Introduction
Allergic rhinitis (AR) is a common inflammatory
disorder of the upper airway, which has an estimated worldwide
incidence rate of 10–20% (1). Over
the last two decades the pathogenesis of AR has been widely
studied, and genetic factors are considered to be major players
affecting the development, severity and treatment of AR (2). The single nucleotide polymorphisms
(SNPs) of important cytokines or genes may predict susceptibility
to or clinical features of AR. Several loci and candidate genes
have been reported to be associated with AR (3–5). Our
recent studies demonstrated associations between polymorphisms in
interleukin (IL)-23R, Fc receptor-like 3 gene and IL-27 with AR
risk in Chinese subjects (6–8).
However, the details of AR pathogenesis currently remain
unclear.
Tumor necrosis factor receptor superfamily 4
(TNFSF4, also known as OX4OL) belongs to the TNF superfamily, and
is expressed on dendritic cells, macrophages, cluster of
differentiation (CD)4+/CD8+ T cells,
activated NK cells and other cells (9,10).
Interaction between TNFSF4 and its binding partner OX40 provides a
costimulatory signal, resulting in T cell proliferation,
differentiation and cytokine production (11,12).
Recent studies have indicated that TNFSF4 and OX40 interaction may
promote the T-helper (Th)2 response, depress IL-17 production and
inhibit the differentiation of regulatory T cells (13–15).
Therefore, TNFSF4 is regarded as an important cytokine in the
pathogenic mechanisms of immune-related disorders.
B cell lymphocyte kinase (BLK) is a tyrosine kinase
of the src family with highly restricted B lymphocyte expression.
BLK participates in signal transduction downstream of the B-cell
receptor; therefore, it may influence the proliferation and
differentiation of B cells (16).
B cells serve critical roles in the pathogenesis of immune-related
disorders via antigen presentation to T cells, antibody production
and cytokine secretion. Therefore, it may be hypothesized that the
BLK protein may have an impact on the immune mechanisms of B cells,
and participate in the adaptive immune response.
Although the pathogenic mechanism of AR is not
completely understood, it is known to be associated with a
dysfunctional immune system, and involves T and B cell responses.
Recent research indicated that gene-level interaction between BLK
and TNFSF4 may have a synergistic effect on T cells and B cells via
the nuclear factor (NF)-κB pathway, and this may have a role in
determining immunologic aberration (17). Furthermore, previous studies have
reported that TNFSF4 and BLK polymorphisms may contribute to the
pathogenesis of further immune-related diseases, including primary
Sjogren's syndrome (18,19) and Systemic Lupus Erythematosus
(SLE) (20).
The present study hypothesized that TNFSF4 and BLK
genes may participate in NF-κB pathway regulation, and may contain
SNPs that are associated with AR risk. Therefore, the association
between TNFSF4 and BLK polymorphisms and AR susceptibility were
examined in a Han Chinese population.
Materials and methods
Ethics statement
The study protocol was approved by the Ethics
Committee of the First Affiliated Hospital of Chongqing Medical
University (Chongqing, China). All participants were from Chongqing
and were of the Han Chinese ethnic origin. Informed consent was
obtained from the next of kin, caretakers or guardians of minors
and children participating in the study.
Subjects
A total of 600 patients (296 men, 304 women; age
range, 6–81 years) were recruited from April 2013 to June 2014. All
patients were enrolled and treated at the outpatient clinic of the
Department of Otolaryngology Head and Neck Surgery at the First
Affiliated Hospital of Chongqing Medical University. AR diagnoses
were based on medical history, symptoms and positive skin prick
test (SPT; Allergopharma GmbH & Co., KG, Reinbek, Germany)
according to ARIA 2008 guidelines (21). A total of 18 inhaled allergens were
tested, including house dust, pollen, grass, tree, mold, food, cat
and dog dander, cockroaches, feathers, cotton, cigarettes,
penicillin, milk, shrimp, egg, soybean and peanut. SPT results were
diagnosed in accordance with the recommendations of the
Subcommittee on Allergen Standardization and Skin Tests of the
European Academy of Allergy and Clinical Immunology (22). AR patients with chronic sinusitis,
asthma, hypertension, diabetes or any other systemic disease were
excluded from the study. A total of 700 healthy volunteers of the
same ethnicity as the patients were recruited as the control group
from the Department of Physical Examination at the First Affiliated
Hospital of Chongqing Medical University (Chongqing, China), from
April 2013 to October 2013. The selection criteria for healthy
volunteers were as follows: No chronic pathology, in particular, no
history of allergy or respiratory pathology, no other systemic
diseases and no family history of allergy. The clinical features of
the study cohort are described in Table I.
 | Table I.Clinical features and demographic
characteristics of the study population. |
Table I.
Clinical features and demographic
characteristics of the study population.
| Characteristic | Value |
|---|
| Allergic rhinitis
(n=600) |
|
| Gender
(male/female) | 296/304 |
| Age
[mean (range)] years | 33.06 (6.5–81) |
| Allergen |
|
| House
dust mite | 377 |
|
Pollen | 94 |
|
Multiple allergens | 129 |
| Control
(n=700) |
|
| Gender
(male/female) | 343/357 |
| Age
[mean (range)] years | 31.28 (9–78) |
SNP selection and DNA extraction
Nine SNPs in the TNFSF4 (rs1234313, rs1234314,
rs1234315, rs12039904, rs844648, rs10912580) and BLK (rs1600249,
rs13277113, rs2254546) genes were analyzed as candidate sites
(Table II). SNPs were selected on
the basis of recent reports, which demonstrated their possible
functional effect in immune-related diseases (23–27).
Genomic DNA was isolated from EDTA-anticoagulated peripheral blood
leukocytes, using a Wizard® Genomic DNA Purification kit
(Promega Corporation, Madison, WI, USA), according to the
manufacturer's protocol. Briefly, 300 µl blood was mixed with cell
lysis solution. Leucocytes were spun down at 11,100 × g for 1 min
at room temperature and lysed with the nuclei lysis solution, and
the pellet was separated using the protein precipitation solution.
Precipitated proteins were removed by centrifugation at 16,000 × g
for 30 sec at room temperature. Two-filar DNA was subsequently
separated out by methyl alcohol. The DNA on the EP tube was
dissolved in 100 µl DNA rehydration solution.
 | Table II.Characteristics of the studied SNPs
in TNFSF4 and BLK genes. |
Table II.
