Introduction
Birth defects and abnormal fetal growth constitute
major fetal diseases associated with adverse pregnancy outcomes,
including fetal structural anomalies, fetal chromosomal disorders,
fetal growth retardation and abnormal fetal brain development or
lung maturation (1). Although
ultrasonographic methods, karyotyping, and developing noninvasive
prenatal test techniques can assist in diagnosing most of the
structural malformations and chromosomal disorders, the monitoring
and diagnosis of other types of fetal developmental diseases in
utero remain a challenge.
Amniotic fluid is the fluid within the amniotic sac
that surrounds the fetus, providing physical protection and
nutritional sources for the fetus (2,3).
Free diffusion occurs bi-directionally between the amniotic fluid
and fetal skin, placenta and umbilical cord. Therefore, the
composition of amniotic fluid is dynamic at different gestational
ages, containing biomolecules from maternal plasma and fetal
metabolism, in addition to cells originating from fetal kidney,
heart, lung, liver and hematopoetic cell lineages (4). In the clinic, amniocentesis is
routinely used to diagnose fetal chromosomal disorders such as Down
syndrome (DS).
Previous studies suggest that amniotic fluid
contains many biologically important molecules such as DNA, RNA and
metabolites that may reflect fetal development and pregnancy
condition, including preterm birth and preeclampsia (2,5). For
example, Larrabee et al (6)
performed the first transcriptomic analysis of human amniotic fluid
and observed that fetal gene expression was dynamic and changed
over the course of gestation. Hui et al (7), using microarray analysis, identified
that the transcriptome in mid-trimester amniotic fluid came from
specific organs and physiological systems. Using proteomic
profiling, Queloz et al (8)
observed that amniotic fluid proteins were observed in differential
abundances in early pregnancy compared with full term; and Romerao
et al (9) analyzed preterm
labor with and without intra-amniotic infection, and identified
different biomarkers for these clinical subgroups comprising
preterm labor.
Metabolomics is a technique that encompasses
individual metabolic profiles and their changes over time due to
physiology, disease, toxicity or other effects (10). Due to the fact that in-vivo
metabolites are the downstream products of gene expression and
protein synthesis, they may more closely represent actual cellular
activity at a functional level and at a specific point in time
(10). At present, the most
commonly used approaches for metabolic studies are liquid
chromatography-coupled mass spectrometry (LC-MS) and nuclear
magnetic resonance (NMR) spectroscopy, which can detect
in-vivo metabolites, including amino acids, oligopeptides,
sugars, steroids, biliary acids, fatty acids and other intermediary
compounds (11). In 1994, Bock
(12) profiled metabolites in the
amniotic fluid by NMR to characterize second-trimester and
third-trimester deliveries. Using ultra performance liquid
chromatography and mass spectrometry (UPLC-MS), Graca et al
(13) identified a decrease in
specific amino acids and an increase in hexose in amniotic fluid
samples taken from preterm deliveries. However, unlike genomic
studies, variability exists in different research laboratories with
respect to metabolomics analyses of fetal growth using amniotic
fluid. The majority of this variability may be due to the
short-term dynamics exhibited by metabolites, heterogeneous study
populations, different metabolomic research approaches, and varied
sampling and analysis protocols. Therefore, it remains a challenge
to identify universal biomarkers such as α-fetoprotein and human
chorionic gonadotrophin for DS screening, which could be
potentially applied for clinical use.
The aim of the current study was to explore whether
stable and highly specific markers during different experiments in
a relatively homogenous population carrying the same fetal disease.
Fetal DS was selected as a model as it is one of the most common
birth defects and is also a risk factor for fetal growth
retardation. Due to the accuracy of DS diagnosis, this model would
be more likely to consist of homogenous subjects and to represent
characteristics of both fetal defects and growth retardation.
