Introduction
Mesenchymal stem cells (MSCs) possess the potential
for self-renewal and may differentiate into multiple connective
tissue cell types, including chondrogenic, adipogenic and
osteogenic lineages (1–3). Tissue engineering using suitable MSCs
has been considered to be a novel treatment for bone deficiency.
Human amnion-derived mesenchymal stem cells (HAMSCs), obtained from
abundant human amniotic membrane, are associated with fewer ethical
issues and low anti-inflammatory properties (4). Previous studies (5,6) have
suggested that HAMSCs are capable of promoting proliferation and
osteogenic differentiation of human bone marrow mesenchymal stem
cells (HBMSCs). However, the complex molecular mechanisms of
HAMSC-derived osteogenic differentiation remain largely
unknown.
During the past decade, the ENCODE project and
large-scale sequencing efforts have revealed that long non-coding
RNAs (lncRNAs) belong to a novel heterogeneous class of ncRNAs that
includes thousands of different species (7,8).
lncRNAs have been proposed to serve crucial roles in a wide range
of biological pathways and cellular processes, including the
reprogramming of stem cell pluripotency (9), inactivation of chromosomes (10), modulation of invasion and apoptosis
(11) and regulation at
transcriptional and post transcriptional levels (12). A previous study (13) demonstrated that the
lncRNA-differentiation antagonizing non-protein coding RNA (DANCR),
an important regulator of osteogenic differentiation, is associated
with the inhibition of runt-related transcription factor 2 (RUNX2)
expression and subsequent osteogenic differentiation in human
osteoblastic cell. Therefore, the potential role of lncRNAs in
HAMSC-derived osteogenic differentiation was investigated.
In the present study, the differential expression
profiles of mRNAs and lncRNAs of HBMSCs co-cultured with or without
HAMSCs during osteogenic differentiation were obtained. RNA
sequencing (RNA-seq) data was validated using the reverse
transcription-quantitative polymerase chain reaction (RT-qPCR).
Based on these results, the present study investigated whether
HAMSCs downregulated the DANCR expression level in HBMSCs, and
whether the effect of HAMSCs on the promotion of RUNX2 expression
in HBMSCs was inhibited by DANCR overexpression. The observations
demonstrated that lncRNAs may possibly provide novel insights into
the mechanisms by which HAMSCs regulate osteogenic
differentiation.
Materials and methods
Cell culture
The HBMSC cell line PTA-1058 was obtained from the
American Type Culture Collection (Manassas, VA, USA). The HAMSCs
were collected from abundant amniotic membrane samples using the
pancreatin/collagenase digestion method as previously described
(14). The two cell lines were
expanded in Dulbecco's modified Eagle's medium (HyClone, Logan, UT,
USA) supplemented with 10%fetal bovine serum (FBS; Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA), 100 U/l penicillin and
100 mg/l streptomycin (both Gibco; Thermo Fisher Scientific, Inc.)
in a humidified atmosphere of 5% CO2 at 37°C. The
confluent cells were transferred to next passage using 0.25%
trypsin for up to five passages and culture medium was changed
every 3 days. The experiments were approved by the Ethics Committee
of Nanjing Medical University (Nanjing, China). Prior informed
consent was obtained from all participants enrolled in the present
study.
Preparation of the co-culture
system
The HBMSCs were seeded at an initial cell density of
5×104 cells/cm2 in 6-well culture plates (EMD
Millipore, Billerica, MA, USA). Transwells (6-Well Millicell
Hanging Cell Culture Inserts, 0.4 µm; EMD Millipore) were placed in
other 6-well culture plates and HAMSCs were seeded at
1.5×105 cells/cm2 in the transwells.
Following the attachment of the cells, transwells containing HAMSCs
were moved into the corresponding wells of the 6-well culture plate
containing HBMSCs to create the HAMSC/HBMSC co-culture system.
HBMSCs in wells without transwells were designated as the control
groups, while HBMSCs with transwells were served as the experiment
groups.
Proliferation assay
The proliferation assay was measured at day 3, 5 and
7 by flow cytometry. The transwells containing HAMSCs were moved
into the corresponding wells containing HBMSCs and then the two
cell lines were treated with serum-free medium for 24 h, following
which the medium was replaced with culture medium containing 10%
FBS. HBMSCs were harvested at day 3, 5 and 7 and fixed with 75%
ice-cold ethanol as described previously (15). DNA content was measured by a
FACScan flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA)
and the cell cycle fractions (G0, G1, S, and G2 M phases) were
processed using CellQuest Pro software (BD Biosciences). Data were
analyzed using Modfit LT 3.2 (Verity Software House, Topsham, ME,
USA).
In vitro osteogenic
differentiation
The transwells containing HAMSCs were moved into the
corresponding wells containing HBMSCs and then the regular medium
in all cultures was replaced with osteogenic medium containing 10
mM β-glycerophosphate (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany), 100 nM ascorbic acid and 100 nM dexamethasone (both
Sigma-Aldrich; Merck KGaA). HBMSCs co-cultured with HAMSCs were
subjected to alkaline phosphatase (ALP) activity assays and
alizarin red staining. ALP activity assay was performed using an
ALP assay kit (Jiancheng Corp, Nanjing, China) according to the
manufacturer's protocols. Alizarin red staining was performed using
40 mM alizarin red S (pH 4.4) for 10 min at room temperature.
