Introduction
Lung cancer is one of the most common cancers
worldwide and the first leading cause of the cancer-related death
(1,2). Despite tremendous efforts made to
improve lung cancer treatment including surgery, chemotherapy and
radiotherapy, a substantial number of patients still face the high
risk of drug resistance and subsequent tumor metastasis (3–5).
Moreover, metastasis is the major contributor to morbidity and
mortality in patients with non-small cell lung cancer (NSCLC).
Therefore, further research into the potential molecular basis of
NSCLC progression is pivotal to improving the treatment and
prognosis of NSCLC.
Long noncoding RNAs (lncRNAs) are an important class
of the noncoding RNA family, longer than 200 nucleotides without
evident protein-coding capabilities (6). Increasing evidence has showed that
lncRNAs act a crucial role in cancer occurrence and progression
(7–10). Some lncRNAs can participate in
diverse biological processes, including cell viability, modulation
of migration and invasion (11–13).
However, compared to the well-characterized microRNA, the functions
of lncRNAs have not been fully unravelled.
Recent studies have identified that lncRNAs are
involved in NSCLC pathogenesis (14,15),
which provide a new insight into the clinical benefit in NSCLC
treatment. TSLC1 (also known as CADM1, IGSF4, SynCAM, SgIGSF and
Necl-2) (16–18) has been proved as a tumor suppressor
gene in various cancers including nasopharyngeal cancer, breast
cancer, gastric cancer, pancreatic cancer, colorectal cancer,
cervical cancer and prostate cancer (19–25).
Intriguingly, recent study has showed that TSLC1 is significantly
down-regulated in NSCLC tissues and cell lines, and elevated
expression of TSLC1 inhibits NSCLC cell viability, migration, and
invasion (26). BRAF is well known
as a genetic mutation that could activate oncogenes, which have
been identified to be associated with NSCLC (27). CYLD and APC are the negative
regulators of Wnt signaling pathway and also reported to be
associated with NSCLC (28,29).
Nonetheless, as the antisense RNA of TSLC1 (30), lncRNA RP11-713B9.1 expression and
its biological role in NSCLC development and progression remain
largely unknown.
In our study, we discovered that the expression of
RP11-713B9.1 in NSCLC samples was significantly lower than that in
the adjacent normal lung tissues. The expression of RP11-713B9.1
and TSLC1 was positively correlated and co-regulated in NSCLC.
Furthermore, overexpression and knockdown experiments were
performed to explore the role of RP11-713B9.1 in NSCLC cells. We
found that RP11-713B9.1 overexpression inhibited the NSCLC cells
viability, whereas RP11-713B9.1 knockdown showed the opposite
results. On the basis of our findings, we speculate that lncRNA
RP11-713B9.1 may serve as a novel potential biomarker in NSCLC
treatment.
Materials and methods
Lung cancer samples
Samples of surgically removed tumors used in this
study were collected from 46 patients with a postoperative
diagnosis of NSCLC from 2010 to 2012 in First Affiliated Hospital
of Yangtz University. Demographic and clinical characteristics of
46 NSCLC patients were shown in Table
I. No patients had received therapy before surgery. Each
participated patient signed a written informed consent, which was
in accordance with our institutional ethical guidelines. The
utilization of tumor tissues for this study was all approved by the
ethical committee of First Affiliated Hospital of Yangtz
University. The 46 NSCLC tumor samples were quick frozen at the
time of resection until analysis.
 | Table I.Demographic and clinical
characteristics of 46 NSCLC patients. |
Table I.
Demographic and clinical
characteristics of 46 NSCLC patients.
| Variables | NSCLC patients n
(%) | Deaths n=22 | MST (months) | Log-rank P | Crude HR (95%
CI) |
|---|
| Age |
| ≤65 | 21 (0.46) | 10 | 25.9 | – | 1.00
(Reference) |
|
>65 | 25 (0.54) | 12 | 24.3 | 0.83 | 1.093
(0.48–2.49) |
| Sex |
| Male | 40 (0.87) | 13 | 30.7 | – | 1.00
(Reference) |
|
Female | 6
(0.13) | 9 | 24.9 | 0.13 | 0.52
(0.22–1.22) |
| Smoking |
|
Never | 7
(0.15) | 2 | 40.1 | – | 1.00
(Reference) |
| Ever | 39 (0.85) | 20 | 31.7 | <0.01 | 14.26
(3.30–61.64) |
| Histology |
|
Adenocarcinoma | 21 (0.46) | 9 | 25.3 | – | 1.00
(Reference) |
|
Squamous cell carcinoma | 15 (0.33) | 8 | 21.9 | 0.39 | 1.52
(0.59–3.95) |
|
Others | 10 (0.21) | 5 | 17.6 | 0.66 | 1.28
(0.43–3.83) |
| Clinical stage |
|
Early | 12 (0.26) | 4 | 58.4 | – | 1.00
(Reference) |
|
Advanced | 34 (0.74) | 18 | 12.8 | 0.003 | 5.36
(1.80–15.93) |
| Surgery |
| No | 29 (0.63) | 14 | 22.7 | – | 1.00
(Reference) |
|
Yes | 17 (0.37) | 8 | 32.1 | 0.004 | 0.19
(0.06–0.59) |
| Chemotherapy |
| No | 11 (0.24) | 10 | 23.9 | – | 1.00
(Reference) |
|
Yes | 35 (0.76) | 12 | 25.2 | 0.98 | 1.01
(0.44–2.34) |
| Radiotherapy |
| No | 25 (0.54) | 11 | 20.4 | – | 1.00
(Reference) |
|
Yes | 21 (0.46) | 11 | 22.9 | 0.58 | 0.79
(0.34–1.83) |
Cancer cell lines
The human normal lung epithelial cell BEAS-2B, the
commercial human NSCLC cell lines H460, H1975, A549 were obtained
from the American Tissue Culture Collection (ATCC, Manassas, VA,
USA), and cell lines were maintained in Dulbecco's modified Eagle's
medium (DMEM) medium (Gibco, Carlsbad, CA, USA) supplemented with
10% fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA, USA) in a
humidified atmosphere of 5% (v/v) CO2 and 95% air at
37°C.
