Introduction
Cubital tunnel syndrome (CuTS), also known as
delayed ulnar neuritis, is the second most common peripheral nerve
compression disease in the upper limb following carpal tunnel
syndrome (1). It is estimated that
25 per 100,000 individuals are affected by CuTS in Italy each year,
and is two times more common in men, compared with women (2). CuTS can occur as a result of ischemia
or mechanical compression by repeated elbow flexion, post-traumatic
scarring, anomalous musculature or direct compression, although the
exact cause may be difficult to identify (3). Early symptoms in patients can include
elbow discomfort, numbness of the little finger, and inflexibility
when writing or using tools. In more severe forms of CuTS, the
strength of the flexor carpi ulnaris, and flexor muscles of the
ring and little fingers are weakened; atrophy of the intrinsic
muscle of the hand and mild claw finger deformity can also occur
(4). Treatment methods include
non-surgical therapies, for example rest, splinting, real-time
visualized ultrasound-guided injection and surgical decompression,
which can include open in situ decompression, anterior
transposition and endoscopic decompression (5–8).
At present, investigations of CuTS are focused
predominantly on surgical methods, and less on the discussion of
pathogenesis and pathology. Although ulnar nerve compression is a
common cause of CuTS, its pathogenesis and pathology remain to be
fully elucidated. The common areas of entrapment of the ulnar nerve
include the arcade of Struthers, the medial intermuscular septum,
insertion of the medial head of the triceps, the leading edge of
the cubital tunnel retinaculum, and the anconeus epitrochlearis
muscle, medial epicondyle, Osborne's ligament and flexor-pronator
group. Hypertrophy of Osborne's ligament is an important cause of
ulnar nerve compression (9).
Extensive investigations have been performed to
characterize the differences between healthy and diseased
ligaments, including alterations in gene expression. MicroRNAs
(miRNAs), defined as endogenously expressed, short non-coding RNAs
(18–25 nucleotides in length), suppress protein translation through
binding to target messenger RNAs (mRNAs). They are expressed at
specific stages of tissue development or cell differentiation, and
have marked effects on the expression of a variety of genes at the
post-transcriptional stage (10).
Previous studies have suggested that miRNAs
contribute to the pathogenesis of fibrotic diseases. Hypertrophy of
the ligamentum flavum (LF) is crucial in lumbar spinal stenosis
(LSS) and is caused primarily by fibrosis. miRNA (miR)-155 is
positively correlated with different fibrotic diseases. Previous
data have shown that the expression levels of miR155 differ between
LF and LSS groups, and between LF and lumbar disc herniation (LDH)
groups. Therefore, the fibrosis-associated miR-155 may be important
in the pathogenesis of hypertrophic LF (11). However, the role of miRNAs in the
development of hypertrophy of the ligaments remains to be
elucidated.
To improve understanding of the pathogenesis of
CuTS, the present collected examined pachyntic Osborne's ligaments
and control tendinous tissues. Global miRNA expression levels in
the pachyntic Osborne's ligaments and control tendinous tissues
were examined using microarray techiques. The results showed
differences in miRNAs expression levels between the Osborne's
ligament and control tendinous tissues from patients with CuTS.
These findings have important implications for revealing the
pathogenesis of CuTS.
Materials and methods
Patients and tissue samples
The patients included in the present study comprised
six patients with CuTS, which was diagnosed according to their
symptoms, neurological examination, electrophysiological
examination and imaging. The patients all experienced numbness of
the little finger and the ring finger, pain in the elbow and
forearm, weakness and clumsiness of the hand and motor deficit,
including Froment's sign. X-ray imaging of the positive and lateral
position of the elbow joint, tangential position of the ulnar
nerve, conventional assay and ECG were included in the basic
examination. Reductions in the velocity of motor nerve conduction
in the patient elbow segments were identified by electromyography.
Local color Doppler ultrasound was used to exclude elbow tumor
lesions.
The Osborne's ligament and control tendinous tissue
samples were obtained from four men and two women with CuTS
(average age, 53.83 years; range, 31–76 years) who underwent
surgery for anterior subcutaneous transposition. Detailed
characteristics for the patients are shown in Table I. During surgery, an arched 6–8 cm
incision with a tourniquet control was made posterior to the medial
epicondyle. The retrocondylar groove, Osborne's ligament and ulnar
nerve were identified. Proximally, the ulnar nerve was exposed to
the medial intermuscular septum, which was divided to avoid
possible future compression. Following division and transection of
the Osborne's ligament from the cubital tunnel retinaculum, the
ulnar nerve was released distally to the two heads of the flexor
carpi ulnaris. Soft loops were used to isolate the ulnar nerve, and
the ulnar nerve was transposed forward to the medial epicondyle. To
stabilize the ulnar nerve, the subcutaneous tissue was sutured
using a fascial flap. As medical waste, the Osborne's ligaments
were collected. In addition ~0.3×1 cm tendinous tissue around the
two heads of the flexor carpi ulnaris was excised as a control
sample. The specimens wesre collected by the same experienced
surgeon in the Department of Orthopedics of Tianjin Medical
University General Hospital (Tianjin, China). The Tianjin Medical
University General Hospital Medical Ethics Committee approved the
consent forms and protocol for evaluating the tissues. Written
informed consent was provided by each patient prior to surgery. The
specimens were frozen in liquid nitrogen and stored at −80°C until
the microarray was performed.
 | Table I.Characteristics of patients. |
Table I.
Characteristics of patients.
| Characteristic | Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 | Patient 6 |
|---|
| Gender | Male | Female | Male | Male | Female | Male |
| Age (years) | 61 | 76 | 64 | 53 | 31 | 38 |
| Surgical method | ASCT | ASCT | ASCT | ASCT | ASCT | ASCT |
| Side of surgery | Left | Right | Right | Bilateral | Left | Left |
| Surgery on dominant
side | No | Yes | Yes | Yes | No | No |
| Electromyography | Positive | Positive | Positive | Positive | Positive | Positive |
| Tinel's sign | Positive | Positive | Positive | Positive | Positive | Positive |
| Sample
application | Microarray | Microarray | Microarray | RT-qPCR | RT-qPCR | RT-qPCR |
RNA isolation
Three groups of stored samples were assayed using an
Affymetrix GeneChip 3000 TG system purchased from Affymetrix, Inc.