Characteristics of the studied SNPs
in TNFSF4 and BLK genes.
| Chromosome | SNP ID | Location | Alleles |
|---|
| TNFSF4 |
| 1 | rs1234313 | Intron region | A/G |
| 1 | rs1234314 | Upstream
region | C/G |
| 1 | rs1234315 | Upstream
region | C/T |
| 1 | rs12039904 | Intron region | C/T |
| 1 | rs844648 | Intron region | A/G |
| 1 | rs10912580 | Intron region | A/G |
| BLK |
| 8 | rs1600249 | Intron region | A/C |
| 8 | rs13277113 | Promoter
region | A/G |
| 8 | rs2254546 | Intergenic
region | A/G |
Genotyping
SNPs were genotyped using the polymerase chain
reaction (PCR)-restriction fragment length polymorphism method.
Amplification was performed using initial denaturation at 95°C for
4 min, followed by 37 cycles of 95°C for 40 sec, 56–60°C for 40 sec
and 72°C for 40 sec, followed by a final extension at 72°C for 4
min. The primer sequences and reaction conditions used in the
present study are provided in Table
III. The PCR products were incubated with restriction enzymes
for ≥4 h. The selected SNP genotyping was performed using the
Sequenom MassARRAY iPLEX Gold platform (Sequenom Laboratories, San
Diego, CA, USA) according to the manufacturer's instructions. To
verify the genotyping results, PCR-amplified DNA samples were
examined by direct sequencing (20% of all the blood samples). The
sequencing PCR reaction system included 30 µl Taq enzyme (Go
Taq® Green Master Mix; Promega Corporation), 20 µl
enzyme free water and 5 µl primer pairs (forward, 2.5 µl; reverse,
2.5 µl). Amplification of the target fragments by PCR and the
results were read by Chromas 2.1.1 software (Technelysium Pty Ltd.,
South Brisbane, Australia). The results of RFLP and direct
sequencing were 100% concordant.
 | Table III.Primer sequences, PCR conditions and
restriction enzymes used for PCR-restriction fragment length
polymorphism analysis of the tumor necrosis factor receptor
superfamily 4 and B-cell lymphocyte kinase polymorphisms. |
Table III.
Primer sequences, PCR conditions and
restriction enzymes used for PCR-restriction fragment length
polymorphism analysis of the tumor necrosis factor receptor
superfamily 4 and B-cell lymphocyte kinase polymorphisms.
| SNP reference | Primer
sequence | Annealing
temperature (°C) | Restriction
enzyme |
|---|
| rs1234313 | F:
5′CCTACCATGTCTCAAACATAATGGCAC-3′ | 60 | TaiI |
|
| R:
5′TGTCTTCCACAGTCCTCTACAATGGTT-3′ |
|
|
| rs1234315 | F:
5′CACCAGGCTGGAAGTTTCAGGC-3′ | 62 | TspRI |
|
| R:
5′TAGCCAGACCTGGTGTTGCGTG-3′ |
|
|
| rs1234314 | F:
5′ACCAGGTACCCTTTACCACTAAAATAAAC-3′ | 60 | MvaI |
|
| R:
5′TCTCCCTCCTTTCTTTACATATCTGCT-3′ |
|
|
| rs12039904 | F:
5′TGCTCATAGTTGCTTAATGC-3′ | 60 | Mn1I |
|
| R:
5′AATAATCAGGCTGTGGAAAC-3′ |
|
|
| rs844648 | F:
5′AGTTACACTATGTGGCGTTTA-3′ | 54 | AseI |
|
| R:
5′CAGGCAGTTCCTCTTTGAT-3′ |
|
|
| rs10912580 | F:
5′GCAAGACCCTGACTCAAAAATAAAATAA-3′ | 60 | AccI |
|
| R:
5′TTGCCTTAGTAGATATCACTGAACGC-3′ |
|
|
| rs1600249 | F:
5′ACAACTAAATGGATTCTATTAAAACAAAGG-3′ | 60 | DraI |
|
| R:
5′GTTCCTGTTGTATAAATACTCCCACC-3′ |
|
|
| rs13277113 | F:
5′ACCATTCCCATTAGGTAACCT-3′ | 58 | BsaBI |
|
| R:
5′CAATAAAGTAAGTGGAAGAAACATAAA-3′ |
|
|
| rs2254546 | F:
5′GAAAATGGCTCTGAGAGAACTCCAAAGT-3′ | 60 | EcoNI |
|
| R:
5′ATTGGCGAAGACTCTGTGGTATTCAGT-3′ |
|
|
Statistical analysis
Statistical analyses were performed using SPSS
version 18.0 (SPSS Inc., Chicago, IL, USA). P<0.05 was
considered to indicate statistical significance. To evaluate the
quality of the genotyping data, the Hardy-Weinberg equilibrium for
SNP genotype frequencies was tested using a Chi-square test
(χ2 test). Allelic and genotypic frequencies between
patients with AR and the control patients were compared using the
χ2 test. The online software platform SHEsis (http://analysis2.bio-x.cn/myanalysis.php) was used to
analyze the haplotype and probability values. The association
between genotypes/alleles and AR risk was estimated by calculating
the odds ratios (OR) and 95% confidence intervals (CI).
Additionally, pairwise linkage disequilibrium (LD) among the SNPs
was calculated according to the genotype correlation coefficient
(r2). The r2 values were calculated using
Haploview v4.2, with default settings (CI for a strong LD was
minimal for upper 0.98 and low 0.7, and maximal for a strong
recombination of 0.9, a fraction of strong LD in informative
comparisons was ≥0.95) (28).
To evaluate the synergistic relationships between
the TNFSF4 and BLK polymorphisms and the risk of AR, the
multifactor dimensionality reduction (MDR) method was used, to
detect and characterize locus-locus and gene-gene interaction
models (27). Each best model was
tested for accuracy, cross-validation consistency and significance
level, determined using permutation testing, testing accuracy and
testing OR (95% CI). Cross-validation consistency (CVC) was defined
as the number of cross-validation replicates (partitions) for which
the same n-locus model was chosen as the best model (i.e., the
number of replicates within which the classification error was
minimized).
Results
Clinical features of the
participants
The clinical characteristics of the subjects are
presented in Table I. The AR
patients and the controls were similar in terms of gender
distribution (P>0.05) and mean age (P>0.05). A total of 377
(62.8%) patients were recorded as sensitive to house dust mite, 94
(15.5%) were sensitive to tree pollen and 129 (21.5%) were
sensitive to multiple allergens.