Real-Time Quantitative PCR
Total RNA was extracted from the NSCLC tumor tissue
and adjacent normal tissue using the Trizol Total RNA Reagent
(Invitrogen, Carlsbad CA, USA) following the manufacturer's
protocol. cDNA synthesis was performed with 2 µg total RNA using
the RevertAidTM H Minus First Strand cDNA Synthesis Kit (Takara,
Ohtsu, Japan). The primers were obtained from GenePharma (Shanghai,
China) and the sequences were shown in Table II. Real-Time Quantitative PCR was
performed using the SYBR PrimeScript Real-Time Quantitative PCR kit
(Takara, Ohtsu, Japan) in an Applied Biosystems 7500 Fluorescent
Quantitative PCR System (Applied Biosystems, Foster City, CA, USA).
The reaction mixtures started at 95°C for 30 sec, followed by 40
amplification cycles of 95°C for 5 sec and 60°C for 34 sec. The
quantification of gene expression was performed using the ΔΔCT
calculation with CT as the threshold cycle.
 | Table II.Primers Real-Time Quantitative PCR
analysis. |
Table II.
Primers Real-Time Quantitative PCR
analysis.
| Gene name | Forward | Reverse |
|---|
| β-actin |
5′-CCACTGGCATCGTGATGGA-3′ |
5′-CGCTCGGTGAGGATCTTCAT-3′ |
| RP11-713B9.1 |
5′-TGACAAAGGCAGGAGGTA-3′ |
5′-GCACTATGGCTGAGGAAA-3′ |
| TSLC1 |
5′-ATGGCGAGTGTAGTGCTGC-3′ |
5′-GATCACTGTCACGTCTTTCGT-3′ |
| CYLD |
5′-TTCACTGACGGGGTGTACCA-3′ |
5′-CAGGACCTGCGTAATCACTTTC-3′ |
| APC |
5′-TCCTCCGCAATGTGTCCAG-3′ |
5′-AGGCTGTGCGAAGTCAGATG-3′ |
| BRAF |
5′-AATACACCAGCAAGCTAGATGC-3′ |
5′-AATCAGTTCCGTTCCCCAGAG-3′ |
Overexpression of RP11-713B9.1 in A549
cell line
pcDNA-RP11-713B9.1 was cloned into BamHI-EcoRI sites
of pcDNA3.1. The RP11-713B9.1 low-expressing A549 cells were
transfected with pcDNA-RP11-713B9.1 using Lipofectamine 2000
(Invitrogen, US) according to the manufacturer's instructions.
Cells were collected after transfection for RNA extraction, MTT
cell viability assay.
Knockdown of RP11-713B9.1 in H460 cell
line
For small interfering RNA (siRNA) analysis, siRNA
for the RP11-713B9.1 sequence and negative-control (NC) siRNA were
given from GenePharma (Shanghai, China). The target sequences of
RP11-713B9.1 siRNA were shown in Table III, which were also used in our
previous literature (30). H460
cells were plated to the 12-well plates with a density of
2×103 cells per well and cultured at least 24 h before
transfection. siRNA transfection was performed with X-tremeGENE
transfection reagent (Roche) according to the manufacturer's
instructions. High-expressing RP11-713B9.1 in H460 cell line was
harvested or fixed 48 h after transfection for RNA extraction, MTT
assay.
 | Table III.siRNA for RP11-713B9.1 sequences
(30). |
Table III.
siRNA for RP11-713B9.1 sequences
(30).
| Name | The sequences |
|---|
|
RP11-713B9.1-si1 | Sense strand
5′-rGrUrArCrCrUrCrCrUrGrCrCrUrUrUrGrUrCrArArGrCrCAA-3′ |
|
| Antisense strand
5′-rUrUrGrGrCrUrUrGrArCrArArArGrGrCrArGrGrArGrGrUrArCrArA-3′ |
|
RP11-713B9.1-si2 | Sense strand
5′-rGrArCrCrUrArUrCrGrArGrArArCrUrGrArGrArGrCrGrACA-3′ |
|
| Antisense strand
5′-rUrGrUrCrGrCrUrCrUrCrArGrUrUrCrUrCrGrArUrArGrGrUrCrArG-3′ |
Cell viability assay
After transfection, cell viability was tested using
a 3-(4,5-Dimethyl-thiazol-2-yl)-2,5-diphen yltetrazolium bromide
(MTT) assay (Promega) according to the manufacturer's instruction.
A549 and H460 cells were seeded at a density of 2×103
cells per well in 96-well plates and maintained in Dulbecco's
modified Eagle's medium (DMEM) supplemented with 10% FBS for 24–68
h prior to transfection with siRNA. Cells were transfected using 25
nM RP11-713B9.1 siRNA or control NC siRNA in serum-free and
antibiotic-free DMEM. After transfection 3–5 days, cells were
analyzed using the MTT reagent for another 4 h. 100 µl DMSO was
added to dissolve the MTT-formazan formed by metabolically viable
cells and then shaken for homogeneity. Optical density was measured
at 24, 48 and 72 h after transfection on a microplate reader with
the wave length of 490 nm.