In the present study, metabolites were analyzed in
two separate experiments that included a discovery set and a
validation set, using amniotic fluid from DS fetuses. It was
hypothesized that the metabolic alterations in amniotic fluid from
DS fetuses cluster in specific metabolic pathways, and are
associated with the altered gene expression observed at chromosome
21 during embryonic development of DS fetuses. To measure
experimental repeatability, amniotic fluid samples were processed
by different individuals separately, using the identical protocol,
and metabolomic fingerprinting was conducted in these separate
experiments using the same analytical approach. By analyzing the
metabolic profiles of different experimental sets and comparing
them to known genomic characteristics of DS fetuses, the present
study aimed to validate them with consistency and repeatability
regarding fetal disease, and to identify universal markers that
would assist us in understanding the molecular mechanisms
underlying fetal disease.
Materials and methods
Study population
The present study was approved by the Institutional
Review Boards of the Third Affiliated Hospital of Guangzhou Medical
University (Guangzhou, China) and 767 pregnant women with singleton
pregnancies between August 2014 and May 2015 each donated 5 ml of
amniotic fluid (AF), having provided written informed consent. The
AF samples were collected from women undergoing amniocentesis for
routine clinical indications (advanced maternal age, abnormal
quadruple/triple test, family history of chromosomal abnormalities,
suspected fetal anomalies or infection and upon maternal request)
at the second trimester of gestation. The clinical outcomes from
ultrasonogaphy, karyotyping and CMA results were additionally
obtained. Pregnancies associated with an obstetric complication
including hypertension, diabetes, premature rupture of the
membranes or uterine infections were excluded from the AF analyses.
Cases were a normal pregnancy with a DS fetus, and the controls
were a normal pregnancy with a normal fetus, matched with the case
samples in a 1:1 ratio based upon maternal age, fetal sex and
gestational age. Finally, the discovery set consisted of 10 DS
samples and 10 matched control samples. The validation set
consisted of 15 DS samples and 15 matched controls.
AF sample collection
Transabdominal amniocentesis was performed with a
21-gauge needle under ultrasound guidance to evaluate the position
of the fetus. A total of 5 ml residual AF was collected for
research purposes, in addition to that taken for diagnostic
testing. Samples were transported immediately in a capped sterile
syringe to the Biobank of the Third Affiliated Hospital of
Guangzhou Medical University. The AF samples were then centrifuged
for 10 min at 500 × g at 4°C, and the supernatant and pellet cells
were separately stored in aliquots at −80°C. The processing time
was <4 h from the time of sample collection to the time of
sample freezing.
Chemicals and columns
LC-MS grade organic solvents, including
acetonitrile, methanol, ammonium acetate, ammonium fluoride,
ammonia, and formic acid, were purchased from Sigma (Sigma-Aldrich;
Merck Millipore, Darmstadt, Germany). The HILIC column was Waters,
ACQUITY UPLC BEH Amide 1.7 µm, 2.1×100 mm column; and the HSS T3
column was ACQUITY UPLC HSS T3 1.8 µm, 2.1×100 mm column (Waters
US, Milford, MA, USA).
Sample preparation
For proteins precipitation, 100 µl of AF sample was
thawed at 4°C, and 400 µl of cold (−20°C) mixture of
methanol/acetonitril (1:1, v/v) was added. Samples were
vortex-mixed and stored at −20°C for 10 min. Following
centrifugation at 14,000 × g for 15 min at 4°C, the supernatant was
filtered using a 0.22 µm nylon filter and dried under vacuum. For
LC-MS analysis, 100 µl acetonitrile/water (1:1, v/v) was added to
each sample, vortexed and centrifuged at 14,000 × g for 15 min at
4°C for final injection. QC samples were prepared by pooling equal
volumes of each sample.
LC-QTOF-MS analysis
The UPLC system (Agilent 1290 Infinity LC System;
Agilent Technologies, Inc., Santa Clara, CA, USA) consisting of a
degasser, two binary pumps and a thermostated autosampler
(maintained at 4°C) coupled with a Triple TOF 5600 mass
spectrometer (AB SCIEX, Framingham, MA, USA) were used for
metabolomic analysis. Briefly, a 4.0 µl sample of extracted AF
samples was injected into a thermostated (25°C) reverse-phase
ACQUITY UPLC HSS T3 C18 column or ACQUITY UPLC BEH Amide column.