Genotype distribution of the TNFSF4
polymorphisms
The genotype distribution of all 9 analyzed SNPs in
the AR group and the controls were revealed to be in Hardy-Weinberg
equilibrium (P>0.05). The TNFSF4 genotype and allele frequencies
are presented in Table IV. The
rs1234315T allele demonstrated a significantly increased prevalence
in AR cases (49.7%) compared with the controls (43.1%),
demonstrating a statistical association between the rs1234315T
allele and AR susceptibility (P=5.51×10−4, OR=1.30, 95%
CI=1.11–1.52). Similarly, the rs1234314G allele was associated with
a higher risk of AR (P=2.64×10−4, OR=1.38, 95%
CI=1.18–1.61). However, the C allele and CC genotypes of rs1234315
and rs123431 demonstrated lower prevalence in AR patients, compared
with the controls (P=5.51×10−3, OR=0.77, 95%
CI=0.66–0.90; P=3.06×10−4, OR=0.58, 95% CI=0.45–0.75;
P=2.64×10−4, OR=0.72, 95% CI=0.62–0.84;
P=5.40×10−4, OR=0.57, 95% CI=0.43–0.74;
respectively).
 | Table IV.The genotype and allele frequencies
of tumor necrosis factor receptor superfamily 4 polymorphisms in AR
patients and controls. |
Table IV.
The genotype and allele frequencies
of tumor necrosis factor receptor superfamily 4 polymorphisms in AR
patients and controls.
| Genotype
allele | AR (%) | Control (%) | χ2 | P-value
(unadjusted) | OR (95% CI) |
|---|
| rs1234313 |
| AA | 222 (37.0) | 294 (42.0) |
3.37 | 1.26 | 0.81
(0.65–1.01) |
| AG | 319 (53.2) | 306 (43.7) | 11.56 | 0.02 | 1.46
(1.17–1.82) |
| GG | 59 (9.8) | 100 (14.3) |
5.97 | 0.27 | 0.65
(0.47–0.92) |
| A | 763 (63.6) | 894 (63.9) |
0.02 | 5.34 | 0.99
(0.84–1.16) |
| G | 437 (36.4) | 506 (36.1) |
0.02 | 5.34 | 1.01
(0.86–1.19) |
| rs1234315 |
|
|
|
|
|
| CC | 135 (22.5) | 233 (33.3) | 18.52 |
3.06×10−4 | 0.58
(0.45–0.75) |
| CT | 334 (55.7) | 330 (47.1) |
9.39 | 0.036 | 1.41
(1.13–1.75) |
| TT | 131 (21.8) | 137 (19.6) |
1.01 | 5.67 | 1.15
(0.88–1.50) |
| C | 604 (50.3) | 796 (56.9) | 11.07 |
5.51×10−3 | 0.77
(0.66–0.90) |
| T | 596 (49.7) | 604 (43.1) | 11.07 |
5.51×10−3 | 1.30
(1.11–1.52) |
| rs1234314 |
|
|
|
|
|
| CC | 106 (17.7) | 192 (27.4) | 17.43 |
5.40×10−4 | 0.57
(0.43–0.74) |
| CG | 302 (50.3) | 328 (46.9) |
1.56 | 3.79 | 1.15
(0.92–1.43) |
| GG | 192 (32.0) | 180 (25.7) |
6.25 | 0.22 | 1.36
(1.07–1.73) |
| C | 514 (42.8) | 712 (50.9) | 16.69 |
2.64×10−4 | 0.72
(0.62–0.84) |
| G | 686 (57.2) | 688 (49.1) | 16.69 |
2.64×10−4 | 1.38
(1.18–1.61) |
| rs12039904 |
|
|
|
|
|
| CC | 282 (47.0) | 380 (54.3) |
6.86 | 0.16 | 0.75
(0.60–0.93) |
| CT | 275 (45.8) | 276 (39.4) |
5.43 | 0.36 | 1.30
(1.04–1.62) |
| TT | 43 (7.2) | 44 (6.3) | 0.4 | 9.47 | 1.15
(0.75–1.78) |
| C | 839 (69.9) | 1036 (74.0) |
5.36 | 0.12 | 0.82
(0.69–0.97) |
| T | 361 (30.1) | 364 (26.0) |
5.36 | 0.12 | 1.23
(1.03–1.45) |
| rs844648 |
|
|
|
|
|
| AA | 110 (18.3) | 152 (21.7) |
2.3 | 2.34 | 0.81
(0.62–1.06) |
| AG | 302 (50.3) | 345 (49.3) |
0.14 | 12.71 | 1.04
(0.84–1.30) |
| GG | 188 (31.3) | 203 (29.0) |
0.84 | 6.48 | 1.12
(0.88–1.42) |
| A | 522 (43.5) | 649 (46.4) |
2.13 | 0.86 | 0.89
(0.76–1.04) |
| G | 678 (56.5) | 751 (53.6) |
2.13 | 0.86 | 1.12
(0.96–1.31) |
| rs10912580 |
|
|
|
|
|
| AA | 354 (59.0) | 383 (54.7) |
2.42 | 2.16 | 1.19
(0.96–1.49) |
| AG | 222 (37.0) | 280 (40.0) |
1.23 | 4.82 | 0.88
(0.70–1.10) |
| GG | 24 (4.0) | 37 (5.3) |
1.19 | 4.93 | 0.75
(0.44–1.26) |
| A | 930 (77.5) | 1046 (74.7) |
2.75 | 0.58 | 1.17
(0.97–1.40) |
| G | 270 (22.5) | 354 (25.3) |
2.75 | 0.58 | 0.86
(0.72–1.03) |
Genotype distribution of the BLK
polymorphisms
The BLK genotype and allele frequencies are
presented in Table V. The
frequencies of rs13277113 and rs1600249 were significantly
different between the AR cases and the controls. An increased
prevalence of the rs13277113G allele and the AG genotype was
observed in the AR group, compared with the controls
(P=9.0×10−3, OR=1.26, 95% CI=1.08–1.48;
P=1.8×10−3, OR=1.51, 95% CI=1.21–1.88; respectively).