(Santa Clara, CA, USA). First frozen samples were homogenized in
QIAzolLysis Reagent (Qiagen GmbH, Hilden, Germany) using the
TissueRuptor. Total RNA was precipitated using chloroform and the
aqueous phase was mixed with 1.5 volumes of 100% Ethanol. Then
total RNA, including miRNAs, was extracted using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
according to the manufacturer's protocol. The concentration of RNA
was measured using a NanoDrop ND-2000 spectrophotometer (NanoDrop;
Thermo Fisher Scientific, Inc., Wilmington, DE, USA), and the
purity of RNA was confirmed using spectrophotometry. The optical
density 260/280 nm ratio was between 1.90 and 2.10 for each RNA
sample.
Microarray hybridization and data
analysis
The RNA quality was assessed using denaturing gel
electrophoresis. The Affymetrix GeneChip system was used for
hybridization, staining and imaging of the arrays, according to the
standard Affymetrix protocol. Following hybridization, the arrays
were washed using a Fluidics Station 450 (Affymetrix; Thermo Fisher
Scientific Inc.) and then scanned using a Scanner 3000 7G 4C
(Affymetrix; Thermo Fisher Scientific Inc.). Quality control
analysis was performed using the Affymetrix miRNA QCTool
(Affymetrix; Thermo Fisher Scientific Inc.). For each miRNA,
multiple probes were spotted on the array, and the mean intensity
of these probes was calculated to represent the expression value of
the miRNAs. In addition, multiple spots were included as negative
controls. For each sample, 1 µl total RNA was hybridized with the
miRNA array and further processed in accordance with the
manufacturer's protocol. Only those miRNAs with significant
(P<0.05) differential expression of ≥2.0-fold change were
reported. Array scanning and data analysis were performed using
Expression Console™ software version 1.4 (Affymetrix; Thermo Fisher
Scientific Inc.), which provides signal estimation and quality
control functionality for the Affymetrix GeneChip and Transcriptome
Analysis Console software version 1.0 (Affymetrix; Thermo Fisher
Scientific Inc.), which performs statistical analysis and provides
a list of differentially expressed miRNAs. miRWalk2.0 (http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/)
(12) was used synthesizing four
existing miRNA-target prediction programs (miRanda, miRDB,
TargetScan and RNA22), as the commonly used web tools for
bioinformatics algorithms, to identify the potential targets of
those miRNAs by combined analysis of the mRNAs in the whole genome
expression microarray. The Bioconductor gene annotation tool
version 3.4 (http://www.bioconductor.org) was used to perform Gene
Ontology (GO) function and Kyoto Encyclopedia of Genes and Genomes
(KEGG) pathway analysis. A Venn diagram was produced to show the
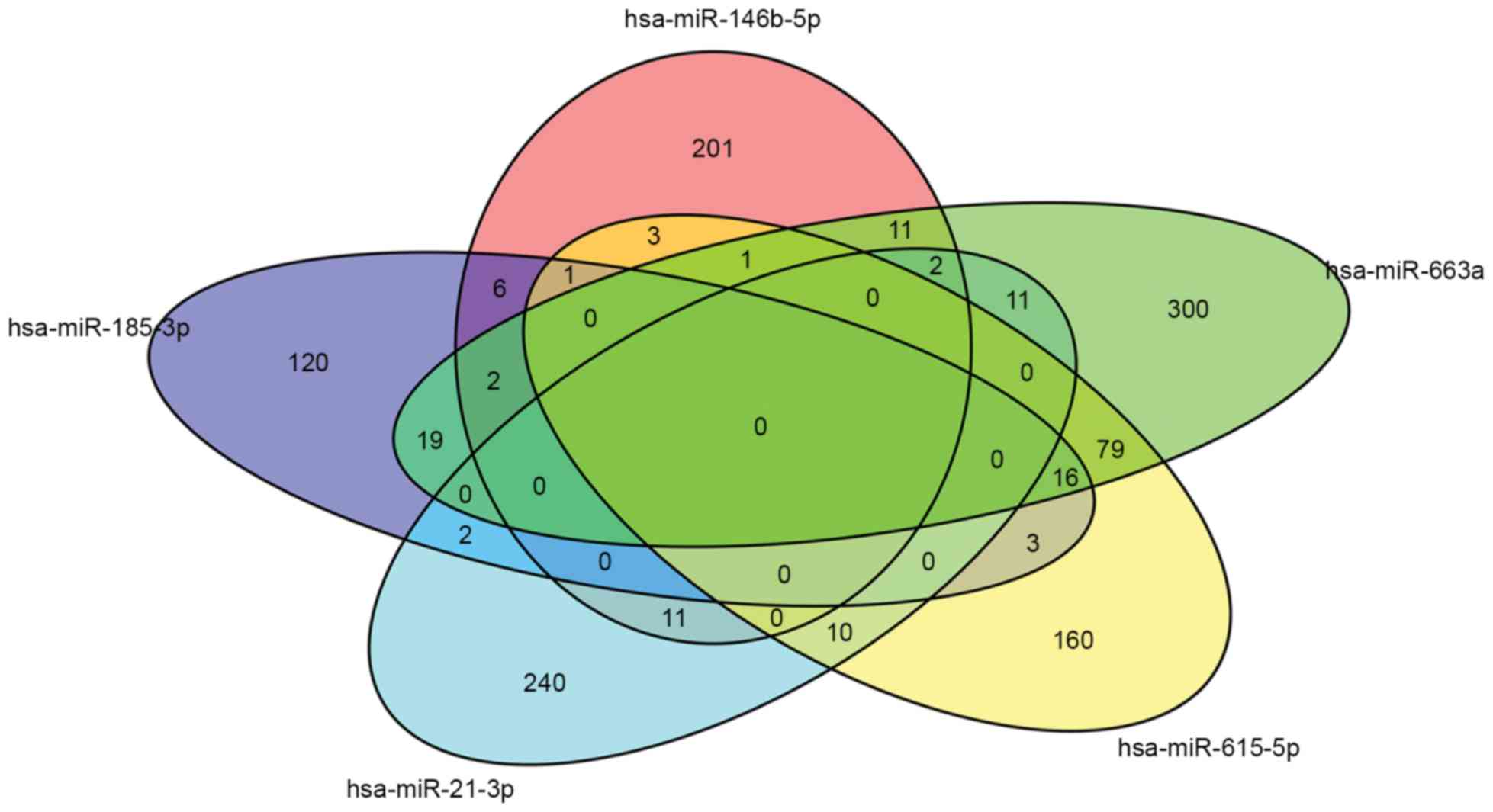
potential common significant GO terms among the five miRNAs
(hsa-miR-146b-5p, hsa-miR-21-3p, hsa-miR-185-3p, hsa-miR-615-5p and
hsa-miR-663a), which included the majority of the predicted target
genes.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
In total, three miRNAs were identified with
significant differences, one differentially upregulated
(hsa-miR-1343-3p) and two differentially downregulated
(hsa-miR-21-3p and hsa-miR-146b-5p). These three miRNAs were
verified using RT-qPCR analysis with the Bio-Rad CFX96 Real-Time
PCR system (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Total
RNA was isolated from the samples of the other three patients with
CuTS using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.), and
this was polyadenylated and reverse-transcribed with a poly (T)
adapter into cDNA by miScript Reverse Transcription kit (Qiagen
GmbH) according to the manufacturer's protocol. Then qPCR was
performed using SYBR green dye in a thermal cycler with the
following parameters: 1 µl buffer, 4 µl total RNA, 2 µl cDNA, 1 µl
specific primers, 4 µl RNase-free water and 10 µl SYBR qPCR Mix. An
initial denaturation step at 95°C for 30 min; 40 cycles at 95°C for
5 sec and 60°C for 30 sec. The complete experimental procedure was
performed for each sample in triplicate. All primers were
synthesized by Shanghai Shenggong Biology Engineering Technology
Service, Ltd. (Shanghai, China) and the miRNA-specific primers are
listed in Table II. All data were
analyzed using the 2-ΔΔCq method (13) to calculate the differences between
the quantification cycle values of the target genes in each sample.