The flow rate was 600 µl/min with solvent A (water with 0.1% formic
acid) and solvent B (acetonitrile with 0.1% formic acid). QC
samples were prepared by pooling equal volumes of each sample and
injected at the beginning and at the end of each analysis, and at
every five samples, to provide a measurement of the system's
stability and performance in addition to reproducibility of the
sample treatment procedure.
For the HILIC column, solvent A was composed of
water and 25 mM ammonium acetate and 25 mM ammonia, and solvent B
was acetonitrile. The chromatographic gradient started at 85% for
phase B for the first minute, decreased to 65% from 1–12 min, and
then to 40% from 12–12.1 min. The phase remained at 40% for solvent
B from 12.1 to 15 min, returned to 85% for phase B from 15–15.1
min, and remained at 85% for phase B for 5 min.
For the positive ion mode of the HSS T3 column,
solvent A was composed of 0.1% formic acid and water, and solvent B
was 0.1% formic acid and acetonitrile. For the negative ion mode of
the HSS T3 column, solvent A was comprised of 0.5 mM ammonium
fluoride, and solvent B was acetonitrile. The chromatographic
gradient started at 1% for phase B for the first 1.5 min, and
increased to 99% from 1.5–13 min. The phase remained at 99% for
solvent B from 13–16.5 min, returned to 1% for phase B from
16.5–16.6 min, and remained at 1% phase for B for 4 min.
The samples were separated by UPLC and analyzed with
a Triple TOF 5600 mass spectrometer, using electrospray ionization
(ESI) for positive ion and negative ion modes, respectively. For
the HILIC column, the ESI source conditions were ion source gas 1
(Gas 1), 60; ion source gas 2 (Gas 2), 60; curtain gas (CUR), 20;
source temperature, 600°C; and ionspray voltage floating (ISVF)
±5,500 V (positive and negative modes). The detector conditions
were TOF MS scan m/z range, 50–1,000 Da; product ion scan m/z
range, 25–1,000 Da; scan accumulation time, 0.20 s/spectra; product
ion scan accumulation time, 0.05 s/spectra; declustering potential
(DP), ±60 V (positive and negative modes); and collision energy,
35±15 eV. MS/MS was acquired using information-dependent
acquisition (IDA) and high-sensitivity mode; the IDA set excluded
isotopes within 4 Da, and 6 candidate ions were monitored per
cycle.
For the HSS T3 column, the ESI source conditions
were Gas 1, 40; Gas 2, 80; CUR, 30; source temperature, 650°C; and
the ISVF±5,000 V (positive and negative ion modes). The detector
conditions were TOF MS scan m/z range, 60–1,000 Da; product ion
scan m/z range, 25–1,000 Da; TOF MS scan accumulation time, 0.20
s/spectra; product ion scan accumulation time, 0.05 s/spectra; DP,
±60 V (positive and negative modes); and collision energy, 35±15
eV. MS/MS was acquired using IDA with high-sensitivity mode; the
IDA set excluded isotopes within 4 Da, and 6 candidate ions were
monitored per cycle.
Bioinformatics analysis
Kyoto Encyclopedia of Genes and Genomes (KEGG)
pathway analysis (http://www.genome.jp/kegg/) was performed with
Database for Annotation, Visualization and Integrated
Discoverybioinformatics website (https://david.ncifcrf.gov/) platform using default
parameters. Gene classes were identified based on Gene Ontology
term categories and the KEGG pathway database.
Compound identification and
statistical analyses
Raw data acquired using the UHPLC-MS system was
processed to mzML format via ProteoWizard software version 3.0.6458
(http://proteowizard.sourceforge.net)
for subsequent data analysis (14). The background cleaning and
alignment of drift (by retention time and mass), and filtering of
the data collected by LC-MS were performed with XCMS (http://metlin.scripps.edu/download) (15). Each compound was described by mass,
retention time and abundance. Identification of the compounds was
confirmed by accurate mass and MS/MS profiles, and confirmation was
performed with standards by comparison of retention time, isotopic
distribution and fragments of commercially available reagents with
those obtained from analyzed samples. For univariate statistical
analysis, data was uniformed using autoscaling and
multi-dimensional analysis, including PCA and PLS-DA, using Metabo
Analysis software version 3.0 (http://www.metaboanalyst.ca) (16). Numerical data are shown as mean ±
standard deviation, and independent samples Student's t-test and
volcano analysis were performed using the R package.