However, the frequencies of the A allele and the AA genotype were
significantly lower in the AR patients (P=9×10−3,
OR=0.79, 95% CI=0.68–0.93; P=2.7×10−4, OR=0.60, 95%
CI=0.48–0.77). A higher frequency of the rs1600249C allele (P=0.04,
OR=1.23, 95% CI=1.05–1.44), and a lower frequency of the A allele
and the AA genotype (P=0.04, OR=0.04, 95% CI=0.69–0.96; P=0.02,
OR=0.71, 95% CI=0.56–0.88) were observed in AR patients, compared
with the controls.
 | Table V.The genotype and allele frequencies
of B-cell lymphocyte kinase polymorphisms in patients with AR and
controls. |
Table V.
The genotype and allele frequencies
of B-cell lymphocyte kinase polymorphisms in patients with AR and
controls.
| Genotype
allele | AR (%) | Control (%) | χ2
value | P-value
(unadjusted) | OR (95% CI) |
|---|
| rs1600249 |
|
|
|
|
|
| AA | 198 (33.0) | 288 (41.1) |
9.15 | 0.02 | 0.71
(0.56–0.88) |
| AC | 330 (55.0) | 338 (48.3) |
5.53 | 0.17 | 1.30
(1.04–1.62) |
| CC | 72 (12.0) | 74 (10.6) |
0.66 | 3.74 | 1.15
(0.82–1.63) |
| A | 726 (60.5) | 914 (65.3) |
6.35 | 0.04 | 0.81
(0.69–0.96) |
| C | 474 (39.5) | 486 (34.7) |
6.35 | 0.04 | 1.23
(1.05–1.44) |
| rs13277113 |
|
|
|
|
|
| AA | 157 (26.2) | 259 (37.0) | 17.43 |
2.7×10−4 | 0.60
(0.48–0.77) |
| AG | 342 (57.0) | 327 (46.7) | 13.68 |
1.8×10−3 | 1.51
(1.21–1.88) |
| GG | 101 (16.8) | 114 (16.3) |
0.07 | 7.12 | 1.04
(0.78–1.40) |
| A | 656 (54.7) | 845 (60.4) |
8.57 |
9×10−3 | 0.79
(0.68–0.93) |
| G | 544 (45.3) | 555 (39.6) |
8.57 |
9×10−3 | 1.26
(1.08–1.48) |
| rs2254546 |
|
|
|
|
|
| AA | 19 (3.2) | 29 (4.1) |
0.87 | 3.17 | 0.76
(0.42–1.36) |
| AG | 225 (37.5) | 225 (32.1) |
4.1 | 0.39 | 1.27
(1.01–1.59) |
| GG | 356 (59.3) | 446 (63.7) |
2.62 | 0.95 | 0.83
(0.66–1.04) |
| A | 263 (21.9) | 283 (20.2) |
1.13 | 0.86 | 1.11
(0.92–1.34) |
| G | 937 (78.1) | 1117 (79.8) |
1.13 | 0.86 | 0.90
(0.75–1.09) |
Frequencies of haplotypes between AR
cases and controls
Plots of the pair-wise LD [r2 and
coefficient of linkage disequilibrium (D')] values for the tag SNPs
and the LD structures for the selected region of the chromosome are
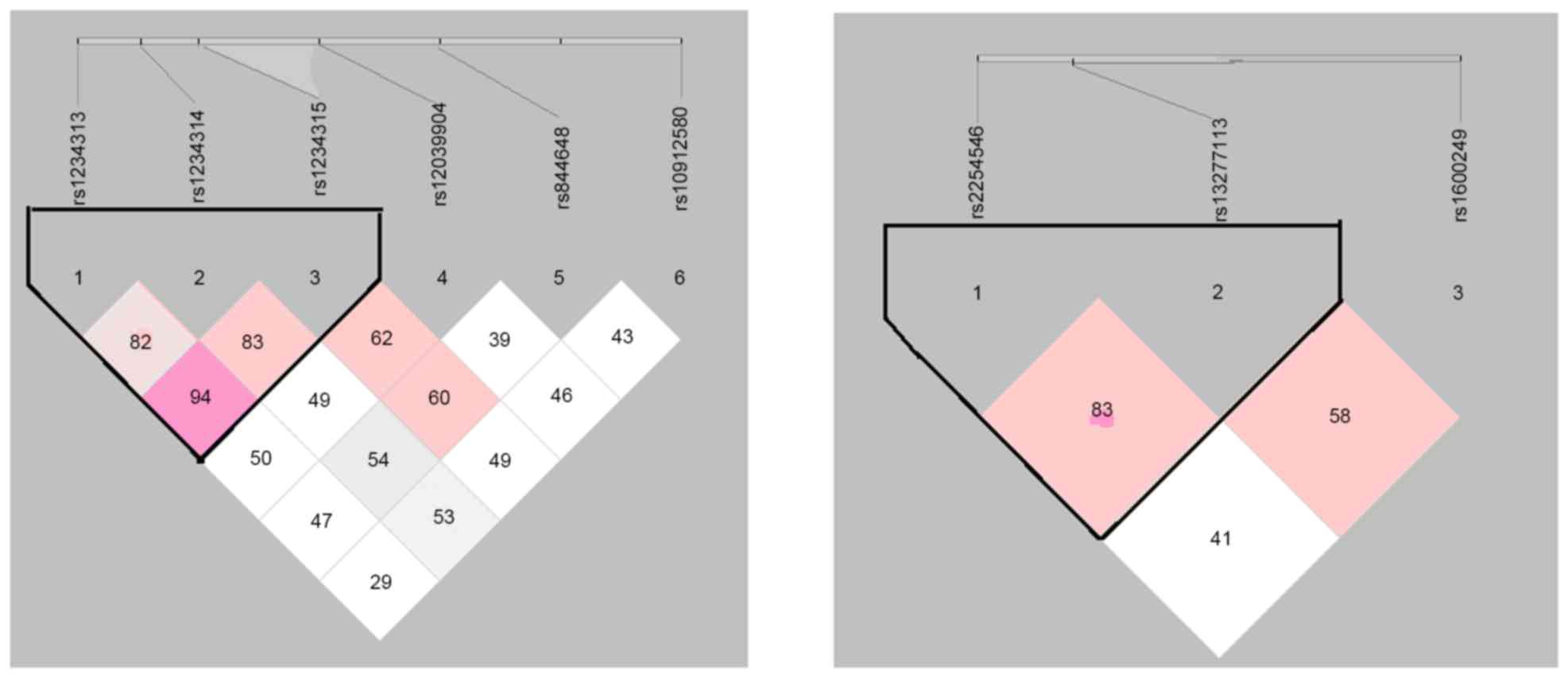
presented in Fig. 1. The
relationships between AR risk and the haplotype frequencies in
blocks rs1234313-rs1234314-rs1234315 and rs2254546-rs13277113 are
summarized in Table VI. The
results indicated that the distribution rate of the GGT haplotype
was markedly increased in AR patients (11.4%), compared with the
controls (6.2%) (P=2.87×10−8, OR=1.940, 95%
CI=1.465–2.570). However, the distribution rate of the ACC
haplotype was significantly reduced in AR cases (12.2%), compared
with controls (17.7%) (P=0.0001, OR=0.650, 95% CI=0.521–0.810). The
AG haplotype demonstrated a significantly higher frequency in AR
patients (9.3%) compared with controls (6.7%) (P=0.013, OR=1.436,
95% CI=1.079–1.912), whereas the distribution rate of the GA
haplotype was significantly reduced (42.1% in cases and 46.8% in
controls; P=0.015, OR=0.825, 95% CI=0.706–0.964).