Following rinsing with PBS, mineralized nodules were visualized
using an inverted microscope (Carl Zeiss AG, Oberkochen, Germany)
and 10 images were captured for each group.
Plasmid construction and cell
transfection
DANCR overexpression was achieved through
pcDNA3.1-DANCR (Guangzhou RiboBio Co., Ltd., Guangzhou, China)
transfection using lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), with an empty pCDNA3.1 vector used as a control.
Plasmid vectors (pcDNA3.1-DANCR and pcDNA3.1) for transfection were
extracted using Midiprep kits (Qiagen GmbH, Hilden, Germany), and
respectively transfected into HBMSCs. Fusion and transfection of
HBMSCs were performed when the cells were cultivated on six-well
plates by lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocols.
RNA isolation and RT-qPCR
Total cellular RNA was isolated from HBMSCs in the
control and treatment groups using TRIzol, according to the
protocols recommended by the manufacturer. The RNA was
reverse-transcribed into cDNA by using a PrimeScript RT Master Mix
kit. Real-time reverse transcription-PCR was performed with a SYBR
Green PCR kit (Toyobo, Osaka, Japan) and ABI 7300 Real-Time PCR
System (Applied Biosystems; Thermo Fisher Scientific, Inc.).
Amplification was performed as follows: Incubation at 95°C for 30
sec, followed by 40 cycles of denaturation at 95°C for 5 sec and
subsequent annealing and extension at 60°C for 34 sec. For each
sample, GAPDH expression was analyzed to normalize target gene
expression. Relative gene expression was calculated using the
2−ΔΔCq method (16).
Each sample was analyzed in triplicate. Primer sequences used in
this study are provided in Table
I.
 | Table I.Primers used in the reverse
transcription-quantitative polymerase chain reaction. |
Table I.
Primers used in the reverse
transcription-quantitative polymerase chain reaction.
| Gene | Sense primer
(5′-3′) | Anti-sense primer
(5′-3′) |
|---|
| SCN7A |
TGCTGGTCAGTGTCCTGAAG |
ATAGGCCATGGCAAGTATGC |
| DANCR |
GCCACTATGTAGCGGGTTTC |
ACCTGCGCTAAGAACTGAGG |
| TSIX |
GTGGTGGCAGGCAACTTAAT |
GCACATTCAGGCTCTCAACA |
| SSTR5-AS1 |
AGCCCCTCATCTTGGCTAAT |
TTTCCCTCTGAGCAGCAAAT |
| XIST |
TTGCATATGTGGGCAAGTGT |
GCTCTTGAGGCTTTTGTTGG |
|
ENST00000433576.1 |
ACACTGTGGGACTTCTTAGCC |
TTCTGTGGTCGTGGTCTTTG |
| LINGO3 |
CGCTGCAATCTCACATCACT |
GCACCAGGTCTCTGAAGGAG |
| MPEG1 |
CCCCAACATGCTACCTGACT |
CTCCTCGTGGATCTGGGATA |
| SLPI |
CTGTGGAAGGCTCTGGAAAG |
AAAGGACCTGGACCACACAG |
| NPPB |
TTCTTGCATCTGGCTTTCCT |
TGTGGAATCAGAAGCAGGTG |
| RUNX2 |
GGTTCCAGCAGGTAGCTGAG |
GCCTACAAAGGTGGGTTTGA |
| MRC1 |
GGCACTTGTGGAGAAGAAGC |
GTGGCCTTGGTGATCTTGTT |
| GAPDH |
GGGCTGCTTTTAACTCTGGT |
GCAGGTTTTTCTAGACGG |
RNA-seq
HBMSCs co-cultured without HAMSCs at day 14 were
regarded as control groups and HBMSCs co-cultured with HAMSCs at
day 14 were regarded as experimental groups. Total RNA was
sequenced using an Illumina HiSeq 2500 at Guangzhou RiboBio Co.,
Ltd. Reads were aligned to the human transcriptome. RNA-seq data
was aligned to the Ensembl v73 transcript annotations using RSEM
v1.1.21 and Bowtie v0.12.7, as previously described (17). The differentially expressed genes
were selected if Benjamini-Hochberg corrected P-values were
<0.05 and if the log2 (fold change) were >2.0 or <-2.0.
Tree visualization of the data was performed with Java Treeview 3.0
software (Stanford University School of Medicine, Stanford, CA,
USA).
Kyoto encyclopedia of genes and
genomes (KEGG) and gene ontology (GO) analysis
Pathway analysis for the differentially expressed
mRNAs was performed based on the most recent KEGG database
(http://www.genome.jp/). This analysis allowed the
investigation of the biological pathways for which a significant
enrichment of differentially expressed mRNAs existed. GO
(http://www.geneontology.org) was also
derived, which provided three structured networks that described
gene product attributes. The-log10 (P-value) demonstrated the
significance of GO Term enrichment in the differentially expressed
mRNA list. All other bioinformatic analysis was performed using
glbase (18).