Results
Patient characteristics
A total of 25 cases of fetal DS and 25 controls were
recruited in the current study, in 2 separate sets. In the
discovery set, the average maternal age of cases and controls was
32 years, and the average gestational ages at amniocentesis were
131 and 130 days, respectively (Table
I). In the validation set, the average maternal ages of cases
and controls were 34 and 33 years, respectively; and the average
gestational age at amniocentesis was 131 days (Table I). The fetal sex rate was identical
in cases and the control group in both sets (Table I).
 | Table I.Demographic characteristics of study
subjects. |
Table I.
Demographic characteristics of study
subjects.
|
| Discovery set | Validation set |
|---|
|
|
|
|
|---|
| Subject
characteristics | Down syndrome | Control | Down syndrome | Control |
|---|
| Number of
samples | 10 | 10 | 15 | 15 |
| Maternal ages,
years | 32 (43–22) | 32 (39–24) | 34 (41–23) | 33 (39–21) |
| Gestational ages,
days | 131 (143–113) | 130 (143–112) | 131 (164–112) | 131 (163–112) |
| Fetal sex | 6 female | 6 female | 6 female | 6 female |
|
| 4 male | 4 male | 9 male | 9 male |
Metabolomic fingerprinting
The UPLC-Q-TOF-MS system was applied to AF
fingerprinting. In total, 20 samples of the discovery set and 30
samples of the validation set were analyzed in 2 separate
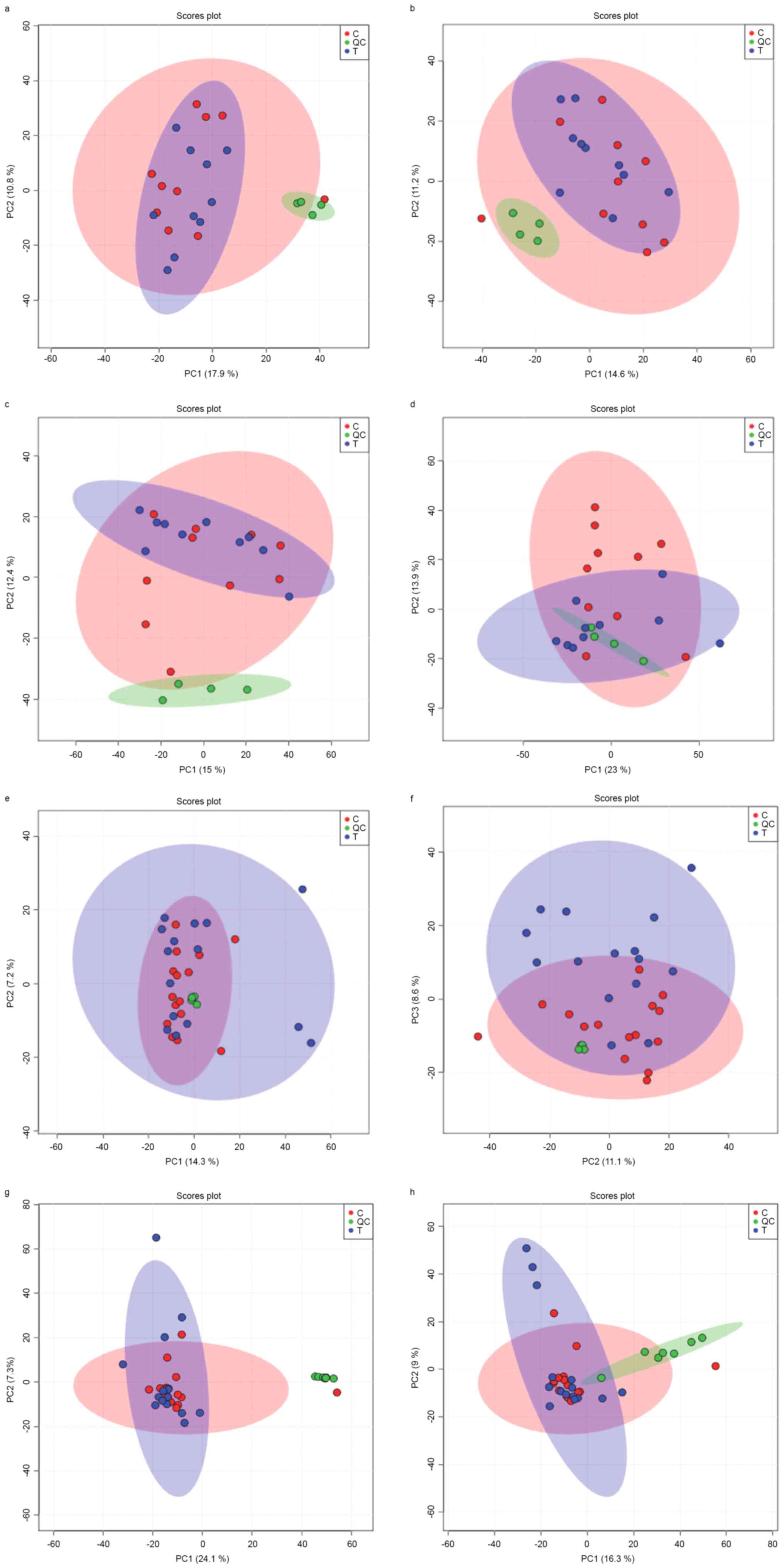
experiments. Unsupervised principal component analysis (PCA-X)
models (Fig. 1) demonstrated good
clustering of all QC samples in all HILLC(+), HILLC(−), HSS(+), and