Statistical analyses
Differences between two groups were carried out
using Student's t-test. One-way ANOVA was used for the comparison
of multiple groups (>2). The data variables were shown as the
mean ± SD. Correlation between genes expression was analyzed using
Pearson's correlation. Statistical analyses were conducted using
SPSS version 18.0 (SPSS, Chicago, IL, USA). For all statistical
analyses, *P<0.05 was considered statistically significant.
Results
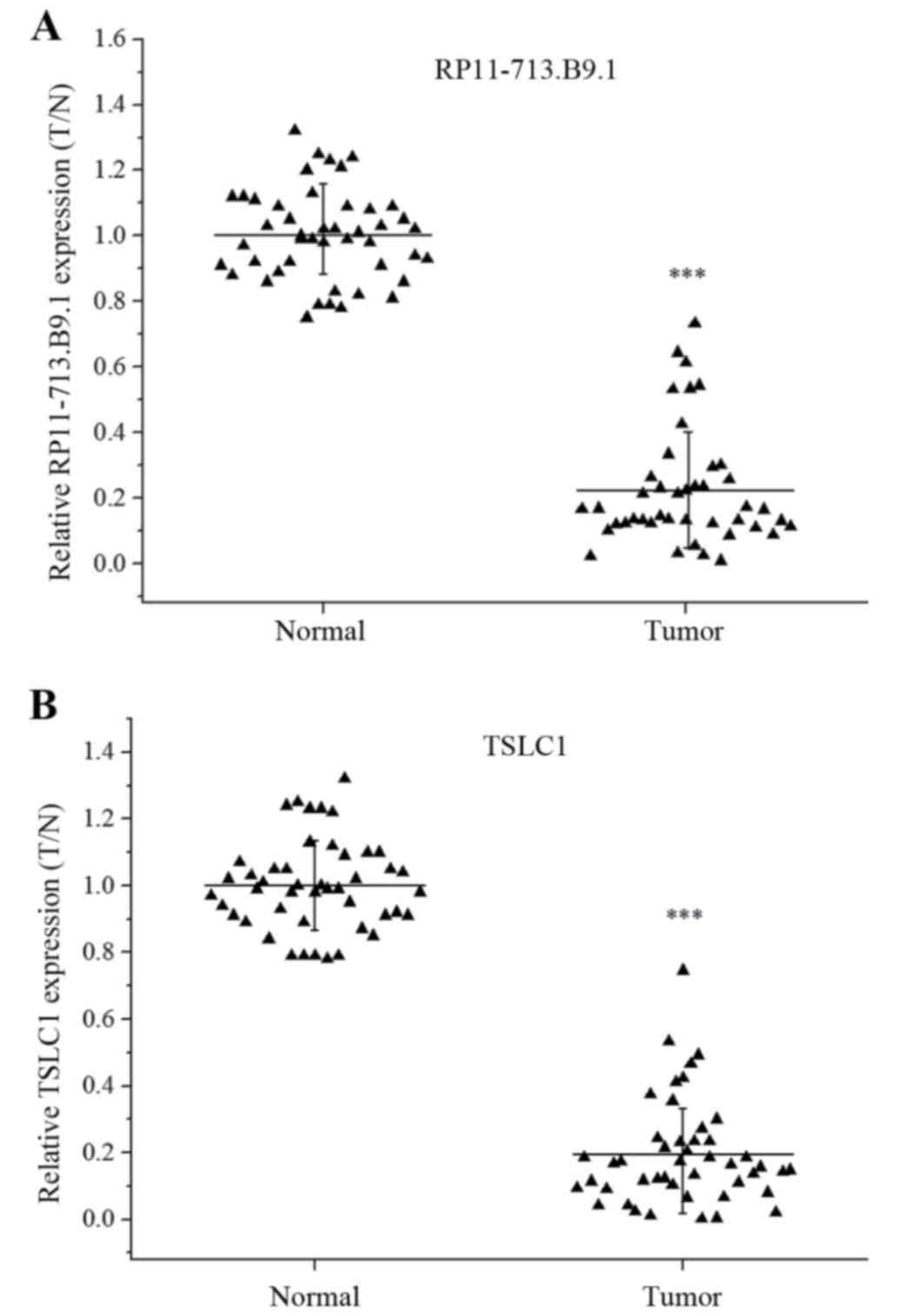
Both RP11-713B9.1 and TSLC1 were
down-regulated in the NSCLC tissue samples
The RP11-713B9.1 and TSLC1 expression levels were
assessed through paired specimen obtained from 46 patients with
lung cancer. And each tumor sample was normalized to the relative
adjacent non-tumor tissue. The data showed that both RP11-713B9.1
and TSLC1 expression were significantly down-regulated in the NSCLC
tissues (P<0.001) (Fig. 1A,
B).