P<0.05 was considered to indicate a statistically significant
difference.
 | Table II.Primers used for reverse
transcription-quantitative polymerase chain reaction analysis. |
Table II.
Primers used for reverse
transcription-quantitative polymerase chain reaction analysis.
| miRNA | miRNA sequence
(5′-3′) | Primer sequence
(5′-3′) | Primer length
(bp) | GC content (%) | Tm (°C) |
|---|
| hsa-miR-21-3p |
CAACACCAGUCGAUGGGCUGU | Forward ATT CAA
CAC | 21 | 52 | 60.2 |
|
|
| CAG TCG ATG GGC |
|
|
|
|
|
| Reverse TAG CTT
ATC |
|
|
|
|
|
| AGA CTG ATG TT |
|
|
|
|
hsa-miR-146b-5p |
UGAGAACUGAAUUCCAUAGGCU | Forward GTG AGA ACT
GAA | 23 | 43 | 59.9 |
|
|
| TTC CAT AGG CT |
|
|
|
|
|
| Reverse GCA CCA
GAA |
|
|
|
|
|
| CTG AGT CCA CA |
|
|
|
|
hsa-miR-1343-3p |
CUCCUGGGGCCCGCACUCUCGC | Forward TTA TTC
TCC | 19 | 63 | 60.6 |
|
|
| TGG GGC CCG C |
|
|
|
|
|
| Reverse ATC CCA
CCA |
|
|
|
|
|
| CTG CCA CC |
|
|
|
Statistical analysis
All data were analyzed using SPSS statistical
software (version 11.5 for Windows; SPSS, Inc., Chicago, IL, USA).
Statistical analysis was performed using two-tailed Student's
t-test. P<0.05 was considered to indicate a statistically
significant difference.
Results
Expression profile of miRNAs in the
ligament
To investigate the miRNA expression profiles in the
pachyntic Osborne's ligament, microarray analysis was performed
using total RNA from pachyntic Osborne's ligaments and control
tendinous tissues obtained from patients with CuTS. The expression
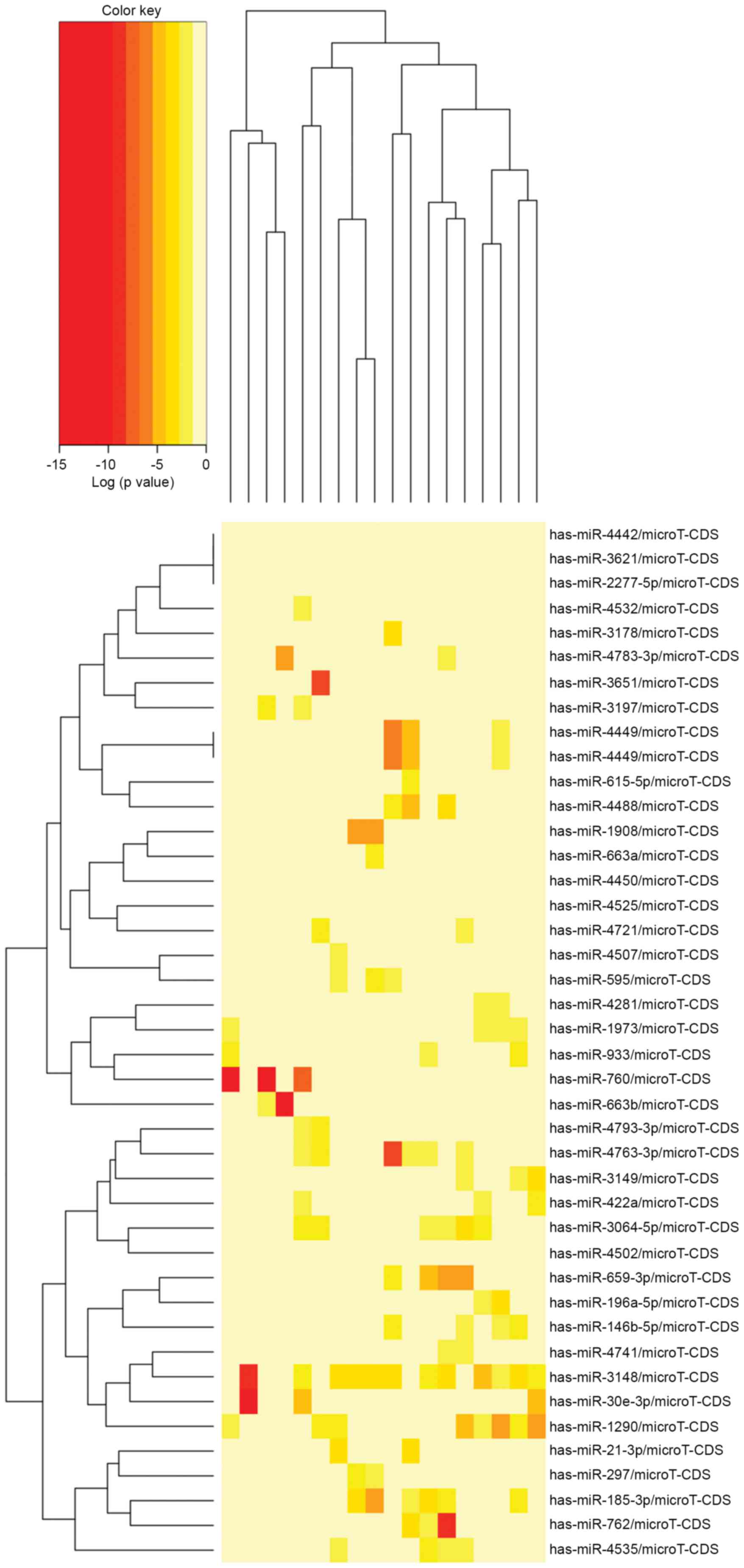
levels of 60 miRNAs showed significant differential expression
between the pachyntic Osborne's ligament group and the control
group (Fig. 1). Among these, only
three miRNAs were upregulated, whereas the remaining 67 were
downregulated (Table III).
 | Table III.Summary of the significantly
differentially expressed miRNAs. |
Table III.