HSS(−) modes in the 2 experimental sets, which indicated that the
system was stable and performance was reliable. The detected peak
numbers were different for the two sets: In the discovery set, the
peak numbers in HILIC(+), HILIC(−), HSS(+), and HSS(−) modes were
4,044, 3,880, 5,877 and 3,970, respectively; while in the
validation set, the peak numbers in HILIC(+), HILIC(−), HSS(+), and
HSS(−) modes were 5,915, 6,029, 4,801 and 5,183, respectively.
In the unsupervised PCA-X analysis, 5 outliers were
observed in the discovery set, and 4 of them were from controls and
1 from cases. Similarly, 8 outliers were observed in the discovery
set, and 3 of them were from controls, and 5 from cases. Therefore,
in later analysis, these outliers were eliminated.
Supervised partial least-squares discriminate
analysis (PLS-DA) was further used to model the differences between
cases and controls. The PLS-DA model demonstrated clear separation
of samples from the 2 groups (Fig.
2). In the discovery set, the explained variance (R2) and
predicted variance (Q2) in HILIC(+), HILIC(−), HSS(+), and HSS(−)
modes were R2=1.00, Q2=0.31, R2=1.00, Q2=0.58, R2=1.00, Q2=0.36,
R2=0.96 and Q2=0.74, respectively. In the validation set, the R2
and Q2 in HILIC(+), HILIC(−), HSS(+), and HSS(−) modes were
R2=0.99, Q2=0.35, R2=0.98, Q2=0.31, R2=0.98, Q2=0.65, R2=0.98 and
Q2=0.69, respectively. Therefore, the HSS(−) mode was the best mode
for PLS-DA analysis in both the discovery and validation sets.