Statistical analysis
All assays were expressed as the mean ± standard
deviation from at least three separate experiments using SPSS 13.0
(SPSS Inc., Chicago, IL, USA). Gene expression levels in osteogenic
differentiation were compared between control and treatment groups.
The Student's t-test was used to compare data between the two
groups. P<0.05 was considered to indicate a statistically
significant difference.
Results
Expression profiles of mRNAs and
lncRNAs in HBMSCs co-cultured with HAMSCs
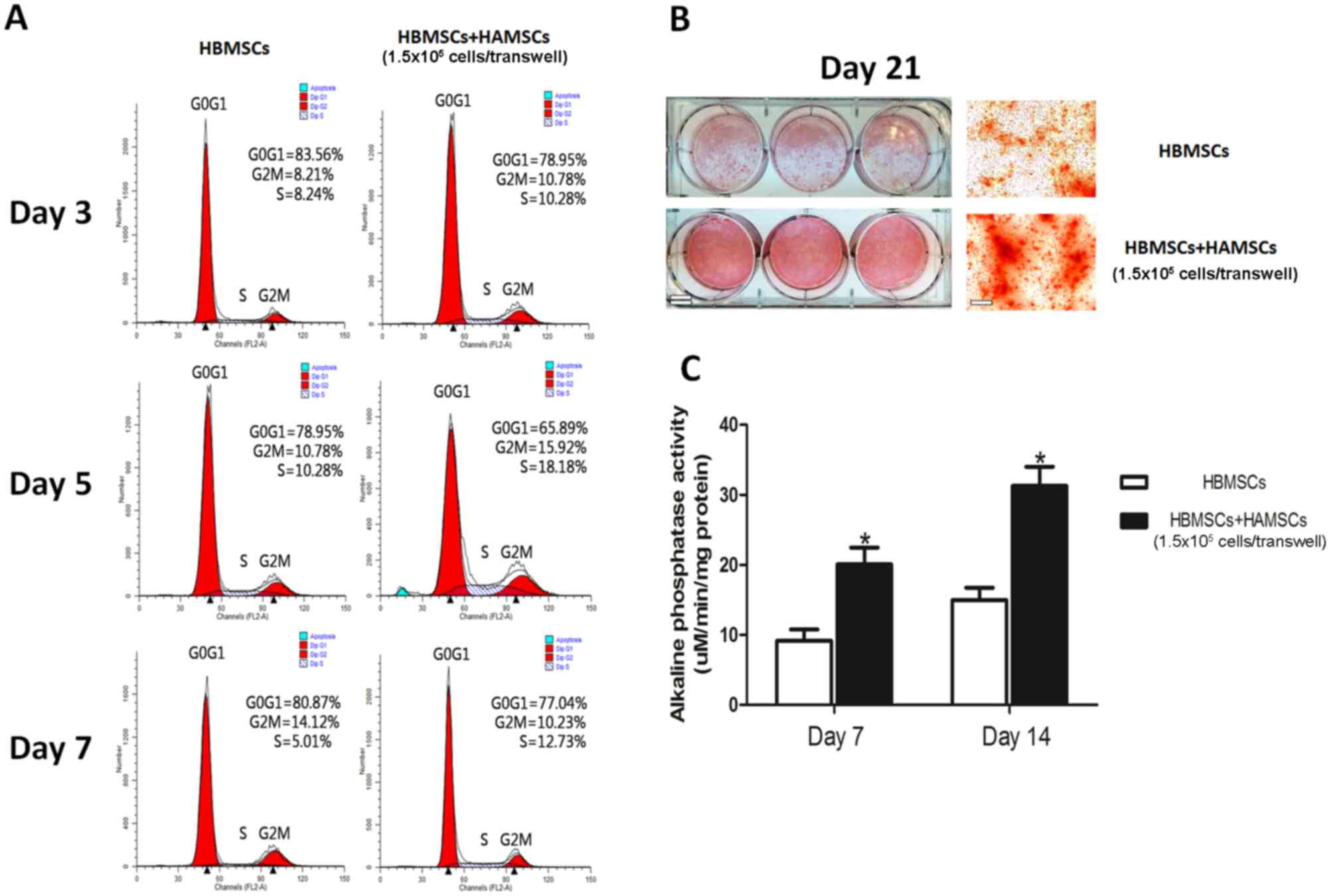
The effects of HAMSCs on HBMSCs are provided in
Fig. 1. Flow cytometric analyses,
ALP activity assays and alizarin red staining demonstrated the
potential of HAMSCs in promoting proliferation and osteogenic
differentiation of HBMSC. The expression profiles of mRNAs and
lncRNAs in HBMSCs co-cultured with or without HAMSCs were