Summary of the significantly
differentially expressed miRNAs.
| miRNA | logFC (fold
change) | P-value | Sequence
(5′-3′) |
|---|
| Upregulated |
|
|
|
|
hsa-miR-7855-5p | 1.045103351 | 0.000425931 |
UUGGUGAGGACCCCAAGCUCGG |
|
hsa-miR-422a | 1.377981007 | 0.000219629 |
ACUGGACUUAGGGUCAGAAGGC |
|
hsa-miR-1343-3p | 0.934668054 | 0.000311621 |
CUCCUGGGGCCCGCACUCUCGC |
| Downregulated |
|
|
|
|
hsa-miR-196a-5p | −6.520640993 | 0.000000057 |
UAGGUAGUUUCAUGUUGUUGGG |
|
hsa-miR-1290 | −3.225990914 | 0.000000895 |
UGGAUUUUUGGAUCAGGGA |
|
hsa-miR-595 | −3.859379525 | 0.000001060 |
GAAGUGUGCCGUGGUGUGUCU |
|
hsa-miR-7110-5p | −3.949821874 | 0.000006230 |
UGGGGGUGUGGGGAGAGAGAG |
|
hsa-miR-3148 | −2.477794005 | 0.000006770 |
UGGAAAAAACUGGUGUGUGCUU |
|
hsa-miR-4532 | −3.334823954 | 0.000008990 |
CCCCGGGGAGCCCGGCG |
|
hsa-miR-3064-5p | −3.049176967 | 0.000009880 |
UCUGGCUGUUGUGGUGUGCAA |
|
hsa-miR-6792-5p | −1.989451562 | 0.000010600 |
GUAAGCAGGGGCUCUGGGUGA |
|
hsa-miR-7844-5p | −2.838409598 | 0.000017300 |
AAAACUAGGACUGUGUGGUGUA |
|
hsa-miR-4535 | −2.017418576 | 0.000018300 |
GUGGACCUGGCUGGGAC |
|
hsa-miR-146b-5p | −5.085472937 | 0.000020700 |
UGAGAACUGAAUUCCAUAGGCU |
|
hsa-miR-3178 | −2.277544498 | 0.000022100 |
GGGGCGCGGCCGGAUCG |
|
hsa-miR-297 | −4.106551942 | 0.000028200 |
AUGUAUGUGUGCAUGUGCAUG |
|
hsa-miR-21-3p | −3.565524376 | 0.000037100 |
CAACACCAGUCGAUGGGCUGU |
|
hsa-miR-4525 | −3.485981804 | 0.000052800 |
GGGGGGAUGUGCAUGCUGGUU |
|
hsa-miR-6789-5p | −1.746637528 | 0.000054700 |
GUAGGGGCGUCCCGGGCGCGCGGG |
|
hsa-miR-6883-5p | −2.808853491 | 0.000061600 |
AGGGAGGGUGUGGUAUGGAUGU |
|
hsa-miR-3149 | −1.879048778 | 0.000066500 |
UUUGUAUGGAUAUGUGUGUGUAU |
|
hsa-miR-4502 | −2.622259256 | 0.000070000 |
GCUGAUGAUGAUGGUGCUGAAG |
|
hsa-miR-933 | −1.918951061 | 0.000081100 |
UGUGCGCAGGGAGACCUCUCCC |
|
hsa-miR-4793-3p | −3.351094169 | 0.000084600 |
UCUGCACUGUGAGUUGGCUGGCU |
|
hsa-miR-6085 | −1.483750019 | 0.000085000 |
AAGGGGCUGGGGGAGCACA |
|
hsa-miR-760 | −1.736503688 | 0.000086000 |
CGGCUCUGGGUCUGUGGGGA |
|
hsa-miR-6075 | −2.645791301 | 0.000090600 |
ACGGCCCAGGCGGCAUUGGUG |
|
hsa-miR-4741 | −1.682786276 | 0.000096200 |
CGGGCUGUCCGGAGGGGUCGGCU |
|
hsa-miR-6875-5p | −3.198242537 | 0.000102689 |
UGAGGGACCCAGGACAGGAGA |
|
hsa-miR-6845-5p | −2.537516128 | 0.000110628 |
CGGGGCCAGAGCAGAGAGC |
|
hsa-miR-1180-3p | −3.290772167 | 0.000120042 |
UUUCCGGCUCGCGUGGGUGUGU |
|
hsa-miR-3620-5p | −1.269433220 | 0.000125189 |
GUGGGCUGGGCUGGGCUGGGCC |
|
hsa-miR-663a | −1.826255642 | 0.000153410 |
AGGCGGGGCGCCGCGGGACCGC |
|
hsa-miR-663b | −3.478527017 | 0.000155267 |
GGUGGCCCGGCCGUGCCUGAGG |
|
hsa-miR-3621 | −1.444956668 | 0.000161666 |
CGCGGGUCGGGGUCUGCAGG |
|
hsa-miR-4721 | −3.187701938 | 0.000188641 |
UGAGGGCUCCAGGUGACGGUGG |
|
hsa-miR-6791-5p | −1.769619706 | 0.000205615 |
CCCCUGGGGCUGGGCAGGCGGA |
|
hsa-miR-1973 | −1.699316842 | 0.000233469 |
ACCGUGCAAAGGUAGCAUA |
|
hsa-miR-659-3p | −1.301640751 | 0.000244329 |
CUUGGUUCAGGGAGGGUCCCCA |
|
hsa-miR-4507 | −1.921339340 | 0.000257577 |
CUGGGUUGGGCUGGGCUGGG |
|
hsa-miR-6740-5p | −2.050797179 | 0.000259650 |
AGUUUGGGAUGGAGAGAGGAGA |
|
hsa-miR-6816-5p | −1.940639258 | 0.000282808 |
UGGGGCGGGGCAGGUCCCUGC |
|
hsa-miR-6836-5p | −3.707920585 | 0.000310829 |
CGCAGGGCCCUGGCGCAGGCAU |
|