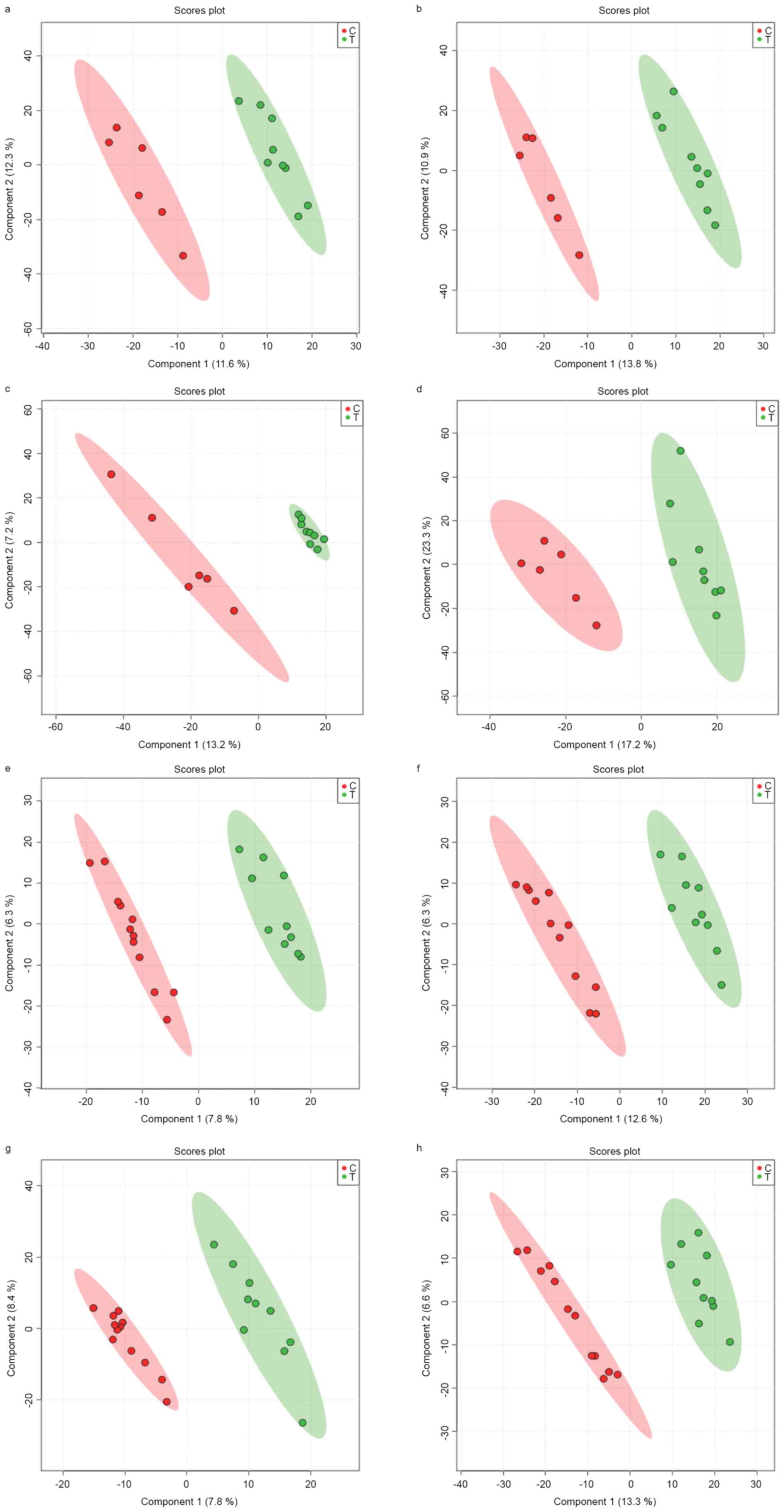 | Figure 2.Supervised PLS-DA model score in
discovery set and validation set. Green circles, cases; red
circles, controls. (A-D) represent the data of discovery set
obtained by PC1 vs. PC2 in HILLC(+), HILLC(−), HSS(+) and HSS(−)
mode. The R2 and Q2 in (A-D) are R2=1.00, Q2=0.31, R2=1.00,
Q2=0.58, R2=1.00, Q2=0.36, R2=0.96 and Q2=0.74, respectively. (E-H)
represent the data of validation set obtained by PC1 vs. PC2 in
HILLC(+), HILLC(−), HSS(+) and HSS(−) mode. The R2 and Q2 in (A-D)
plots are R2=0.99, Q2=0.35, R2=0.98, Q2=0.31, R2=0.98, Q2=0.65,
R2=0.98 and Q2=0.69, respectively. PLS-DA, supervised partial
least-squares discriminate analysis; HILLC, hydrophilic interaction
chromatography; HSS, silica-based bonded phase. |
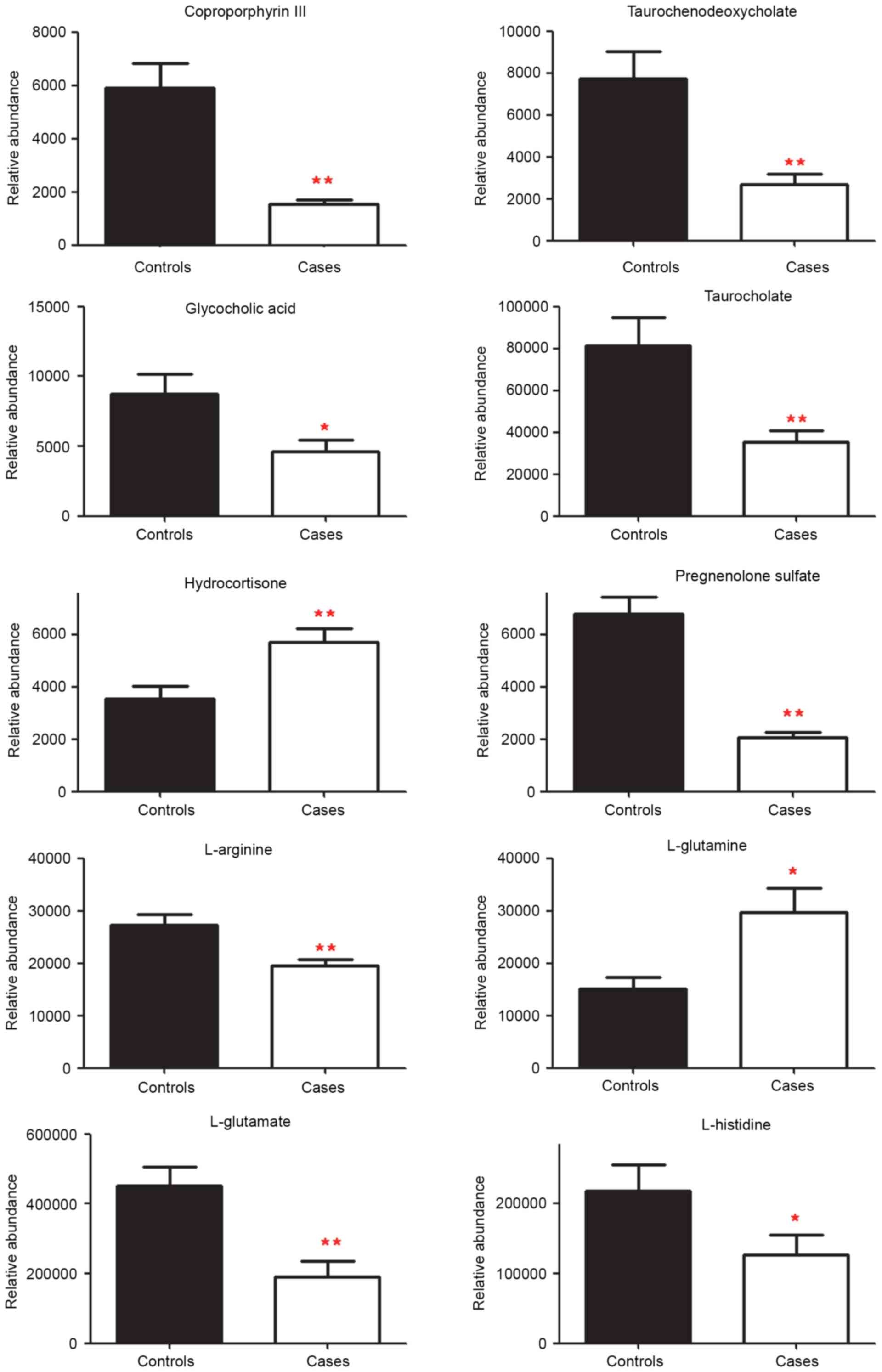
Metabolite analysis
Peaks that were significantly different in the 4
modes of the 2 experimental sets were initially screened, and some
of them had been identified and validated using standards. Those
identified in both the discovery set and validation set, and that
had a similar change in tendency and in significance (P<0.05 or
P<0.1 for both experiments) were chosen as the final identified
markers (Table II). These were
coproporphyrin-III, pregnenolone sulfate, taurochenodeoxycholate,
L-arginine, taurocholate, hydrocortisone (cortisol), L-histidine,
glycocholic acid, L-glutamate and L-glutamine. Among these markers,
amounts of hydrocortisone and L-glutamine were increased
significantly, however the quantities of the other metabolites were
decreased in cases when compared with the controls. When the
quantities were assessed in individual samples for both the
discovery set and the validation set, 10 metabolites demonstrated a
significant difference in cases vs. controls (Fig. 3). The amounts of
coproporphyrin-III, L-glutamate, pregnenolone sulfate,
taurochenodeoxycholate, L-arginine and taurocholate were
significantly decreased in cases compared with the controls
(P<0.01), as were L-histidine and glycocholic acid (P<0.05).