 | Table VI.Frequencies of the haplotypes formed
by the rs1234313-rs1234314-rs1234315 and the rs2254546-rs13277113
SNPs in patients with AR and healthy controls. |
Table VI.
Frequencies of the haplotypes formed
by the rs1234313-rs1234314-rs1234315 and the rs2254546-rs13277113
SNPs in patients with AR and healthy controls.
| Haplotype | AR (%) | Control (%) | χ2
value | P value | OR (95% CI) |
|---|
| ACC | 146.72 (0.122) | 247.19 (0.177) | 14.819 | 0.0001 | 0.650
(0.521–0.810) |
| ACT | 171.01 (0.143) | 193.91 (0.139) | 0.086 | 0.7694 | 1.034
(0.828–1.290) |
| AGC | 245.69 (0.205) | 241.68 (0.173) | 4.375 | 0.0365 | 1.234
(1.013–1.503) |
| AGT | 199.58 (0.166) | 211.23 (0.151) | 1.158 | 0.2819 | 1.123
(0.909–1.386) |
| GCC | 107.09 (0.089) | 158.72 (0.113) | 4.1 | 0.0429 | 0.766
(0.592–0.992) |
| GCT | 89.18 (0.074) | 112.18 (0.080) | 0.306 | 0.5804 | 0.922
(0.690–1.231) |
| GGC | 104.50 (0.087) | 148.41 (0.106) | 2.635 | 0.1046 | 0.804
(0.618–1.047) |
| GGT | 136.23 (0.114) | 86.69 (0.062) | 21.953 |
2.87×10−8 | 1.940
(1.465–2.570) |
| AA | 150.99 (12.6) | 189.36 (13.5) | 0.505 | 0.477 | 0.920
(0.732–1.157) |
| AG | 112.01 (9.3) | 93.64 (6.7) | 6.209 | 0.013 | 1.436
(1.079–1.912) |
| GA | 505.01 (42.1) | 655.64 (46.8) | 5.892 | 0.015 | 0.825
(0.706–0.964) |
| GG | 431.99 (36.0) | 461.36 (33.0) | 2.656 | 0.103 | 1.144
(0.973–1.346) |
Locus-locus and gene-gene
interactions
The best interaction models determined by the MDR
analysis for all 9 SNPs analyzed in TNFSF4 and BLK are presented in
Table VII. Following
cross-validation and permutation tests of the gene-gene
interactions in relation to AR, 3 best models were revealed, and
these demonstrated interactive effects. The best models included a
two-marker model (rs13277113-rs1600249, testing accuracy=60.63%;
CVC=9/10; P=0.037), a three-marker model
(rs1234314-rs13277113-rs1600249, testing accuracy=61.65%; CVC=6/10;
P=0.022), and a four-marker model
(rs1234314-rs10912580-rs13277113-rs1600249, testing
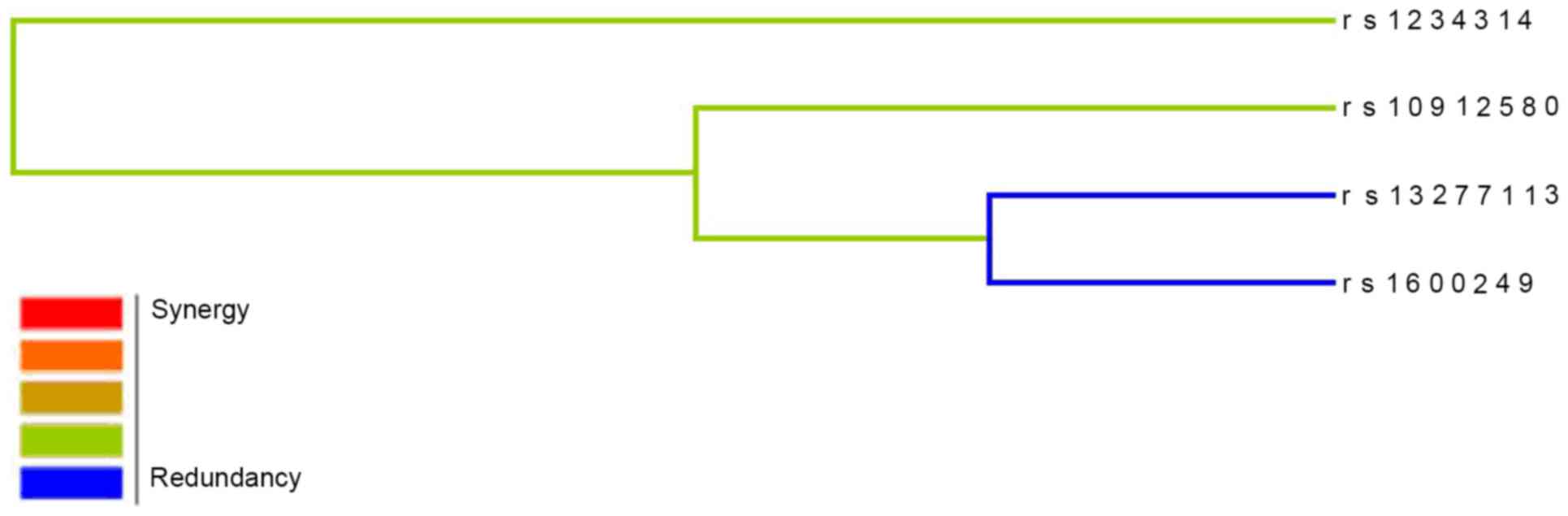
accuracy=62.69%; CVC=10/10; P=0.013). A dendogram of the markers in
the four-marker model is presented in Fig. 2. The presence of the blue line
indicates that the four-locus model may have a redundancy
interaction effect on modulating the risk of AR.
 | Table VII.MDR analysis of gene-gene
interactions in relation to allergic rhinitis. |
Table VII.