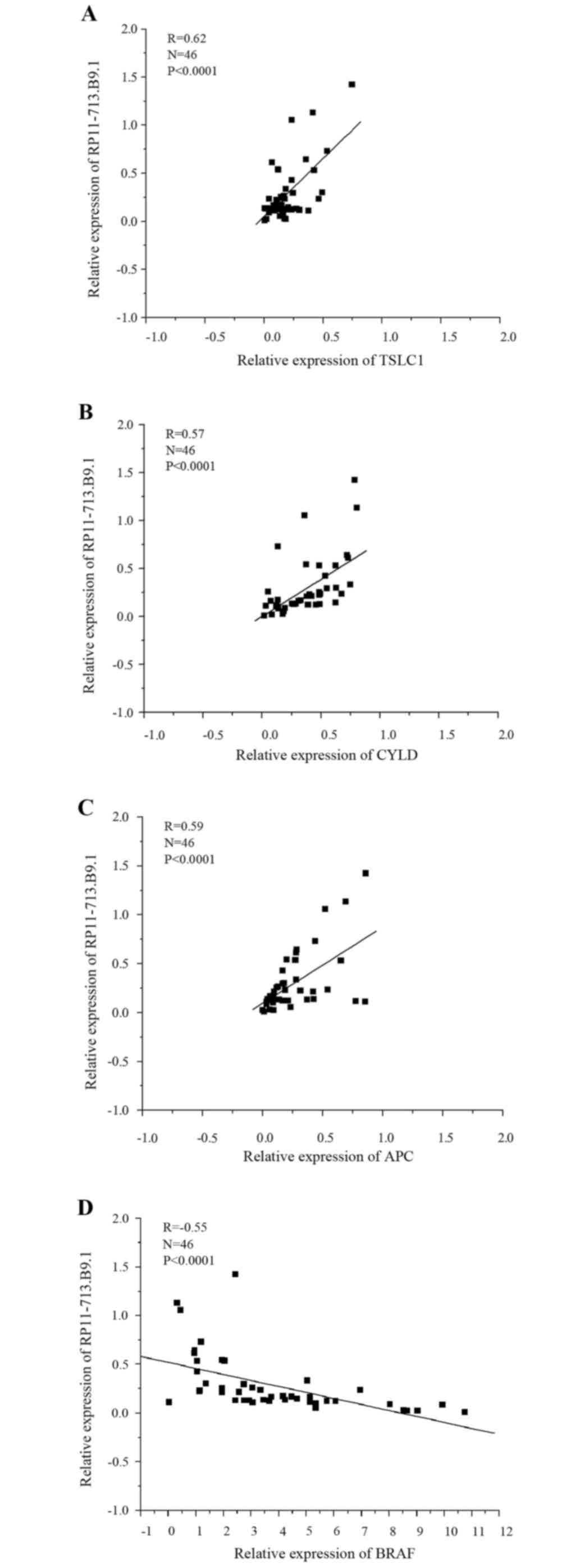
Expression of RP11-713B9.1 was
correlated with TSLC1 and other tumor related genes
The correlation of RP11-713B9.1 expression with
TSLC1 was assessed using Real-Time Quantitative PCR. The data
suggested that RP11-713B9.1 expression was positively correlated
with TSLC1 expression (R=0.62, P<0.0001, Fig. 2A). In addition, the connections
between RP11-713B9.1 expression and other tumor regulating genes in
NSCLC were also analyzed. The results confirmed that RP11-713B9.1
expression was positively correlated with tumor suppressors CYLD
(R=0.57, P<0.0001, Fig. 2B) and
APC (R=0.59, P<0.0001, Fig.
2C), and negatively correlated with the oncogene BRAF (R=−0.55,
P<0.0001, Fig. 2D).
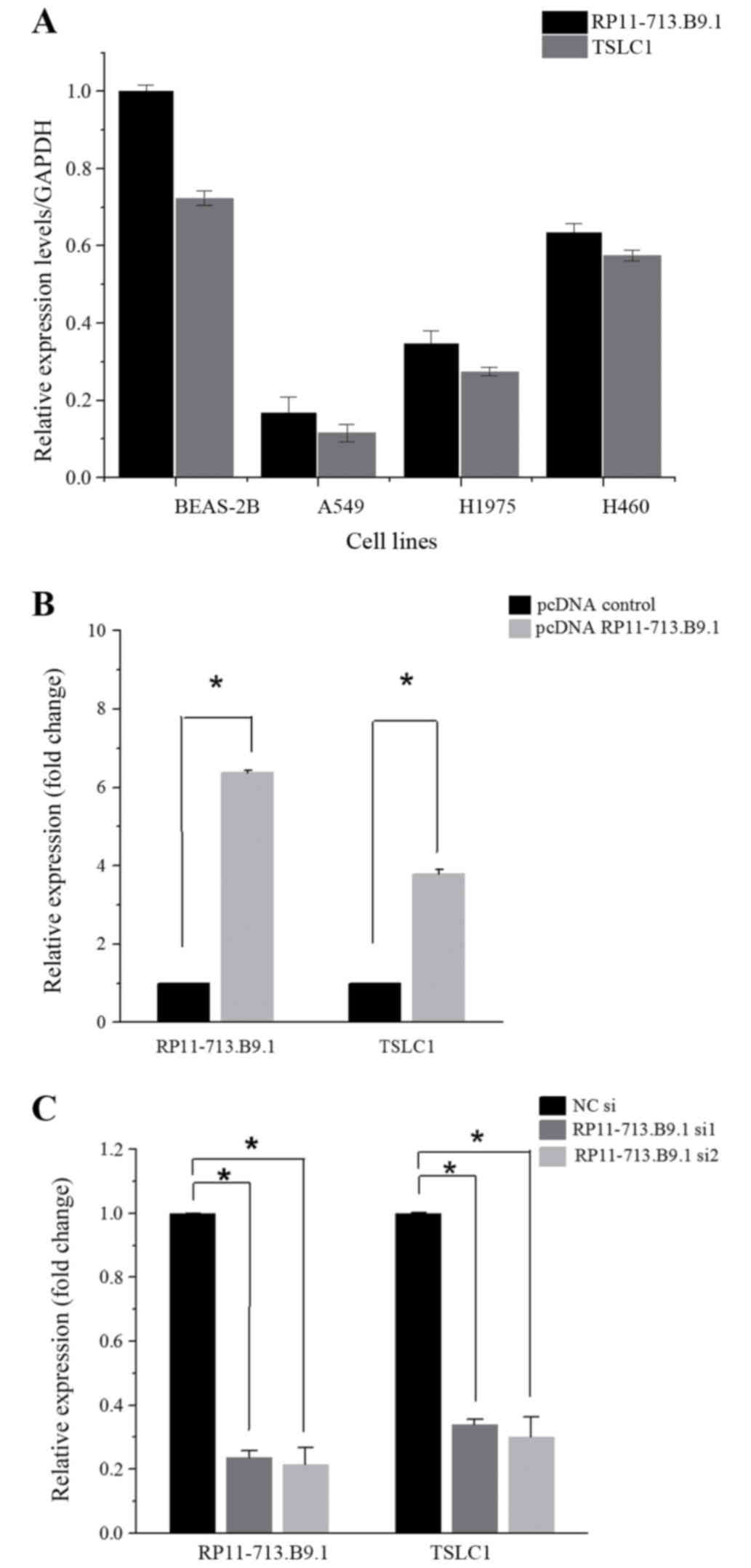
Co-regulation of TSLC1 expression with
RP11-713B9.1 overexpression or knockdown in NSCLC cell lines
Through Real-Time Quantitative PCR, we found that
both RP11-713B9.1 and TSLC1 expression were the highest in H460 and
the lowest in A549 among the three human NSCLC cell lines (Fig. 3A). In the overexpression