hsa-miR-185-3p | −2.848826390 | 0.000322188 |
AGGGGCUGGCUUUCCUCUGGUC |
|
hsa-miR-4763-3p | −1.844484822 | 0.000334158 |
AGGCAGGGGCUGGUGCUGGGCGGG |
|
hsa-miR-30e-3p | −1.602759453 | 0.000335463 |
CUUUCAGUCGGAUGUUUACAGC |
|
hsa-miR-1237-5p | −1.676336572 | 0.000348059 |
CGGGGGCGGGGCCGAAGCGCG |
|
hsa-miR-6084 | −1.326538904 | 0.000356453 |
UUCCGCCAGUCGGUGGCCGG |
|
hsa-miR-4488 | −1.219288223 | 0.000357825 |
AGGGGGCGGGCUCCGGCG |
|
hsa-miR-4450 | −2.075168762 | 0.000360412 |
UGGGGAUUUGGAGAAGUGGUGA |
|
hsa-miR-3651 | −4.214617171 | 0.000361525 |
CAUAGCCCGGUCGCUGGUACAUGA |
|
hsa-miR-4449 | −4.741198585 | 0.000393060 |
CGUCCCGGGGCUGCGCGAGGCA |
|
hsa-miR-762 | −1.666761846 | 0.000444374 |
GGGGCUGGGGCCGGGGCCGAGC |
|
hsa-miR-6765-5p | −1.765624284 | 0.000463632 |
GUGAGGCGGGGCCAGGAGGGUGUGU |
|
hsa-miR-2277-5p | −0.827734961 | 0.000465336 |
AGCGCGGGCUGAGCGCUGCCAGUC |
|
hsa-miR-3197 | −5.698464480 | 0.000488920 |
GGAGGCGCAGGCUCGGAAAGGCG |
|
hsa-miR-615-5p | −3.033691421 | 0.000502284 |
GGGGGUCCCCGGUGCUCGGAUC |
|
hsa-miR-4783-3p | −2.084261251 | 0.000519843 |
CCCCGGUGUUGGGGCGCGUCUGC |
|
hsa-miR-1343-5p | −1.042812782 | 0.000541584 |
UGGGGAGCGGCCCCCGGGUGGG |
|
hsa-miR-6815-5p | −2.100053152 | 0.000542802 |
UAGGUGGCGCCGGAGGAGUCAUU |
Prediction of miRNA target genes and
functional analysis
To elucidate the functionality of the regulated
miRNAs, miRNA gene target prediction was performed for the 60
differentially expressed miRNAs using the online freely available
software, miRWalk2.0. A total of 1,804 predicted target genes of
the three upregulated miRNAs and 67 downregulated miRNAs were
found, respectively. Functional annotation of the major target
genes were from seven miRNAs (hsa-miR-146b-5p, hsa-miR-21-3p,
hsa-miR-185-3p, hsa-miR-615-5p, hsa-miR-659-3p, hsa-miR-663a and
hsa-miR-760), which were analyzed by GO enrichment analysis to
further evaluate the biological implications of the differentially
expressed miRNAs. The possible regulatory pathways of the major
target genes were analyzed based on KEGG pathway terms. The GO
categories included protein metabolic process, regulation of cell
growth, cell cycle process, regulation of cell division, cellular
metabolic process and signal transmission. The Venn diagram, which
was constructed using the Bioconductor tool, showed that there were
certain common significant GO terms among the different miRNAs
(Fig. 2). According to the
analysis of enriched KEGG pathways for the targets identified from
the differentially expressed miRNAs, the predicted target genes of
the differentially expressed miRNAs were associated with adherent
junction, focal adhesion, axon guidance, lysine degradation, other
glycan degradation, cell adhesion molecules (CAMs),
mitogen-activated protein kinase (MAPK) signaling pathway,
retrograde endocannabinoid signaling, ErbB signaling pathway,
glycosaminoglycan biosynthesis-heparin sulfate/heparin and
neurotrophic signaling pathway (Table
IV). KEGG pathway analysis revealed a number of underlying
biological processes, which may be involved in pachynsis of
Osborne's ligament and may provide useful clues for further
investigating the miRNA targets.
 | Table IV.KEGG pathway analysis for the
predicted miRNA targets. |
Table IV.
KEGG pathway analysis for the
predicted miRNA targets.