MDR analysis of gene-gene
interactions in relation to allergic rhinitis.
| Best candidate
model | Testing balanced
accuracy (%) | Testing OR (95%
CI) | Testing
χ2 value | P-value | Cross-validation
Consistency |
|---|
| rs13277113 | 54.84 | 1.51
(0.659–3.440) | 0.944 | 0.331 | 7/10 |
| rs13277113 +
rs1600249 | 60.63 | 2.37
(1.046–5.387) | 4.345 | 0.037 | 9/10 |
| rs1234314 +
rs13277113 + rs1600249 | 61.65 | 2.59
(1.136–5.884) | 5.227 | 0.022 | 6/10 |
| rs1234314 +
rs10912580 + rs13277113 + rs1600249 | 62.69 | 2.83
(1.236–6.457) | 6.203 | 0.013 | 10/10 |
Discussion
The present study demonstrated a novel contribution
of TNFSF4 and BLK polymorphisms towards the risk of AR in a Han
Chinese population. The results demonstrated that the CC
(rs1234314, rs1234315) and AA (rs1600249, rs13277113) genotypes
were statistically associated with protective effects against AR.
However, the AG genotype (rs13277113) presented a risk factor for
AR. The ACC haplotype in block rs1234313-rs1234314-rs1234315 and
the GA haplotype in block rs2254546-rs13277113 significantly
decreased the risk of AR, whereas the GGT and AG haplotypes served
protective roles. Our results suggest that specific SNPs in the
TNFSF4 and BLK genes may modify the risk of suffering from AR.
TNFSF4 regulates the differentiation and
proliferation of Th cells in different cytokine microenvironments
(29). The interaction between
TNFSF4 and OX40 serves an important role at critical
immunoregulatory checkpoints, which are likely involved in the
development of immunorelated diseases, such as inflammatory and
autoimmune diseases and tumors. Previous studies have demonstrated
that TNFSF4 gene polymorphisms are associated with SLE, systemic
sclerosis, breast cancer and myocardial infarction (30–34).
The present study demonstrated an association between the rs1234314
and rs1234315 SNPs and AR susceptibility. These results are mostly
congruent with previous studies concerning immune-related diseases.
In Caucasian populations, the rs1234315 and rs1234314 SNPs in the
5′untranslated region of TNFSF4 were correlated with susceptibility
to primary Sjögren's syndrome (35). Furthermore, an association between
the rs1234315 allele of TNFSF4 and SLE in Asians was revealed in a
meta-analysis (4). In Caucasian
patients, the strongest associated SLE variants were rs844648 and
rs12039904 (36,37), whereas the present results
demonstrated no significant associations between AR and rs844648
and rs12039904. Our study indicates that the genetic background of
AR may be partially similar to those of other immune-related
diseases. However, the disparities of these findings suggest that
the risk of developing AR is determined by a complex interaction
amongst several genes. Furthermore, there may be genetic
heterogeneity of AR amongst different populations.
BLK serves a role in the signal transduction of B
cells. The present study demonstrated a relationship between SNPs
rs13277113 and rs1600249 in the BLK gene and AR susceptibility. The
rs13277113 SNP may be directly involved in AR susceptibility. Our
data indicated that the AG genotype of rs13277113 increases the
risk of AR by 1.51-fold, and the G allele of rs13277113 is related
to a 1.26-fold increase in AR risk. However, the AA genotype and
the A allele decreased the risk of AR by 0.60- and 0.79-fold,
respectively. These results indicate that the G allele is likely to
result in AR susceptibility, and individuals with the G allele in
the rs13277113 SNP of the BLK gene may be more likely to develop
AR. By contrast, the A allele may protect against AR development.
Previous studies have indicated that a BLK rs13277113 A/G
polymorphism is associated with RA susceptibility (38) and associated with the development
of SLE in European (39), Japanese
(40) and Chinese (41) populations. The risk allele appears
to be involved in decreased BLK mRNA expression (39). Based on these studies and the
results of the present study, it may be speculated that the
difference between the A and G alleles in rs13277113 influences AR
susceptibility. This alteration may impact on gene splicing,
transcription factor binding or the non-coding RNA sequence, and
might thereby influence the expression of certain proteins.
Haplotype analysis revealed that the GGT haplotype
in block rs1234313-rs1234314-rs1234315 and the AG haplotype in
block rs2254546-rs13277113 are positively correlated with AR,
however, the ACC and GA haplotypes were negatively correlated with
AR. It is therefore possible that subjects with the GGT and/or AG
haplotypes are at a higher risk of developing AR. By contrast,
individuals with the ACC and/or GA haplotypes may be more resistant
to AR, suggesting that these two haplotypes may serve a role in
protecting against AR.
The present study was carefully designed to minimise
the influence of confounding factors on the results. AR patients
and controls were selected using strict guidelines and the
genotyping results were confirmed by direct sequencing. However,
there are a few limitations that need to be considered. The protein
levels of TNFSF4 and BLK were not measured and functional
experiments were not performed. Furthermore, detailed information
about AR severity was not obtained, and this restricted the
analyses. Finally, gene-gene interactions and environmental factors
are critical for AR development, therefore, more intensive studies
investigating the gene-gene or gene-environment interactions are
needed to clarify the genetic influence of TNFSF4 and BLK in the
pathogenesis of AR.