identified for 14 days following osteogenic induction using
RNA-seq. Expression profiles of mRNAs and lncRNAs were
significantly altered between HBMSCs co-cultured with and without
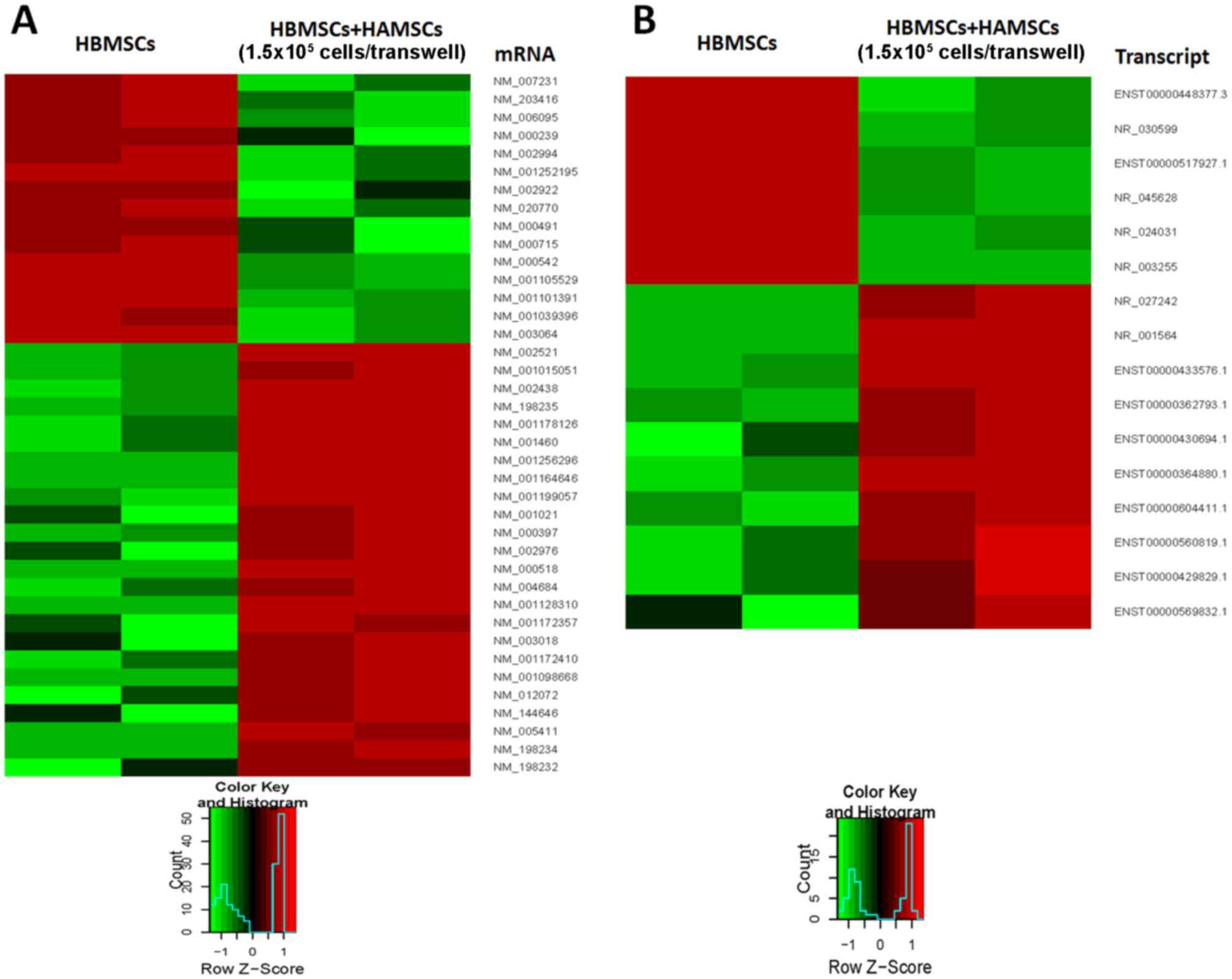
HAMSCs. A total of 415 differentially expressed mRNAs were
identified [log2 (fold change) >2.0 or <-2.0, P<0.05].
Among these, 156 mRNAs were identified to be downregulated more
than 2-fold in the control groups compared with experiment groups,
while 259 mRNAs were upregulated more than 2-fold (P<0.05).
Meanwhile, 339 differentially expressed lncRNAs were identified
[log2 (fold change) >2.0 or <-2.0, P<0.05], consisting of
131 downregulated and 208 upregulated lncRNAs (P<0.05). When the
cut-off was set at 6-fold, 17 mRNAs were downregulated and 24 were
upregulated, and 6 lncRNAs were downregulated and 10 were
upregulated (P<0.05; Fig.
2).