| KEGG pathway | P-value | Genes (n) | miRNAs (n) |
|---|
| Prion diseases | 0.000000 | 3 | 2 |
| Systemic lupus
erythematosus | 0.000000 | 31 | 3 |
| Alcoholism | 0.000000 | 39 | 4 |
| Adherens
junction | 0.000000 | 30 | 9 |
| Axon guidance | 0.000000 | 51 | 9 |
| Other glycan
degradation | 0.000003 | 3 | 2 |
| Lysine
degradation | 0.000111 | 19 | 9 |
| Transcriptional
misregulation in cancer | 0.000165 | 65 | 9 |
| Cell adhesion
molecules | 0.000215 | 23 | 8 |
| Mitogen-activated
protein kinase signaling pathway | 0.000567 | 86 | 8 |
| ErbB signaling
pathway | 0.005328 | 29 | 8 |
| Circadian
entrainment | 0.007704 | 35 | 6 |
| Neurotrophin
signaling pathway | 0.017656 | 47 | 7 |
| Prostate
cancer | 0.01956 | 32 | 5 |
| Focal adhesion | 0.025566 | 67 | 7 |
| Long-term
potentiation | 0.032181 | 25 | 6 |
| Retrograde
endocannabinoid signaling | 0.032885 | 33 | 4 |
| Glycosaminoglycan
biosynthesis-heparan sulfate/heparin | 0.041110 | 8 | 6 |
Validation of miRNA expression by
RT-qPCR analysis
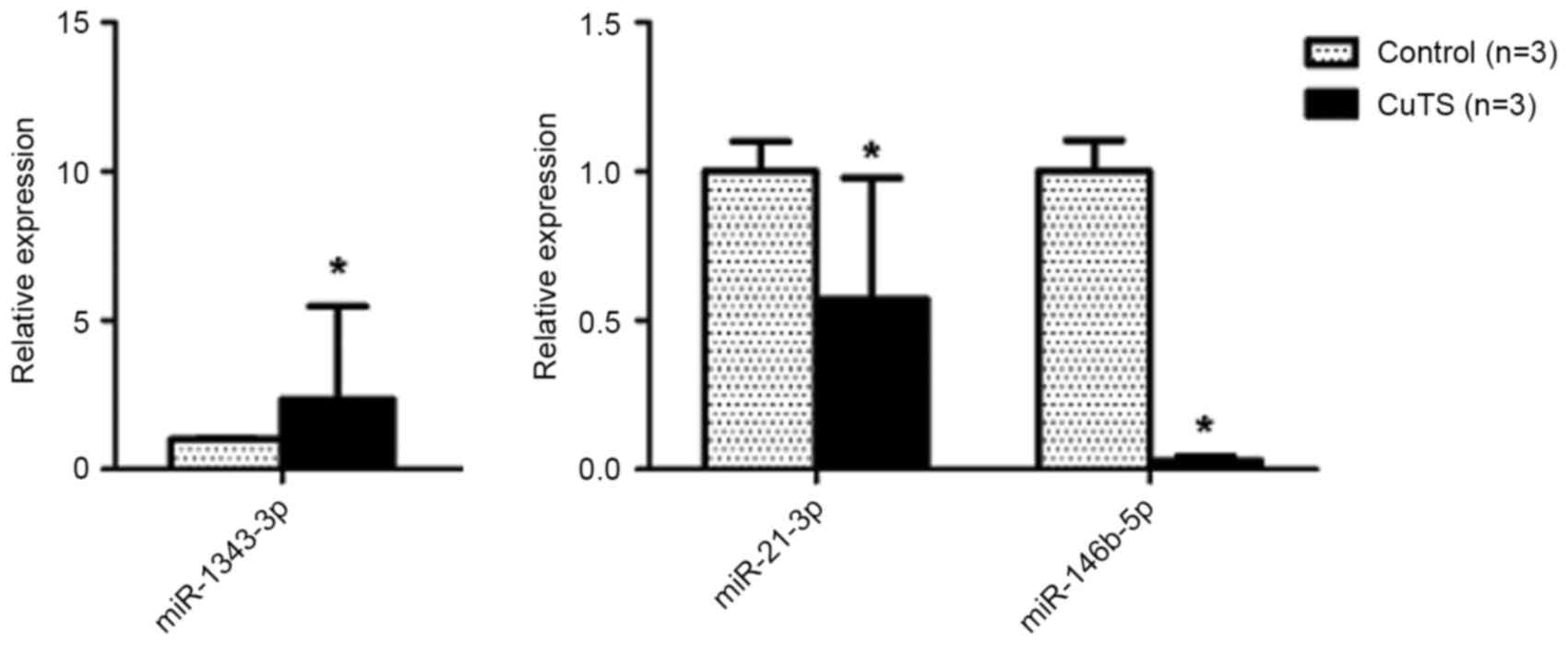
In addition to validating the microarray results,
RT-qPCR was used to quantify particular miRNAs in the pachyntic
Osborne's ligament, including the one differentially upregulated
miRNA (hsa-miR-1343-3p) and two differentially downregulated miRNAs
(hsa-miR-21-3p and hsa-miR-146b-5p), which were closely associated
with fibrotic disease following the online database search. As
shown in Fig. 3, the expression
patterns of hsa-miR-1343-3p, hsa-miR-21-3p and hsa-miR-146-5p
detected using RT-qPCR were consistent with the microarray data,
with significance (P<0.05).
Discussion
In the present study, the patients examined were all
engaged in heavy physical work. Chronic repetitive strain can cause
hypertrophy of Osborne's ligament in the cubital tunnel, and lead
to compression of the ulnar nerve and degenerative ulnar neuritis,
which is the most common etiology of CuTS. Naran and Imbriglia
(14) showed that 55% of patients
engaged in activities, which placed repetitive strain or
compression on the ulnar nerve, and 48% percent of patients worked
as heavy manual laborers in the population demographics. Another
study indicated that the incidence of ulnar nerve entrapment at the
elbow was associated with one biomechanical risk factor, which was
repetitively holding a tool in position (15). Although several other factors,
including cubitus valgus, anomalous musculature and direct
compression, can also lead to CuTS, the present study focused on
the molecular pathogenesis of degenerative ulnar neuritis triggered
by hypertrophy of Osborne's ligament (4).
Emerging evidence has demonstrated that
dysregulation of miRNAs may contribute to the etiology and
pathophysiology of fibrosis and scarring, For example, miR-155 is a
fibrosis-associated miRNA, which increases the expression of
collagen I and collagen III in fibroblasts, and may be involved in
the pathogenesis of LF hypertrophy (11). The inhibition of miR-145 assists in
preventing or reducing hypertrophic scarring of the skin by
reducing skin myofibroblast activity, and decreasing the expression
of α1 type I collagen and secretion of transforming growth
factor-β1 (TGF-β) (16). miRNA-21
regulates systemic sclerosis fibrosis via directly targeting TGF-β
signaling (17). miR-200b, which
regulates the proliferation and apoptosis of human hypertrophic
scar fibroblasts, has previously been reported to be associated
with hypertrophic scarring by affecting the synthesis of collagen I
and III, the expression of fibronectin, and TGF-β1/α-smooth muscle
actin signaling (18). However,
few studies have examined the effects of miRNAs in hypertrophy of
Osborne's ligament.
In the present study, the miRNA expression profiles
showed differentially expressed miRNAs in pachyntic Osborne's
ligaments and control tendinous tissues of patients with CuTS,
identifying miRNAs and the downstream signaling pathway involved in
the mechanism of hypertrophy of Osborne's ligament. In total, three
upregulated and 67 downregulated miRNAs were found. The three
miRNAs (hsa-miR-1343-3p, hsa-miR-21-3p and hsa-miR-146b-5p) were
verified using RT-qPCR analysis, and their expression patterns were
in accordance with the microarray data, with significance
(P<0.05). These miRNAs have been reported in other fields of
investigation. miR-146b-5p and let-7f were validated as miRNA
markers differentiating papillary thyroid cancer from other
clinical conditions (19). In
solid tumors, miR-146b-5p is frequently downregulated, including in
prostate cancer, pancreatic cancer and glioblastoma; in
glioblastoma cell lines, miR-146b-5p is overexpressed, leading to
the silencing of matrix metalloproteinase (MMP) 16 mRNA,
inactivation of MMP2, and inhibition of tumor cell migration and
invasion (20). Another finding
suggested that miR-146b-5p may be associated with pancreatic cancer
cell migration and invasion by MMP16, which is a downstream target
of miR-146b-5p (21). Osborne's
ligament is a well-defined fibrous band, which consists of
connective tissue cells within collagen fibrils (22). Hypertrophy of Osborne's ligament
may be associated with abnormal variations of elastic and collagen
fibers. MMPs, a large family of endopeptidases, are involved in
tissue remodeling and the degradation of extracellular matrix,
regulating various processes, including chronic inflammation,
metastasis and embryonic implantation. MMP-3 (MT3-MMP; MMP-16) in
addition to being fibroblast-associated, it is also involved in the
regulation of MMP-2 and in direct matrix turnover, and is also
widely expressed and capable of degrading several extracellular
matrix components, including type I and III collagen (23–25).