experiment, when pcDNA RP11-713B9.1 was conducted and transfected
into the A549, RP11-713B9.1 expression was remarkably elevated and
TSLC1 expression level was also apparently up-regulated (P<0.05,
Fig. 3B), whereas in the
knock-down experiment, RP11-713B9.1 expression was markedly
decreased in the H460 cells after RP11-713B9.1 siRNA transfection.
Quantification analysis showed that RP11-713B9.1 expression level
was nearly knocked down 90% in RP11-713B9.1 siRNA group and TSLC1
expression level was also down-regulated (P<0.05, Fig. 3C). It indicated that TSLC1
expression may be modulated by RP11-713B9.1 since TSLC1 expression
was co-regulated together with the RP11-713B9.1 in NSCLC cells.
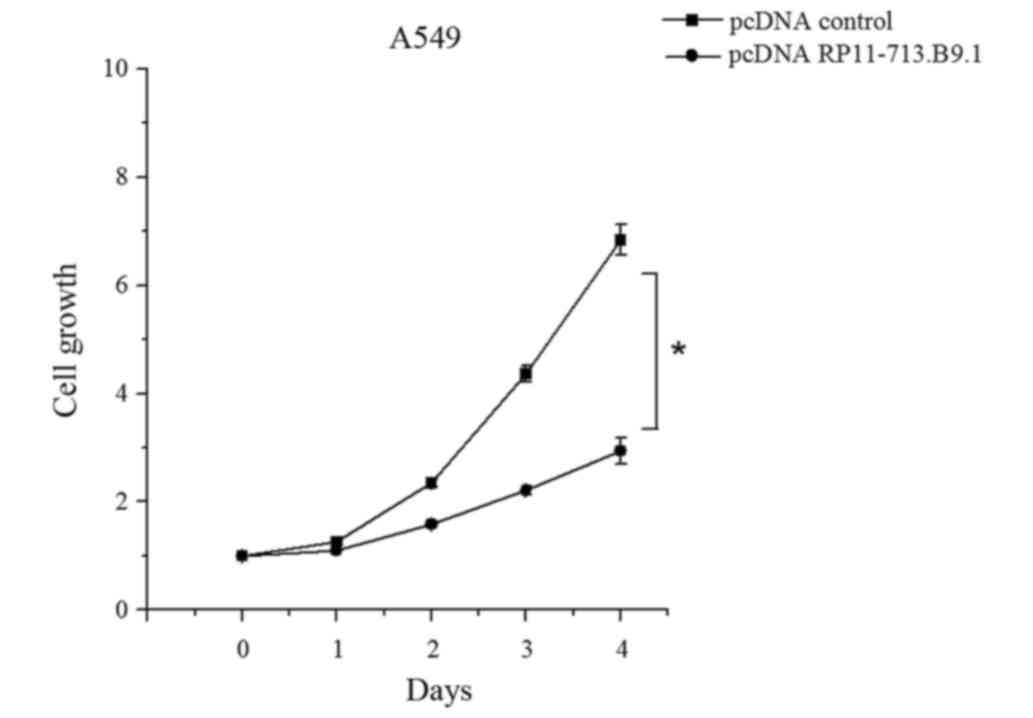
A549 cell viability was inhibited by
RP11-713B9.1 overexpression
To further analyze the role of RP11-713B9.1 in A549
cells, functional assays were conducted by transfecting pcDNA
RP11-713B9.1 and pcDNA control. The results showed that the A549
cells grew evidently slower in the pcDNA RP11-713B9.1 group than
those in pcDNA control group (P<0.05, Fig. 4).
