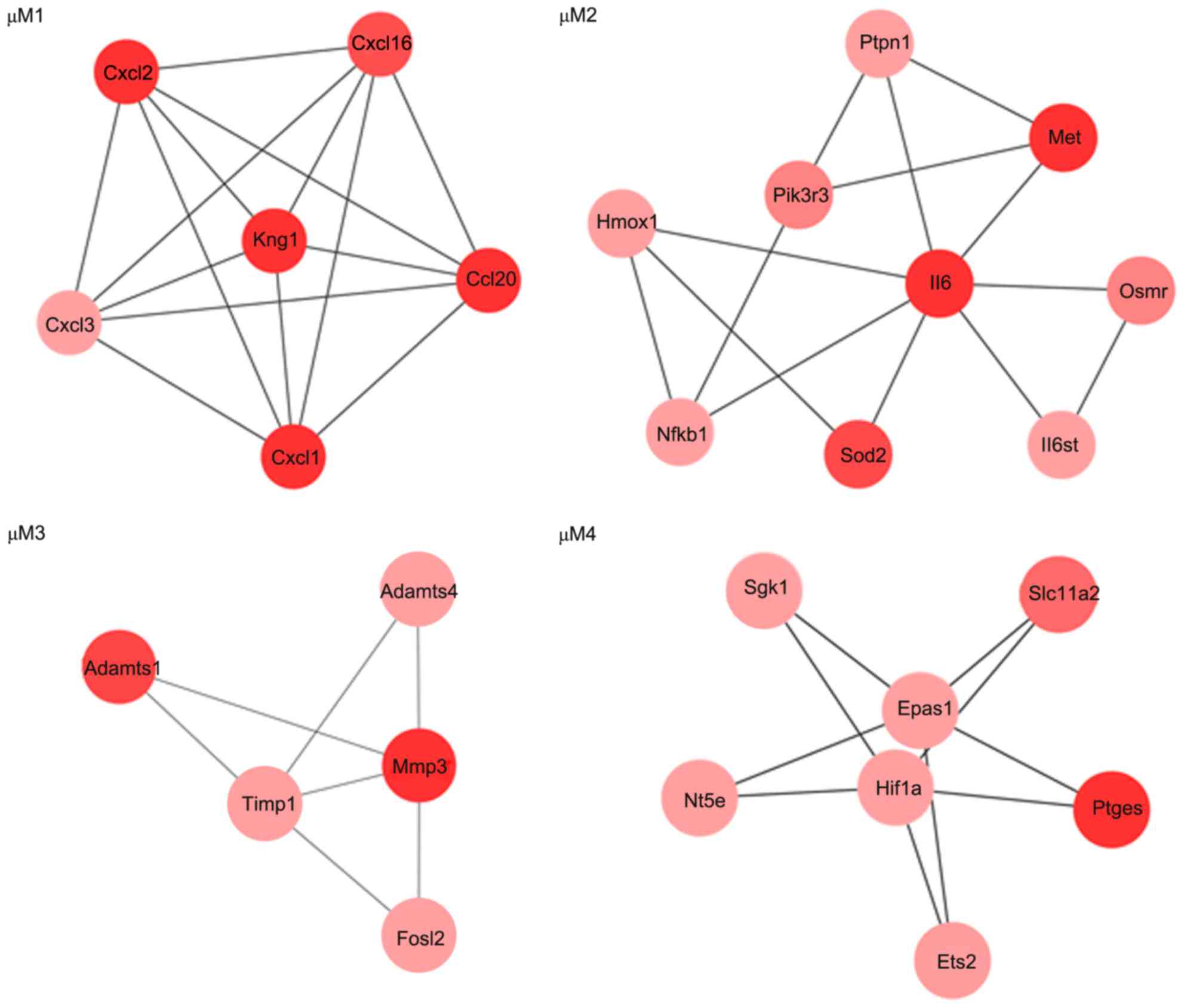
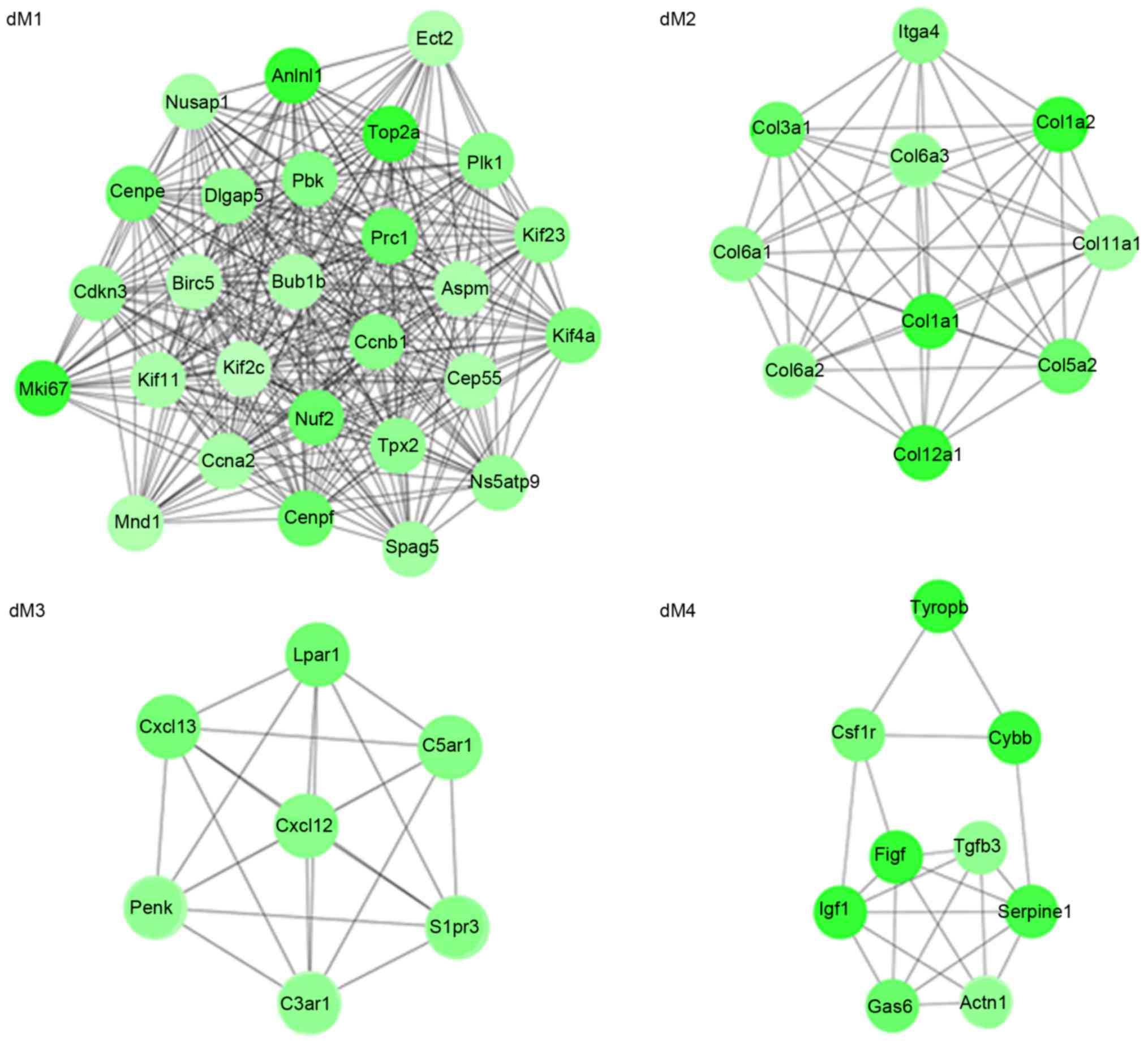
|
1
|
Sakai D, Mochida J, Yamamoto Y, Nomura T,
Okuma M, Nishimura K, Nakai T, Ando K and Hotta T: Transplantation
of mesenchymal stem cells embedded in Atelocollagen gel to the
intervertebral disc: A potential therapeutic model for disc
degeneratio. Biomaterials. 24:3531–3541. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Le Maitre CL, Pockert A, Buttle DJ,
Freemont AJ and Hoyland JA: Matrix synthesis and degradation in
human intervertebral disc degeneration. Biochem Soc Trans.