RT-qPCR analysis of lncRNA and mRNA
expression
According to gene locus and fold difference, a
number of candidate lncRNAs and mRNAs for validation of the RNA-seq
data were investigated. Therefore, six lncRNAs (sodium
voltage-gated channel α subunit 7, DANCR, TSIX, SSTR5-AS1, X
inactive specific transcript and ENST00000433576.1) and six mRNAs
(leucine rich repeat and Ig domain containing 3, macrophage
expressed 1, SLPI, natriuretic peptide B, RUNX2 and mannose
receptor C-type 1) were further examined using RT-qPCR. The results
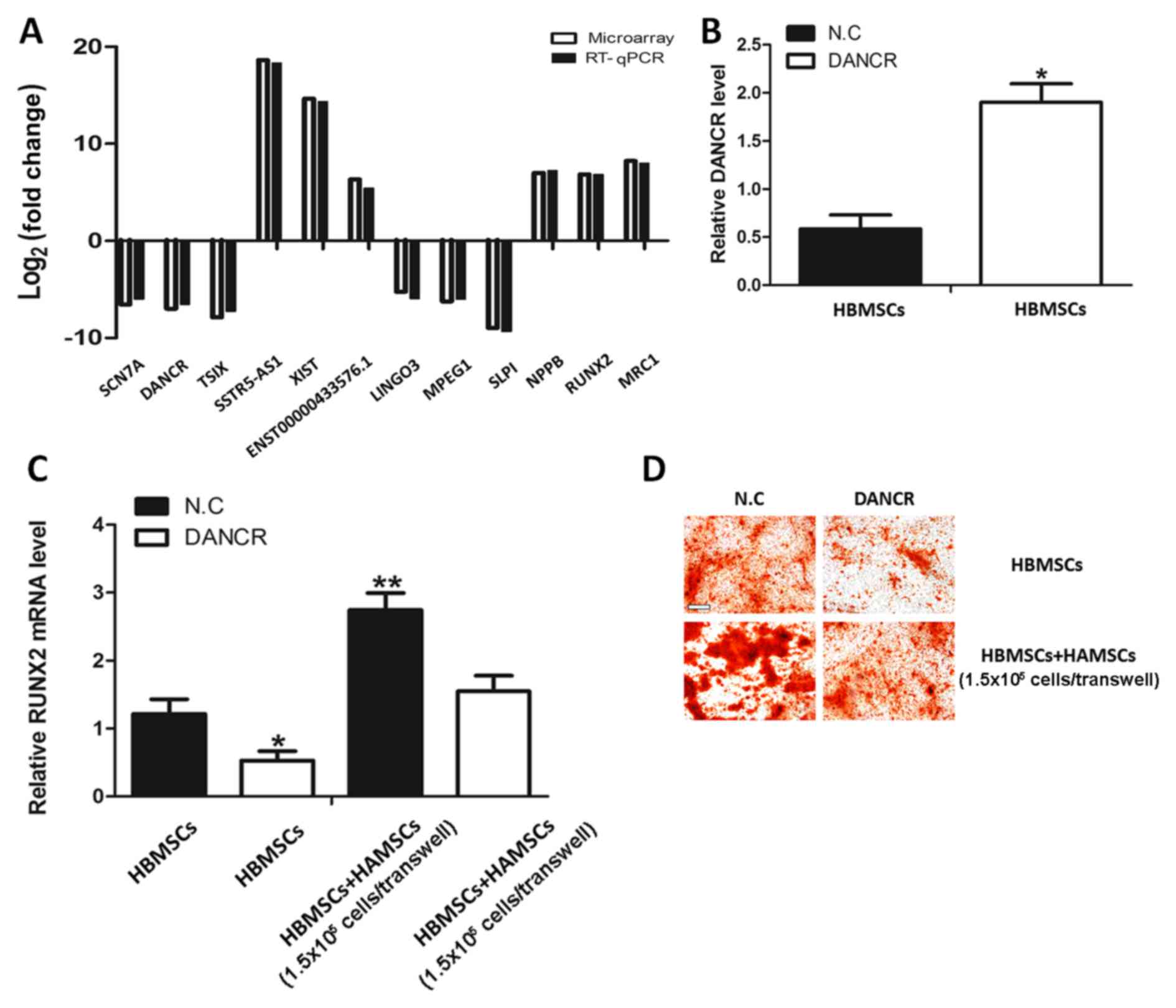
of the RT-qPCR were consistent with the RNA-seq (Fig. 3A).
The upregulation of DANCR inhibits
RUNX2 expression and osteogenic differentiation in HBMSCs
co-cultured with HAMSCs
Previous studies have indicated that DANCR is an
essential mediator of the osteogenic differentiation of mesenchymal
stem cells and that overexpression of DANCR results in the
inhibition of RUNX2 expression and subsequent osteogenic
differentiation (13). To
investigate whether HAMSCs regulates osteogenic differentiation in
HBMSCs by influencing DANCR, the DANCR levels in HBMSCs co-cultured
with HAMSCs were first analyzed. It was identified that the DANCR
levels were decreased and the RUNX2 expression was increased
(Fig. 3A). To further investigate
the biological role of DANCR in the regulation of HAMSCs-derived
osteogenic differentiation, HBMSCs treated with pcDNA-DANCR were
analyzed. Fig. 3B demonstrates
that the pcDNA-DANCR increased endogenous DANCR expression in
HBMSCs. Overexpression of DANCR significantly inhibited the effects
of HAMSCs in promoting RUNX2 expression and osteogenic
differentiation of HBMSC (Fig. 3C and
D). These observations suggested that the HAMSCs are likely to
regulate differentiation process in HBMSCs by downregulating the
DANCR.