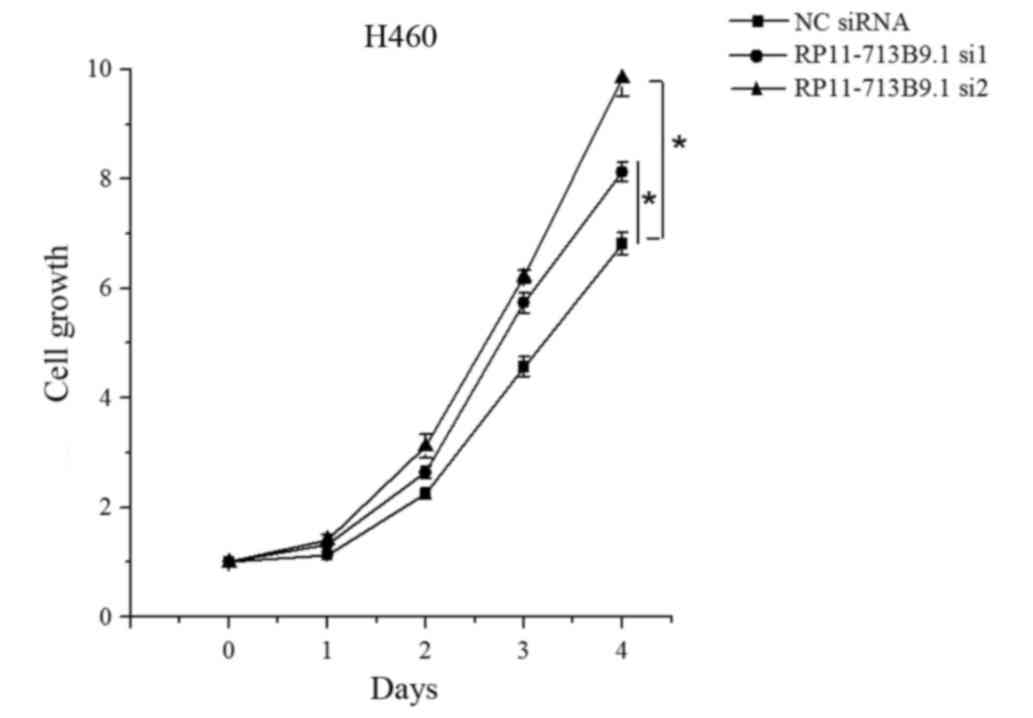
H460 cells viability was enhanced by
RP11-713B9.1 knockdown
To further analyze the role of RP11-713B9.1 in H460
cells, RP11-713B9.1 knockdown experiments were conducted by
transfecting RP11-713B9.1 siRNA and NC siRNA groups. The H460 cells
viability was markedly promoted in the RP11-713B9.1 siRNA group
when compared with the NC siRNA group (P<0.05, Fig. 5).
Discussion
LncRNAs have been shown to exert a crucial role in
tumorigenesis and contributes to a diverse of biological functions
in human cancers (7–10). In recent years, aberrantly
expressed lncRNAs have been reported to be implicated in regulating
cell viability, migration and invasion (11–13).
However, little is known about the expression and function of
specified lncRNA in NSCLC cells. Furthermore, the biological roles
of many lncRNAs in NSCLC initiation and progression are far from
being well elucidated. Due to the fact that tumor metastasis is the
major contributor to NSCLC related death, seeking a specific lncRNA
involved in metastasis may provide novel opportunities to identify
effective therapy against NSCLC.
Recent studies have identified that TSLC1 as a new
tumor suppressor is often abnormally expressed in multiple cancers,
including NSCLC (19–26). Intriguingly, RP11-713B9.1 is the
antisense transcript of TSLC1. Despite RP11-713B9.1 as a novel
lncRNA, its expression profile and functional role in NSCLC has not
yet investigated, which generated our interest.
In our study, we observed that the expression of
RP11-713B9.1 and TSLC1 were down-regulated in NSCLC tumors compared
with matched normal tissues. Numerous protein-coding genes in the
human genome have their antisense transcript partners, most of
which are noncoding, just like RP11-713B9.1 and TSLC1, as a type of
sense-antisense RNA duplex formation. Generally, the antisense
lncRNA can modulate the sense mRNA either in a discordant or a
concordant manner (31–33). For instance, HIF1α together with
its antisense partner aHIF has been recognized as a concordant
sense-antisense RNA couple (34).
Hence, we explored the correlation between RP11-713B9.1 and TSLC1
and it was demonstrated that they were positively correlated, which
indicated TSLC1 may be regulated by RP11-713B9.1. Recent studies
have reported some new genetic mutations that activate oncogenes,
such as BRAF (27) and others that
contribute to the loss of tumor suppressor function, such as CYLD,
APC in NSCLC (28,29). We performed the further work to
investigate the correlation of RP11-713B9.1 with BRAF, APC and
CYLD. Impressively, we found that the expression of RP11-713B9.1
was positively correlated with tumor suppressors APC and CYLD, and
negatively correlated with the oncogene BRAF.
Moreover, we discovered that up-regulating the
RP11-713B9.1 expression in low-expressing A549 cells remarkably
inhibited the viability of NSCLC cells, while down-regulating the
RP11-713B9.1 expression in high-expressing H460 cells showed the
opposite effect. Collectively, we probably firstly found that
RP11-713B9.1 was significantly down-regulated in NSCLC tissues when
compared with nontumorous tissues. In addition, we also observed
that RP11-713B9.1 expression was positively associated with tumor
suppressors TSLC1 and CYLD, negatively correlative with oncogene
BRAF. More important, loss-of and gain-of-function experiments were
conducted through MTT assay, we determined that RP11-713B9.1
expression had evident effect on cell viability.