In conclusion, the present study indicated that
polymorphisms in the TNFSF4 and BLK genes may be correlated with
susceptibility to AR in a Han Chinese population. However, further
studies are needed to elucidate the complex gene-gene and
gene-environment interactions in AR. The results of the present
study provide novel biomarkers that may be investigated as
predictive factors for AR susceptibility in a Han Chinese
population.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 81500774) and the
National Key Clinical Specialties Construction Program of China
Glossary
Abbreviations
Abbreviations:
|
AR
|
allergic rhinitis
|
|
BLK
|
B cell lymphocyte kinase
|
|
TNFSF4
|
tumor necrosis factor receptor
superfamily 4
|
|
SNP
|
single nucleotide polymorphism
|
|
SLE
|
Systemic Lupus Erythematosus
|
|
SPT
|
skin prick test
|
|
PCR-RFLP
|
polymerase chain reaction-restriction
fragment length polymorphism
|
|
HWE
|
Hardy-Weinberg equilibrium
|
|
OR
|
odds ratios
|
|
CI
|
confidence interval
|
|
LD
|
linkage disequilibrium
|
References
|
1
|
Togias A: Rhinitis and asthma: Evidence
for respiratory system integration. J Allergy Clin Immunol.
111:1171–1184. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Vercelli D: Genetic polymorphism in
allergy and asthma. Curr Opin Immunol. 15:609–613. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nilsson D, Henmyr V, Halldén C, Säll T,
Kull I, Wickman M, Melén E and Cardell LO: Replication of
genomewide associations with allergic sensitization and allergic
rhinitis. Allergy. 69:1506–1514. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bønnelykke K, Matheson MC, Pers TH,
Granell R, Strachan DP, Alves AC, Linneberg A, Curtin JA,
Warrington NM, Standl M, et al: Meta-analysis of genome-wide
association studies identifies ten loci influencing allergic
sensitization. Nat Genet. 45:902–906. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fuertes E, Brauer M, MacIntyre E, Bauer M,
Bellander T, von Berg A, Berdel D, Brunekreef B, Chan-Yeung M,
Gehring U, et al: Childhood allergic rhinitis, traffic-related air
pollution, and variability in the GSTP1, TNF, TLR2, and TLR4 genes:
Results from the TAG Study. J Allergy Clin Immunol. 132:342–352.e2.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gu Z, Hong SL, Ke X, Shen Y, Wang XQ, Hu
D, Hu GH and Kang HY: FCRL3 gene polymorphisms confer autoimmunity
risk for allergic rhinitis in a chinese han population. PLoS One.
10:e01164192015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hu D, Hu G, Zhu J, Shen Y, Kang H and Hong
S: Association between polymorphisms of the IL-23R gene and
allergic rhinitis in a Chinese Han population. PLoS One.
8:e638582013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shen Y, Yuan XD, Hu D, Ke X, Wang XQ, Hu
GH, Hong SL and Kang HY: Association between interleukin-27 gene
polymorphisms and susceptibility to allergic rhinitis. Hum Immunol.
75:991–995. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Murata K, Ishii N, Takano H, Miura S,
Ndhlovu LC, Nose M, Noda T and Sugamura K: Impairment of
antigen-presenting cell function in mice lacking expression of OX40
ligand. J Exp Med. 191:365–374. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kashiwakura J, Yokoi H, Saito H and
Okayama Y: T cell proliferation by direct cross-talk between OX40
ligand on human mast cells and OX40 on human T cells: Comparison of
gene expression profiles between human tonsillar and lung-cultured
mast cells. J Immunol. 173:5247–5257. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yan J, Su H, Xu L and Wang C: OX40-OX40L
interaction promotes proliferation and activation of lymphocytes
via NFATc1 in ApoE-deficient mice. PLoS One. 8:e608542013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Duan W, So T and Croft M: Antagonism of
airway tolerance by endotoxion/lipopolysaccharide through promoting
OX40L and suppressing antigen-specific Foxp3+ T
regulatory cells. J Immunol. 181:8650–8659. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jenkins SJ, Perona-Wright G, Worsley AG,
Ishii N and MacDonald AS: Dendritic cell expression of OX40 ligand
acts as a costimulatory, not polarising, signal for optimal Th-2
priming and memory induction in vivo. J Immunol. 179:3515–3523.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li J, Li L, Shang X, Benson J, Elloso M
Merle, Schantz A, Bracht M, Orlovsky Y and Sweet R: Negative
regulation of IL-17 production by OX40/OX40L interaction. Cell
Immunol. 253:31–37. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xiao X, Kroemer A, Gao W, Ishii N, Demirci
G and Li XC: OX40/OX40L costimulation affects induction of
Foxp3+ regulatory T-cells in part by expanding memory
T-cells in vivo. J Immunol. 181:3193–3201. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
International Consortium for Systemic
Lupus Erythematosus Genetics (SLEGEN), . Harley JB,
Alarcón-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL,
Tsao BP, Vyse TJ, Langefeld CD, et al: Genome-wide association scan
in women with systemic lupus erythematosus identifies
susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat
Genet. 40:204–210. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhou XJ, Lu XL, Nath SK, Lv JC, Zhu SN,
Yang HZ, Qin LX, Zhao MH, Su Y, Shen N, et al: Gene-gene
interaction of BLK, TNFSF4, TRAF1, TNFAIP3, and REL in systemic
lupus erythematosus. Arthritis Rheum. 64:222–231. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sun F, Li P, Chen H, Wu Z, Xu J, Shen M,
Leng X, Shi Q, Zhang W, Tian X, Li Y and Zhang F: Association
studies of TNFSF4, TNFAIP3 and FAM167A-BLK polymorphisms with
primary Sjogren's syndrome in Han Chinese. J Hum Genet. 58:475–479.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nordmark G, Kristjansdottir G, Theander E,
Appel S, Eriksson P, Vasaitis L, Kvarnström M, Delaleu N, Lundmark
P and Lundmark A: Association of EBF1, FAM167A(C8orf13)-BLK and
TNFSF4 gene variants with primary Sjögren's syndrome. Genes Immun.
12:100–109. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zuo XB, Sheng YJ, Hu SJ, Gao JP, Li Y,
Tang HY, Tang XF, Cheng H, Yin XY, Wen LL, et al: Variants in
TNFSF4, TNFAIP3, TNIP1, BLK, SLC15A4 and UBE2L3 interact to confer
risk of systemic lupus erythematosus in Chinese population.
Rheumatol Int. 34:459–464. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bousquet J, Khaltaev N, Cruz AA, Denburg
J, Fokkens WJ, Togias A, Zuberbier T, Baena-Cagnani CE, Canonica
GW, van Weel C, et al: Allergic rhinitis and its impact on asthma
(ARIA) 2008 update (in collaboration with the World Health
Organization, GA(2)LEN and AllerGen). Allergy. 63:(Suppl 86).