Therefore, it was hypothesized that miR-146b-5p may affect the
expression of its target, MMP-16, and thus increase collagen fibers
to result in hypertrophy of Osborne's ligament. miR-21-3p has been
found to act as a fibroblast exosomal-derived miRNA, which can
induce cardiomyocyte hypertrophy through silencing of its targets,
sorbin and SH3 domain containing 2 or PDZ and LIM domain 5
(26). Furthermore, miR-21-3p has
previously been reported to be associated with the diagnosis of
fibromyalgia (27). Therefore,
fibroblast-derived miR-21-3p may be involved in the hypertrophy of
Osborne's ligament. The present study is the first, to the best of
our knowledge, to identify the association between the three miRNAs
(hsa-miR-1343-3p, hsa-miR-21-3p and hsa-miR-146-5p) and hypertrophy
of Osborne's ligament, which may provide novel clues in
investigations of hypertrophy of Osborne's ligament.
According to the results of the GO analysis in the
present study, the predicted target genes were involved in protein
metabolic process, regulation of cell growth, cell cycle process,
regulation of cell division, cellular metabolic process and signal
transmission, among others. These biological processes are likely
to be important for the development of hypertrophy of Osborne's
ligament. KEGG pathway analysis showed that CAMs, the MAPK
signaling pathway, retrograde endocannabinoid signaling, the ErbB
signaling pathway, glycosaminoglycan biosynthesis-heparin
sulfate/heparin and the neurotrophic signaling pathway were among
the most relevant pathways for the predicted target genes. Certain
signaling pathways of these may be important in the pachyntic
mechanism of Osborne's ligament, and these results provide a novel
theoretical foundation for identifying potential therapeutic
targets for CuTS.
TGF-β is a member of a large family of
disulfide-bonded cytokines. The TGF-β superfamily members include
TGF-β1-3, activin, nodal, bone morphogenetic protein (BMP)-2, -4
and -7, anti-Müllerian hormone/Müllerian inhibiting substance
(AMH/MIS) and growth differentiation factor (GDF) 5. The TGF-β
superfamily members signal through a unique pair of transmembrane
serine-threonine kinases, known as type I and type II receptors, to
mediate intracellular small mothers against decapentaplegic (Smad)
signaling. The TGF-β/activin/nodal subfamily binds to ALK4, 5 and
7, and activates Smad2/3; whereas the BMP/GDF/MIS subfamily
generally binds to ALK1, 2, 3 or 6, and activates Smad1/5/8.
Activated Smad2/3 and Smad1/5/8 form a complex with Smad4 and enter
the nucleus, where they regulate target gene expression. TGFβ1
signaling has previously been reported to be associated with tissue
fibrosis, wound healing and scarring in humans (28–30).
In addition, the overexpression of miR-146b-5p can decrease levels
of SMAD4 by disrupting TGF-β signal transduction in thyroid cancer
(31). These findings, together
with those of the present study, support the hypothesis that
TGFβ1/Smad signaling may be key in the pathogenesis of pachyntic
Osborne's ligament and potential therapeutic targets of CuTS, even
though this signaling pathway was not predicted by KEGG
analysis.
As it is not possible to obtain normal Osborne's
ligaments from healthy individuals, normal tendinous tissues around
the two heads of the flexor carpi ulnaris were collected in
patients with CuTS in the present study. It was not possible to
confirm that the normal control tissues were entirely normal
tendinous tissues. The majority of patients with degenerative ulnar
neuritis have different degrees of elbow joint degeneration;
therefore, the soft tissues around the bone structure, particularly
the tendon, are likely to be degenerated, and this remains to be
clarified. These clinical limitations may have affected the
experimental results of the present study. In addition, the
prediction of miRNA targets by biological analysis alone is
insufficient and further experiments are required to verify
predicted miRNA targets, for example through the use of miRNA
transfection/knockdown and luciferase assays. Therefore, future
investigations aim to perform these experiments to confirm the
predicted miRNA targets in present study. The identification of the
function of the differentially expressed miRNAs and their
corresponding predicted target genes is likely to contribute to
current understanding of hypertrophy of Osborne's ligament.
In conclusion, the miRNA expression profiles of the
pachyntic Osborne's ligaments in patients with CuTS were
significantly different, compared with those of control tendinous
tissues. Altered miRNAs may lead to the excessive activation or
inactivation of signaling pathways in Osborne's ligament. The
present study also indicated that differentially expressed miRNAs
may be involved in the pathogenesis of CuTS and provided a
theoretical foundation for identifying novel clinical treatments
for CuTS. The etiology of CuTS is complex and remains to be fully
elucidated, however, the present study on the miRNAs of pachyntic
Osborne's ligaments provided novel information for revealing the
pathogenesis and pathology of CuTS.
Acknowledgements
This study was supported by the State Program of
National Natural Science Foundation of China (grant no. 81371957),
the State Key Program of the National Natural Science Foundation of
China (grant no. 81330042), the Special Program for Sino-Russian
Joint Research sponsored by the Ministry of Science and Technology,
China (grant no. 2014DFR31210) and the Key Program sponsored by the
Tianjin Science and Technology Committee, China (grant nos.
13RCGFSY19000 and 14ZCZDSY00044).
References
|
1
|
Wojewnik B and Bindra R: Cubital tunnel
syndrome-Review of current literature on causes, diagnosis and
treatment. J Hand Microsurg. 1:76–81. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mondelli M, Giannini F, Ballerini M,
Ginanneschi F and Martorelli E: Incidence of ulnar neuropathy at
the elbow in the province of Siena (Italy). J Neurol Sci. 234:5–10.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ilker U, Derya B, Ozay O, Orman C and Akan
M: Ulnar nerve compression at the elbow caused by the
epitrochleoanconeus muscle: A case report and surgical approach.