To the best of our knowledge, our data may provide
the first evidence for clarifying the functional role of
RP11-713B9.1 in NSCLC. These novel findings inspire us to speculate
that the RP11-713B9.1 may serve as a potential biomarker and
therapeutic target for NSCLC treatment in the future. Further
analyses of additional clinical samples of NSCLC patients are
needed as only a fraction of patient samples were analyzed in our
study because of the difficulty in collection. In addition, more
functional experiments like cell proliferation, migration, invasion
and apoptosis will be performed to further investigate the
functional roles of RP11-713B9.1 in NSCLC as we are aware that the
functional experiments in our current study may not be sufficient
due to the limited time. Analyzing and exploring the molecular
mechanism of RP11-713B9.1 in NSCLC will be a priority for all on
the basis of this paper. Above mentioned future research directions
will underscore the significance of this study in near future,
which will provide new insight into the effective therapy for
NSCLC.
In summary, we discovered that RP11-713B9.1 and
TSLC1 were down-regulated in NSCLC tissues. Meanwhile, we found
that RP11-713B9.1 was positively correlated with tumor suppressors
TSLC1, CYLD and APC, negatively correlated with oncogene BRAF gene.
Further MTT assay showed that RP11-713B9.1 overexpression inhibited
cell viability while knockdown showed the opposite effect. The data
provides a new insight into the expression and functional role of
RP11-713B9.1 in NSCLC. Taken together, we propose that the
RP11-713B9.1 may play a crucial role in the occurrence and
progression of NSCLC and has the potential to be a promising
diagnostic biomarker, novel prognostic factor and therapeutic
target for NSCLC in future treatment.
Acknowledgements
The authors would like to thank their colleagues for
their insight and technical support.
References
|
1
|
Xu YJ, Du Y and Fan Y: Long noncoding RNAs
in lung cancer: What we know in 2015. Clin Transl Oncol.
18:660–665. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang HH, Pang M, Dong W, Xin JX, Li YJ,
Zhang ZC, Yu L, Wang PY, Li BS and Xie SY: miR-511 induces the
apoptosis of radioresistant lung adenocarcinoma cells by triggering
BAX. Oncol Rep. 31:1473–1479. 2014.PubMed/NCBI
|
|
4
|
Ma Y, Li X, Cheng S, Wei W and Li Y:
MicroRNA-106a confers cisplatin resistance in non-small cell lung
cancer A549 cells by targeting adenosine triphosphatase-binding
cassette A1. Mol Med Rep. 11:625–32. 2015.PubMed/NCBI
|
|
5
|
Saito M, Shiraishi K, Matsumoto K,
Schetter AJ, Ogata-Kawata H, Tsuchiya N, Kunitoh H, Nokihara H,
Watanabe S, Tsuta K, et al: A three-microRNA signature predicts
responses to platinum-based doublet chemotherapy in patients with
lung adenocarcinoma. Clin Cancer Res. 20:4784–4793. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rinn JL and Chang HY: Genome regulation by
long noncoding RNAs. Annu Rev Biochem. 81:145–166. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Guttman M, Amit I, Garber M, French C, Lin
MF, Feldser D, Huarte M, Zuk O, Carey BW and Cassady JP: Chromatin
signature reveals over a thousand highly conserved large non-coding
RNAs in mammals. Nature. 458:223–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Huarte M and Rinn JL: Large non-coding
RNAs: Missing links in cancer? Hum Mol Genet. 19:R152–R161. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Spizzo R, Almeida MI, Colombatti A and
Calin GA: Long non-coding RNAs and cancer: A new frontier of
translational research? Oncogene. 31:4577–4587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tsai MC, Spitale RC and Chang HY: Long
intergenic noncoding RNAs: New links in cancer progression. Cancer
Res. 71:3–7. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang F, Yuan JH, Wang SB, Yang F, Yuan SX,
Ye C, Yang N, Zhou WP, Li WL, Li W and Sun SH: Oncofetal long
noncoding RNA PVT1 promotes proliferation and stem cell-like
property of hepatocellular carcinoma cells by stabilizing NOP2.
Hepatology. 60:1278–1290. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ,
Tao QF, Liu F, Pan W, Wang TT, Zhou CC, et al: A Long Noncoding RNA
Activated by TGF-β Promotes the Invasion-Metastasis Cascade in
Hepatocellular Carcinoma. Cancer Cell. 25:666–681. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhou X, Liu S, Cai G, Kong L, Zhang T, Ren
Y, Wu Y, Mei M, Zhang L and Wang X: Long non coding RNA MALAT1
promotes tumor growth and metastasis by inducing
epithelial-mesenchymal transition in oral squamous cell carcinoma.