S8–S160. 2008. View Article : Google Scholar
|
|
22
|
No authors listed: Position paper:
Allergen standardization and skin tests. The European Academy of
Allergology and Clinical Immunology. Allergy. 48:(14 Suppl).
S48–S82. 1993.
|
|
23
|
Graham DS Cunninghame, Graham RR, Manku H,
Wong AK, Whittaker JC, Gaffney PM, Moser KL, Rioux JD, Altshuler D,
Behrens TW and Vyse TJ: Polymorphism at the TNF superfamily gene
TNFSF4 confers susceptibility to systemic lupus erythematosus. Nat
Genet. 40:83–89. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kong F, Li JX, Li P, Li YZ, Zhang FC and
Zhang J: Association of TNFSF4 polymorphisms with susceptibility to
primary Sjögren's syndrome and primary biliary cirrhosis in a
Chinese Han population. Clin Exp Rheumatol. 31:546–551.
2013.PubMed/NCBI
|
|
25
|
Chen S, Wu W, Li J, Wang Q and Li Y, Wu Z,
Zheng W, Wu Q, Wu C, Zhang F and Li Y: Single nucleotide
polymorphisms in the FAM167A-BLK gene are associated with
polymyositis/dermatomyositis in the Han Chinese population. Immunol
Res. 62:153–162. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Deng FY, Lei SF, Zhu H, Zhang YH and Zhang
ZL: Integrative analyses for functional mechanisms underlying
associations for rheumatoid arthritis. J Rheumatol. 40:1063–1081.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ritchie MD, Hahn LW, Roodi N, Bailey LR,
Dupont WD, Parl FF and Moore JH: Multifactor-dimensionality
reduction reveals high-order interactions among estrogen-metabolism
genes in sporadic breast cancer. Am J Hum Genet. 69:138–147. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Barrett JC, Fry B, Maller J and Daly MJ:
Haploview: Analysis and visualization of LD and haplotype maps.
Bioinformatics. 21:263–265. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu YJ, Soumelis V, Watanabe N, Ito T,
Wang YH, Malefyt Rde W, Omori M, Zhou B and Ziegler SF: TSLP: An
epithelial cell cytokine that regulates T cell differentiation by
conditioning dendritic cell maturation. Annu Rev Immunol.
25:193–219. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gourh P, Arnett FC, Tan FK, Assassi S,
Divecha D, Paz G, McNearney T, Draeger H, Reveille JD, Mayes MD and
Agarwal SK: Association of TNFSF4 (OX40L) polymorphisms with
susceptibility to systemic sclerosis. Ann Rheum Dis. 69:550–555.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ria M, Lagercrantz J, Samnegård A, Boquist
S, Hamsten A and Eriksson P: A common polymorphism in the promoter
region of the TNFSF4 gene is associated with lower allele-specific
expression and risk of myocardial infarction. PLoS One.
6:e176522011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bossini-Castillo L, Broen JC, Simeon CP,
Beretta L, Vonk MC, Ortego-Centeno N, Espinosa G, Carreira P, Camps
MT, Navarrete N, et al: A replication study confirms the
association of TNFSF4 (OX40L) polymorphisms with systemic sclerosis
in a large European cohort. Ann Rheum Dis. 70:638–641. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Delgado-Vega AM, Abelson AK, Sánchez E,
Witte T, D'Alfonso S, Galeazzi M, Jiménez-Alonso J, Pons-Estel BA,
Martin J and Alarcón-Riquelme ME: Replication of the TNFSF4 (OX40L)
promoter region association with systemic lupus erythematosus.
Genes Immun. 10:248–253. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Weiguang Y, Dalin L, Lidan X, Yonggang C,
Shuang C, Yanhong L, Fengyan X, Zhenkun F, Da P and Dianjun L:
Association of OX40L polymorphisms with sporadic breast cancer in
northeast Chinese Han population. PLoS One. 7:e412772012.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Nordmark G, Kristjansdottir G, Theander E,
Appel S, Eriksson P, Vasaitis L, Kvarnström M, Delaleu N, Lundmark
P, Lundmark A, et al: Association of EBF1, FAM167A(C8orf13)-BLK and
TNFSF4 gene variants with primary Sjögren's. Genes Immun.
12:100–109. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Graham DS Cunninghame, Graham RR, Manku H,
Wong AK, Whittaker JC, Gaffney PM, Moser KL, Rioux JD, Altshuler D,
Behrens TW and Vyse TJ: Polymorphism at the TNF superfamily gene
TNFSF4 confers susceptibility to systemic lupus erythematosus. Nat
Genet. 40:83–89. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Delgado-Vega AM, Abelson AK, Sánchez E,
Witte T, D'Alfonso S, Galeazzi M, Jiménez-Alonso J, Pons-Estel BA,
Martin J and Alarcón-Riquelme ME: Replication of the TNFSF4 (OX40L)
promoter region association with systemic lupus erythematosus.
Genes Immun. 10:248–253. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ito I, Kawasaki A, Ito S, Kondo Y,
Sugihara M, Horikoshi M, Hayashi T, Goto D, Matsumoto I, Tsutsumi
A, et al: Replication of association between FAM167A(C8orf13)-BLK
region and rheumatoid arthritis in a Japanese population. Ann Rheum
Dis. 69:936–937. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hom G, Graham RR, Modrek B, Taylor KE,
Ortmann W, Garnier S, Lee AT, Chung SA, Ferreira RC, Pant PV, et
al: Association of systemic lupus erythematosus with C8orf13-BLK
and ITGAM-ITGAX. N Engl J Med. 358:900–909. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ito I, Kawasaki A, Ito S, Hayashi T, Goto
D, Matsumoto I, Tsutsumi A, Hom G, Graham RR, Takasaki Y, et al:
Replication of the association between the C8orf13-BLK region and
systemic lupus erythematosus in a Japanese population. Arthritis
Rheum. 60:553–558. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang Z, Zhu KJ, Xu Q, Zhang XJ, Sun LD,
Zheng HF, Han JW, Quan C, Zhang SQ, Cai LQ, et al: The association
of the BLK gene with SLE was replicated in Chinese Han. Arch
Dermatol Res. 302:619–624. 2010. View Article : Google Scholar : PubMed/NCBI
|