35:652–655. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Richardson SM, Walker RV, Parker S, Rhodes
NP, Hunt JA, Freemont AJ and Hoyland JA: Intervertebral disc
cell-mediated mesenchymal stem cell differentiation. Stem Cells.
24:707–716. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Battié MC, Videman T and Parent E: Lumbar
disc degeneration: Epidemiology and genetic influences. Spine.
29:2679–2690. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sakai D, Mochida J, Iwashina T, Hiyama A,
Omi H, Imai M, Nakai T, Ando K and Hotta T: Regenerative effects of
transplanting mesenchymal stem cells embedded in atelocollagen to
the degenerated intervertebral disc. Biomaterials. 27:335–345.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pockert AJ, Richardson SM, Le Maitre CL,
Lyon M, Deakin JA, Buttle DJ, Freemont AJ and Hoyland JA: Modified
expression of the ADAMTS enzymes and tissue inhibitor of
metalloproteinases 3 during human intervertebral disc degeneration.
Arthritis Rheum. 60:482–491. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gruber HE, Norton HJ, Ingram JA and Hanley
EN Jr: The SOX9 transcription factor in the human disc: Decreased
immunolocalization with age and disc degeneration. Spine (Phila Pa
1976). 30:625–630. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ponnappan RK, Markova DZ, Antonio PJ,
Murray HB, Vaccaro AR, Shapiro IM, Anderson DG, Albert TJ and
Risbud MV: An organ culture system to model early degenerative
changes of the intervertebral disc. Arthritis Res Ther.
13:R1712011. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Weiler C, Nerlich AG, Bachmeier BE and
Boos N: Expression and distribution of tumor necrosis factor alpha
in human lumbar intervertebral discs: A study in surgical specimen
and autopsy controls. Spine (Phila Pa 1976). 30:44–54. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bachmeier BE, Nerlich AG, Weiler C,
Paesold G, Jochum M and Boos N: Analysis of tissue distribution of
TNF-alpha, TNF-alpha-receptors and the activating
TNF-alpha-converting enzyme suggests activation of the TNF-alpha
system in the aging intervertebral disc. Ann N Y Acad Sci.
1096:44–54. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Le Maitre CL, Freemont AJ and Hoyland JA:
The role of interleukin-1 in the pathogenesis of human
intervertebral disc degeneration. Arthritis Res Ther. 7:R732–R745.
2005. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hoyland J, Le Maitre C and Freemont A:
Investigation of the role of IL-1 and TNF in matrix degradation in
the intervertebral disc. Rheumatology (Oxford). 47:809–814. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Le Maitre CL, Hoyland JA and Freemont AJ:
Catabolic cytokine expression in degenerate and herniated human
intervertebral discs: IL-1beta and TNFalpha expression profile.
Arthritis Res Ther. 9:R772007. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Markova DZ, Kepler CK, Addya S, Murray HB,
Vaccaro AR, Shapiro IM, Anderson DG, Albert TJ and Risbud MV: An
organ culture system to model early degenerative changes of the
intervertebral disc II: Profiling global gene expression changes.
Arthritis Res Ther. 15:R1212013. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Smyth GK: Limma: Linear Models for
Microarray Data. Bioinformatics and Computational Biology Solutions
Using R and Bioconductor Springer. 397–420. 2005. View Article : Google Scholar
|
|
17
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J Royal Statistical Soci. 57:289–300. 1995.
|
|
18
|
Huang DW, Sherman BT, Tan Q, Kir J, Liu D,
Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC and Lempicki RA:
DAVID bioinformatics resources: Expanded annotation database and
novel algorithms to better extract biology from large gene lists.