GO and pathway analysis of mRNAs
differentially expressed between HBMSCs co-cultured with and
without HAMSCs
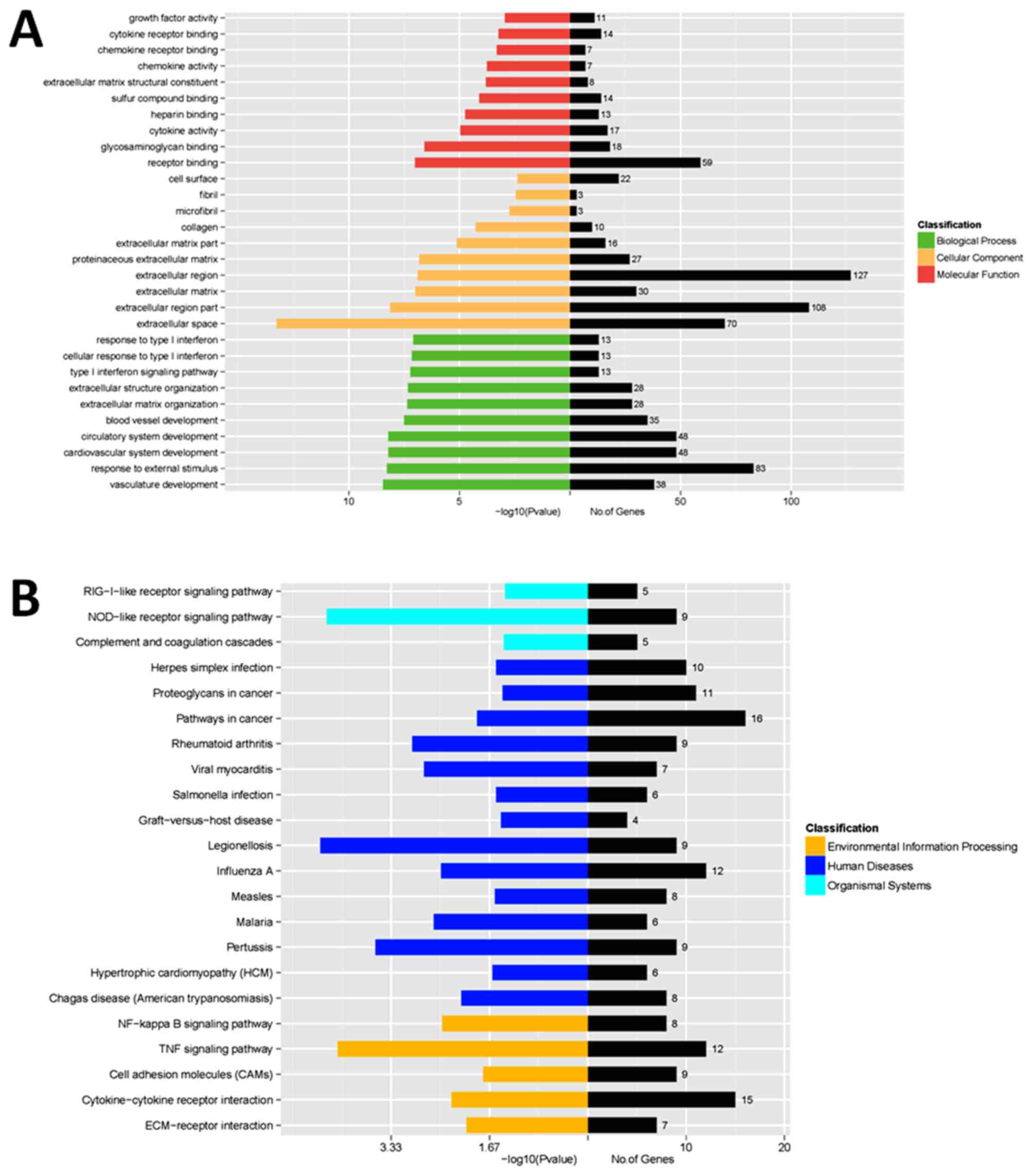
GO analysis demonstrated that the most significant
biological processes consisted of growth factor or chemokine
activity, cytokine or chemokine receptor binding, extracellular
region, extracellular matrix, response to type I interferon and
other functions. Pathway analysis demonstrated that the most
significant pathways consisted of retinoic acid-induced protein
I-like receptor signaling pathway, nucleotide oligomerization
domain-like receptor signaling pathway, nuclear factor kB signaling
pathway and pathways in cancer (Fig.
4). These results aid the improved identification of the
mechanisms of HAMSC-derived osteogenic differentiation.
Discussion
Stem cells are present in a variety of mesenchymal
tissues. They possess self-renewable capacities and
multi-directional differentiation potentials (19,20).
Given appropriate culture conditions, mesenchymal stem cells (MSCs)
are capable of differentiating into adipocytes (21), chondrocytes (22,23),
endothelial cells (24),
osteocytes (25) and
cardiomyocytes (26). Recently,
tissue engineering, using suitable MSCs that mimic the natural
healing process, has demonstrated great potential in treating bone
deficiency. Human amniotic membrane, composed of a single layer of
epithelial cells, an avascular collagenous stroma and underlying
fibroblasts, is a readily available and highly abundant tissue
(27). HAMSCs, obtained from
discarded human amniotic membrane, possess low anti-inflammatory
properties and fewer ethical concerns compared with other sources
of MSCs (4). Previous studies have
demonstrated that although HAMSC osteogenesis was lower compared
with other MSCs, they are capable of driving proliferation and
osteogenic differentiation of HBMSCs (5). However, the regulatory mechanisms of
MSC-derived osteogenic differentiation remains to be
elucidated.
lncRNAs are evolutionarily conserved ncRNAs that
lack protein encoding capacity and are >200 nucleotides in
length. They are abundantly encoded in mammalian genomes, numbering
in the tens of thousands. The ratio of lncRNAs in total ncRNAs is
>80% (28,29). Recently, numerous lncRNAs have been
characterized and a number of pieces of evidence suggest that their
transcriptional activity may serve a major biological role in cell
differentiation and human diseases (30,31).