Sci Rep. 5:159722015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sun M, Liu XH, Wang KM, Nie FQ, Kong R,
Yang JS, Xia R, Xu TP, Jin FY, Liu ZJ, et al: Downregulation of
BRAF activated non-coding RNA is associated with poor prognosis for
non-small cell lung cancer and promotes metastasis by affecting
epithelial-mesenchymal transition. Mol Cancer. 13:682014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gutschner T, Hämmerle M, Eissmann M, Hsu
J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, et al:
The noncoding RNA MALAT1 is a critical regulator of the metastasis
phenotype of lung cancer cells. Cancer Res. 73:1180–1189. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nowacki S, Skowron M, Oberthuer A, Fagin
A, Voth H, Brors B, Westermann F, Eggert A, Hero B, Berthold F and
Fischer M: Expression of the tumour suppressor gene CADM1 is
associated with favourable outcome and inhibits cell survival in
neuroblastoma. Oncogene. 27:3329–3338. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fischer M, Oberthuer A, Brors B, Kahlert
Y, Skowron M, Voth H, Westermann F, Eggert A, Hero B, Berthold F
and Fischer M: Differential expression of neuronal genes defines
subtypes of disseminated neuroblastoma with favorable and
unfavorable outcome. Clin Cancer Res. 12:5118–5128. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Watabe K, Ito A, Koma YI and Kitamura Y:
IGSF4: A new intercellular adhesion molecule that is called by
three names, TSLC1, SgIGSF and SynCAM, by virtue of its diverse
function. Histol Histopathol. 18:1321–1329. 2003.PubMed/NCBI
|
|
19
|
Lung HL, Cheung AK, Xie D, Cheng Y, Kwong
FM, Murakami Y, Guan XY, Sham JS, Chua D, Protopopov AI, et al:
TSLC1 is a tumor suppressor gene associated with metastasis in
nasopharyngeal carcinoma. Cancer Res. 66:9385–9392. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Heller G, Geradts J, Ziegler B, Newsham I,
Filipits M, Markis-Ritzinger EM, Kandioler D, Berger W, Stiglbauer
W, Depisch D, et al: Downregulation of TSLC1 and DAL-1 expression
occurs frequently in breast cancer. Breast Cancer Res Treat.
103:283–291. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Honda T, Tamura G, Waki T, Jin Z, Sato K,
Motoyama T, Kawata S, Kimura W, Nishizuka S and Murakami Y:
Hypermethylation of the TSLC1 gene promoter in primary gastric
cancers and gastric cancer cell lines. Jpn J Cancer Res.
93:857–860. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jansen M, Fukushima N, Rosty C, Walter K,
Altink R, Heek TV, Heek TV, Hruban R, Offerhaus JG and Goggins M:
Aberrant methylation of the 5-CpG island of TSLC1 is common in
pancreatic ductal adenocarcinoma and is first manifest in
high-grade PanlNs. Cancer Biol Ther. 1:293–296. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen K, Wang G, Peng L, Liu S, Fu X, Zhou
Y, Yu H, Li A, Li J, Zhang S, et al: CADM1/TSLC1 inactivation by
promoter hypermethylation is a frequent event in colorectal
carcinogenesis and correlates with late stages of the disease. Int
J Cancer. 128:266–273. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang YX, Yang AH, Yang ZJ, Wang ZR and Xia
XH: Involvement of tumor suppressor in lung cancer 1 gene
expression in cervical carcinogenesis. Int J Gynecol Cancer.
16:1868–1872. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Williams YN, Masuda M, Sakurai-Yageta M,
Maruyama T, Shibuya M and Murakami Y: Cell adhesion and prostate
tumor-suppressor activity of TSLL2/IGSF4C, an immunoglobulin
superfamily molecule homologous to TSLC1/IGSF4. Oncogene.
25:1446–1453. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kuramochi M, Fukuhara H, Nobukuni T, Kanbe
T, Maruyama T, Ghosh HP, Pletcher M, Isomura M, Onizuka M, Kitamura
T, et al: TSLC1 is a tumor suppressor gene in human non-small cell
lung cancer. Nat Genet. 27:427–430. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nguyen-Ngoc T, Bouchaab H, Adjei AA and
Peters S: BRAF alterations as therapeutic targets in non-small-cell
lung cancer. J Thorac Oncol. 10:1396–1403. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Deng LL, Shao YX, Lv HF, Deng HB and Lv
FZ: Over-expressing CYLD augments antitumor activity of TRAIL by
inhibiting the NF-κB survival signaling in lung cancer cells.
Neoplasma. 59:18–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Guo S, Tan L, Pu WL, Wu J, Xu K, Wu J, Li
Q, Ma Y, Xu J, Jin L and Wang J: Quantitative assessment of the
diagnostic role of APC promoter methylation in non-small cell lung
cancer. Clin Epigenetics. 6:52014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Qin X, Yao J, Geng P, Fu X, Xue J and
Zhang Z: LncRNA TSLC1-AS1 is a novel tumor suppressor of glioma.
Int J Clin Exp Pathol. 7:3065–3072. 2014.PubMed/NCBI
|
|
31
|
Wahlestedt C: Natural antisense and
noncoding RNA transcripts as potential drug targets. Drug Discov
Today. 11:503–508. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Faghihi MA and Wahlestedt C: Regulatory
roles of natural antisense transcripts. Nat Rev Mol Cell Biol.
1:637–643. 2009. View
Article : Google Scholar
|
|
33
|
Yelin R, Dahary D, Sorek R, Levanon EY,
Goldstein O, Shoshan A, Diber A, Biton S, Tamir Y, Khosravi R, et
al: Widespread occurrence of antisense transcription in the human
genome. Nat Biotechnol. 21:379–386. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Thrash-Bingham CA and Tartof KD: aHIF: A
natural antisense transcript overexpressed in human renal cancer
and during hypoxia. J Natl Cancer Inst. 91:143–151. 1999.
View Article : Google Scholar : PubMed/NCBI
|