Nucleic Acids Res. 35:W169–W175. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Carbon S, Ireland A, Mungall CJ, Shu S,
Marshall B and Lewis S: AmiGO Hub; Web Presence Working Group:
AmiGO: Online access to ontology and annotation data.
Bioinformatics. 25:288–289. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kanehisa M, Araki M, Goto S, Hattori M,
Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T
and Yamanishi Y: KEGG for linking genomes to life and the
environment. Nucleic Acids Res. 36:(Database issue). D480–D484.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Szklarczyk D, Franceschini A, Kuhn M,
Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork
P, et al: The STRING database in 2011: Functional interaction
networks of proteins, globally integrated and scored. Nucleic Acids
Res. 39:(Database issue). D561–D568. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Noponen-Hietala N, Virtanen I, Karttunen
R, Schwenke S, Jakkula E, Li H, Merikivi R, Barral S, Ott J,
Karppinen J and Ala-Kokko L: Genetic variations in IL6 associate
with intervertebral disc disease characterized by sciatica. Pain.
114:186–194. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fujita N, Imai J, Suzuki T, Yamada M,
Ninomiya K, Miyamoto K, Iwasaki R, Morioka H, Matsumoto M, Chiba K,
et al: Vascular endothelial growth factor-A is a survival factor
for nucleus pulposus cells in the intervertebral disc. Biochem
Biophys Res Commun. 372:367–372. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sato J, Sakuma Y, Yamauchi K, Orita S,
Kubota G, Oikawa Y, Inage K, Sainoh T, Fujimoto K, Takahashi K, et
al: Elevated VEGF in degenerative intervertebral discs in rats with
injured intervertebral discs of the caudal vertebrae. Global Spine
J. 4:po. 165. 2014. View Article : Google Scholar
|
|
27
|
Haro H, Kato T, Komori H, Osada M and
Shinomiya K: Vascular endothelial growth factor (VEGF)-induced
angiogenesis in herniated disc resorption. J Orthop Res.
20:409–415. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gruber HE, Ingram JA and Hanley EN Jr:
Immunolocalization of thrombospondin in the human and sand rat
intervertebral disc. Spine (Phila Pa 1976). 31:2556–2561. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hirose Y, Chiba K, Karasugi T, Nakajima M,
Kawaguchi Y, Mikami Y, Furuichi T, Mio F, Miyake A, Miyamoto T, et
al: A functional polymorphism in THBS2 that affects alternative
splicing and MMP binding is associated with lumbar-disc herniation.
Am J Med Genet. 82:1122–1129. 2008.
|
|
30
|
Lawler J, Sunday M, Thibert V, Duquette M,
George EL, Rayburn H and Hynes RO: Thrombospondin-1 is required for
normal murine pulmonary homeostasis and its absence causes
pneumonia. J Clin Invest. 101:982–992. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tilkeridis C, Bei T, Garantziotis S and
Stratakis CA: Association of a COL1A1 polymorphism with lumbar disc
disease in young military recruits. J Med Genet. 42:e442005.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yang Z, Chen X, Zhang Q, Cai B, Chen K,
Chen Z, Bai Y, Shi Z and Li M: Dysregulated COL3A1 and RPL8, RPS16,
and RPS23 in disc degeneration revealed by bioinformatics methods.
Spine (Phila Pa 1976). 40:E745–E751. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Noponen-Hietala N, Kyllönen E, Männikkö M,
Ilkko E, Karppinen J, Ott J and Ala-Kokko L: Sequence variations in
the collagen IX and XI genes are associated with degenerative
lumbar spinal stenosis. Ann Rheum Dis. 62:1208–1214. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mio F, Chiba K, Hirose Y, Kawaguchi Y,
Mikami Y, Oya T, Mori M, Kamata M, Matsumoto M, Ozaki K, et al: A
functional polymorphism in COL11A1, which encodes the alpha 1 chain
of type XI collagen, is associated with susceptibility to lumbar
disc herniation. Am J Med Genet. 81:1271–1277. 2007.
|