Among them, certain lncRNAs express differently during lineage
commitment and cell development (32). Previous studies suggested that
lncRNAs serve a key role in the circuitry controlling cell state
(33) and also modulate
pluripotency in mouse embryonic stem cells (34). The lncRNA DANCR is highly expressed
in the epidermis and its loss eradicates the normal blockade of
differentiation in the progenitor-containing compartment; it is
fundamental to maintaining the undifferentiated cell state within
the epidermis. Enhancer of zeste homolog 2 (EZH2), an important
epigenetic regulatory factor and highly expressed in MSCs, mediates
modifications in histone methylation resulting in the regulation of
the differentiation of MSCs into different cell lineages, including
hepatocytes, neurons and osteoblasts (35,36).
A previous study demonstrated that DANCR is an important regulator
of osteogenic differentiation in human fetal osteoblastic cells. By
associating with EZH2, DANCR inhibits RUNX2 expression and
subsequent osteogenic differentiation (13). However, the role of lncRNAs in the
complex mechanisms of MSC-derived osteogenic differentiation has
not been studied.
Based on the observation that HAMSCs may promote
proliferation and osteogenic differentiation in HBMSCs by providing
a preferential environment, the present study first demonstrated
lncRNA and mRNA expression profiles, during the osteogenic
differentiation of HBMSCs co-cultured with or without HAMSCs using
RNA-seq. The RNA-seq data was validated by RT-qPCR. Bioinformatic
analyses were subsequently applied to further study these
differentially expressed lncRNAs and mRNAs, including GO and
pathway analysis.
It was identified that during osteogenic
differentiation, the DANCR lncRNA level was significantly decreased
and the RUNX2 mRNA level was significantly increased in HBMSCs
co-cultured with HAMSCs. Based on these results, the biological
role of DANCR in the mechanisms of HAMSC-derived osteogenic
differentiation was further investigated. It was identified that
the overexpression of DANCR in HBMSCs neutralized the positive
effects of HAMSCs on the promotion of RUNX2 expression and
osteogenic differentiation. These results suggested that HAMSCs are
likely to exercise their function by downregulating DANCR
expression and upregulating RUNX2 expression.
In conclusion, the present study revealed, for first
time to the authors' knowledge, the roles of lncRNAs in the
mechanisms of osteogenic differentiation derived by HAMSCs. By
bioinformatics analysis, certain target genes were obtained that
correlated with the biological processes and signaling pathways of
the candidate lncRNAs. Furthermore, it was demonstrated that the
lncRNA DANCR is an important regulator in the course of osteogenic
differentiation and this may provide a guide for further
investigation into the complicated regulatory mechanisms of
HAMSC-derived osteogenic differentiation. Further studies in the
functional analysis of more lncRNAs involved this system may help
clarify the regulatory mechanisms underlying this process and
provide more conclusive evidence for HAMSCs in the field of tissue
engineering.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 81271109) and
Project Funded by the Priority Academic Program Development of
Jiangsu Higher Education Institutions (grant no. KYZZ15_0266).
References
|
1
|
Frith JE, Thomson B and Genever PG:
Dynamic three-dimensional culture methods enhance mesenchymal stem
cell properties and increase therapeutic potential. Tissue Eng Part
C Methods. 16:735–749. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang T and Xu Z: miR-27 promotes
osteoblast differentiation by modulating Wnt signaling. Biochem
Biophys Res Commun. 402:186–189. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kapinas K, Kessler C, Ricks T, Gronowicz G
and Delany AM: miR-29 modulates Wnt signaling in human osteoblasts
through a positive feedback loop. J Biol Chem. 285:25221–25231.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Leyva-Leyva M, Barrera L, López-Camarillo
C, Arriaga-Pizano L, Orozco-Hoyuela G, Carrillo-Casas EM,
Calderón-Pérez J, López-Díaz A, Hernandez-Aguilar F,
González-Ramírez R, et al: Characterization of mesenchymal stem
cell subpopulations from human amniotic membrane with dissimilar
osteoblastic potential. Stem Cells Dev. 22:1275–1287. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang Y, Yin Y, Jiang F and Chen N: Human
amnion mesenchymal stem cells promote proliferation and osteogenic
differentiation in human bone marrow mesenchymal stem cells. J Mol
Histol. 46:13–20. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang Y, Jiang F, Liang Y, Shen M and Chen
N: Human amnion-derived mesenchymal stem cells promote osteogenic
differentiation in human bone marrow mesenchymal stem cells by
influencing the ERK1/2 signaling pathway. Stem Cells Int.
2016:48510812016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Batista PJ and Chang HY: Long noncoding
RNAs: Cellular address codes in development and disease. Cell.
152:1298–1307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Djebali S, Davis CA, Merkel A, Dobin A,
Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F,
et al: Landscape of transcription in human cells. Nature.