Turk Neurosurg. 24:266–271. 2014.PubMed/NCBI
|
|
4
|
Assmus H, Antoniadis G, Bischoff C,
Hoffmann R, Martini AK, Preissler P, Scheglmann K, Schwerdtfeger K,
Wessels KD and Wüstner-Hofmann M: Cubital tunnel syndrome - a
review and management guidelines. Cent Eur Neurosurg. 72:90–98.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shah CM, Calfee RP, Gelberman RH and
Goldfarb CA: Outcomes of rigid night splinting and activity
modification in the treatment of cubital tunnel syndrome. J Hand
Surg Am. 38:1125–1130, e1. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Choi CK, Lee HS, Kwon JY and Lee WJ:
Clinical implications of real-time visualized ultrasound-guided
injection for the treatment of ulnar neuropathy at the elbow: A
pilot study. Ann Rehabil Med. 39:176–182. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bacle G, Marteau E, Freslon M, Desmoineaux
P, Saint-Cast Y, Lancigu R, Kerjean Y, Vernet E, Fournier J, Corcia
P, et al: Cubital tunnel syndrome: Comparative results of a
multicenter study of 4 surgical techniques with a mean follow-up of
92 months. Orthop Traumatol Surg Res. 100:(4 Suppl). S205–S208.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu CH, Chen CX, Xu J, Wang HL, Ke XB,
Zhuang ZY, Lai ZL, Wu ZQ and Lin Q: Anterior subcutaneous versus
submuscular transposition of the ulnar nerve for cubital tunnel
syndrome: A systematic review and meta-analysis. PLoS One.
10:e01308432015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Brown JM, Mokhtee D, Evangelista MS and
Mackinnon SE: Scratch collapse test localizes osborne's Band as the
point of maximal nerve compression in cubital tunnel syndrome. Hand
(N Y). 5:141–147. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen J, Liu Z, Zhong G, Qian L, Li Z, Qiao
Z, Chen B and Wang H: Hypertrophy of ligamentum flavum in lumbar
spine stenosis is associated with increased miR-155 level. Dis
Markers. 2014:7865432014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dweep H and Gretz N: miRWalk2.0: A
comprehensive atlas of microRNA-target interactions. Nat Methods.
12:6972015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Naran S, Imbriglia JR, Bilonick RA, Taieb
A and Wollstein R: A demographic analysis of cubital tunnel
syndrome. Ann Plast Surg. 64:177–179. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Descatha A, Leclerc A, Chastang JF and
Roquelaure Y: Study Group on Repetitive Work: Incidence of ulnar
nerve entrapment at the elbow in repetitive work. Scand J Work
Environ Health. 30:234–240. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gras C, Ratuszny D, Hadamitzky C, Zhang H,
Blasczyk R and Figueiredo C: miR-145 contributes to hypertrophic
scarring of the skin by inducing myofibroblast activity. Mol Med.
21:296–304. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhu H, Luo H, Li Y, Zhou Y, Jiang Y, Chai
J, Xiao X, You Y and Zuo X: MicroRNA-21 in scleroderma fibrosis and
its function in TGF-β-regulated fibrosis-related genes expression.
J Clin Immunol. 33:1100–1109. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li P, He QY and Luo CQ: Overexpression of
miR-200b inhibits the cell proliferation and promotes apoptosis of
human hypertrophic scar fibroblasts in vitro. J Dermatol.
41:903–911. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Geraldo MV, Fuziwara CS, Friguglieti CU,
Costa RB, Kulcsar MA, Yamashita AS and Kimura ET: MicroRNAs
miR-146-5p and let-7f as prognostic tools for aggressive papillary
thyroid carcinoma: A case report. Arq Bras Endocrinol Metabol.
56:552–557. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lin F, Wang X, Jie Z, Hong X, Li X, Wang M
and Yu Y: Inhibitory effects of miR-146b-5p on cell migration and
invasion of pancreatic cancer by targeting MMP16. J Huazhong Univ
Sci Technolog Med Sci. 31:509–514. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li Y, Wang Y, Yu L, Sun C, Cheng D, Yu S,
Wang Q, Yan Y, Kang C, Jin S, et al: miR-146b-5p inhibits glioma
migration and invasion by targeting MMP16. Cancer Lett.
339:260–269. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Simsek S, Er U, Demirci A and Sorar M:
Operative illustrations of the Osborne's ligament. Turk Neurosurg.
21:269–270. 2011.PubMed/NCBI
|
|
23
|
Jung JC, Wang PX, Zhang G, Ezura Y, Fini
ME and Birk DE: Collagen fibril growth during chicken tendon
development: Matrix metalloproteinase-2 and its activation. Cell
Tissue Res. 336:79–89. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Van Doren SR: Matrix metalloproteinase
interactions with collagen and elastin. Matrix Biol. 44–46:224–231.
2015. View Article : Google Scholar
|
|
25
|
Singh D, Srivastava SK, Chaudhuri TK and
Upadhyay G: Multifaceted role of matrix metalloproteinases (MMPs).
Front Mol Biosci. 2:192015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bang C, Batkai S, Dangwal S, Gupta SK,
Foinquinos A, Holzmann A, Just A, Remke J, Zimmer K, Zeug A, et al:
Cardiac fibroblast-derived microRNA passenger strand-enriched
exosomes mediate cardiomyocyte hypertrophy. J Clin Invest.
124:2136–2146. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cerdá-Olmedo G, Mena-Durán AV, Monsalve V
and Oltra E: Identification of a microrna signature for the
diagnosis of fibromyalgia. PLoS One. 10:e01219032015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Miyazono K, Suzuki H and Imamura T:
Regulation of TGF-beta signaling and its roles in progression of
tumors. Cancer Sci. 94:230–234. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kamato D, Burch ML, Piva TJ, Rezaei HB,
Rostam MA, Xu S, Zheng W, Little PJ and Osman N: Transforming
growth factor-β signalling: Role and consequences of Smad linker
region phosphorylation. Cell Signal. 25:2017–2024. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Finnson KW, Mclean S, Di Guglielmo GM and
Philip A: Dynamics of transforming growth factor beta signaling in
wound healing and scarring. Adv Wound Care (New Rochelle).
2:195–214. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Geraldo MV, Yamashita AS and Kimura ET:
MicroRNA miR-146b-5p regulates signal transduction of TGF-β by
repressing SMAD4 in thyroid cancer. Oncogene. 31:1910–1922. 2012.
View Article : Google Scholar : PubMed/NCBI
|