489:101–108. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Thomson CS and Forman D: Cancer survival
in England and the influence of early diagnosis: What can we learn
from recent EUROCARE results? Br J Cancer. 101 Suppl 2:S102–S109.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lee JT and Bartolomei MS: X-inactivation,
imprinting, and long noncoding RNAs in health and disease. Cell.
152:1308–1323. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ørom UA and Shiekhattar R: Long noncoding
RNAs usher in a new era in the biology of enhancers. Cell.
154:1190–1193. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li X, Wu Z, Fu X and Han W: Long noncoding
RNAs: Insights from biological features and functions to diseases.
Med Res Rev. 33:517–553. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhu L and Xu PC: Downregulated LncRNA-ANCR
promotes osteoblast differentiation by targeting EZH2 and
regulating Runx2 expression. Biochem Biophys Res Commun.
432:612–617. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang D, Jiang M and Miao D: Transplanted
human amniotic membrane-derived mesenchymal stem cells ameliorate
carbon tetrachloride-induced liver cirrhosis in mouse. PLoS One.
6:e167892011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang Y, Yan M, Yu Y, Wu J, Yu J and Fan Z:
Estrogen deficiency inhibits the odonto/osteogenic differentiation
of dental pulp stem cells via activation of the NF-kB pathway. Cell
Tissue Res. 352:551–559. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pike KA, Hutchins AP, Vinette V, Théberge
JF, Sabbagh L, Tremblay ML and Miranda-Saavedra D: Protein tyrosine
phosphatase 1B is a regulator of the interleukin-10-induced
transcriptional program in macrophages. Sci Signal. 7:ra432014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hutchins AP, Jauch R, Dyla M and
Miranda-Saavedra D: glbase: A framework for combining, analyzing
and displaying heterogeneous genomic and high-throughput sequencing
data. Cell Regen (Lond). 3:12014.PubMed/NCBI
|
|
19
|
Bianco P and Robey PG: Stem cells in
tissue engineering. Nature. 414:118–121. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jones E and McGonagle D: Human bone marrow
mesenchymal stem cells in vivo. Rheumatology (Oxford). 47:126–131.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Karagianni M, Brinkmann I, Kinzebach S,
Grassl M, Weiss C, Bugert P and Bieback K: A comparative analysis
of the adipogenic potential in human mesenchymal stromal cells from
cord blood and other sources. Cytotherapy. 15:76–88. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Macchiarini P, Jungebluth P, Go T, Asnaghi
MA, Rees LE, Cogan TA, Dodson A, Martorell J, Bellini S, Parnigotto
PP, et al: Clinical transplantation of a tissue-engineered airway.
Lancet. 372:2023–2030. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jungebluth P, Alici E, Baiguera S, Le
Blanc K, Blomberg P, Bozóky B, Crowley C, Einarsson O, Grinnemo KH,
Gudbjartsson T, et al: Tracheobronchial transplantation with a
stem-cell-seeded bioartificial nanocomposite: A proof-of-concept
study. Lancet. 378:1997–2004. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Crisan M: Transition of mesenchymal
stem/stromal cells to endothelial cells. Stem Cell Res Ther.
4:952013. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Heino TJ and Hentunen TA: Differentiation
of osteoblasts and osteocytes from mesenchymal stem cells. Curr
Stem Cell Res Ther. 3:131–145. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhao JW, Zhang MR, Ji QY, Xing FJ, Meng LJ
and Wang Y: The role of slingshot-1L (SSH1L) in the differentiation
of human bone marrow mesenchymal stem cells into cardiomyocyte-like
cells. Molecules. 17:14975–14994. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bourne GL: The microscopic anatomy of the
human amnion and chorion. Am J Obstet Gynecol. 79:1070–1073. 1960.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gibb EA, Brown CJ and Lam WL: The
functional role of long non-coding RNA in human carcinomas. Mol
Cancer. 10:382011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wilusz JE, Sunwoo H and Spector DL: Long
noncoding RNAs: Functional surprises from the RNA world. Genes Dev.
23:1494–1504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kung JT, Colognori D and Lee JT: Long
noncoding RNAs: Past, present, and future. Genetics. 193:651–669.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Guttman M, Donaghey J, Carey BW, Garber M,
Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al:
lincRNAs act in the circuitry controlling pluripotency and
differentiation. Nature. 477:295–300. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mohamed J Sheik, Gaughwin PM, Lim B,
Robson P and Lipovich L: Conserved long noncoding RNAs
transcriptionally regulated by Oct4 and Nanog modulate pluripotency
in mouse embryonic stem cells. RNA. 16:324–337. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yu YL, Chou RH, Chen LT, Shyu WC, Hsieh
SC, Wu CS, Zeng HJ, Yeh SP, Yang DM, Hung SC and Hung MC: EZH2
regulates neuronal differentiation of mesenchymal stem cells
through PIP5K1C-dependent calcium signaling. J Biol Chem.
286:9657–9667. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Juan AH, Kumar RM, Marx JG, Young RA and
Sartorelli V: Mir-214-dependent regulation of the polycomb protein
Ezh2 in skeletal muscle and embryonic stem cells. Mol Cell.
36:61–74. 2009. View Article : Google Scholar : PubMed/NCBI
|