The levels of hydrocortisone and L-glutamine were significantly
increased in cases compared with controls (P<0.01 and P<0.05,
respectively).
 | Table II.Altered KEGG pathways and involved
metabolite numbers in two sets. |
Table II.
Altered KEGG pathways and involved
metabolite numbers in two sets.
| Name | Discovery | Validation |
|---|
| Metabolic
pathways | 26 | 15 |
| Biosynthesis of
amino acids | 9 | 4 |
| Biosynthesis of
antibiotics | 7 | 4 |
| Bile secretion | 6 | 4 |
| Tryptophan
metabolism | 5 | 3 |
| Two-component
system | 4 | 2 |
| Purine
metabolism | 2 | 2 |
| ABC
transporters | 8 | 5 |
| Biosynthesis of
secondary metabolites | 14 | 4 |
| Central carbon
metabolism in cancer | 10 | 4 |
| Protein digestion
and absorption | 9 | 4 |
| Aminoacyl-tRNA
biosynthesis | 8 | 4 |
| Biosynthesis of
alkaloids derived from ornithine, lysine and nicotinic acid | 7 | 2 |
| Microbial
metabolism in diverse environments | 5 | 4 |
| Alanine, aspartate
and glutamate metabolism | 4 | 2 |
| Glyoxylate and
dicarboxylate metabolism | 4 | 2 |
| Alcoholism | 3 | 2 |
| Primary bile acid
biosynthesis | 3 | 3 |
| GABAergic
synapse | 2 | 2 |
| Galactose
metabolism | 2 | 2 |
| Glutamatergic
synapse | 2 | 2 |
| Histidine
metabolism | 2 | 3 |
| Glutathione
metabolism | 2 | 2 |
| Nitrogen
metabolism | 2 | 2 |
| Porphyrin and
chlorophyll metabolism | 3 | 2 |
| Neuroactive
ligand-receptor interaction | 3 | 3 |
| Secondary bile acid
biosynthesis | 3 | 3 |
| Arginine and
proline metabolism | 3 | 4 |
| D-Glutamine and
D-glutamate metabolism | 2 | 2 |
| Proximal tubule
bicarbonate reclamation | 4 | 2 |
| Amyotrophic lateral
sclerosis | 2 | 2 |
| Taurine and
hypotaurine metabolism | 2 | 2 |
KEGG analysis
Using KEGG pathway analysis, it was identified that
there were at least 30 KEGG pathways altered in cases compared with
the controls (Table III),
including biosynthesis of amino acids, ABC transporters, alanine,
aspartate and glutamate metabolism, bile secretion, arginine and
proline metabolism, histidine metabolism, taurine and hypotaurine
metabolism. The majority of these molecules are involved largely
with amino acid metabolism, liver function, growth hormone and
neural development. Notably, using KEGG pathways, certain molecules
were shared with genes on chromosome 21. There were 9 KEGG pathways
altered in AF from DS fetuses associated with genes on chromosome
21 (Table IV). These were
galactose metabolism, purine metabolism, histidine metabolism, ABC
transporters, neuroactive ligand-receptor interaction, Parkinson's
disease, ALS, Huntington's disease and pathways in cancer.
 | Table III.Identified common markers in two
sets. |
Table III.
Identified common markers in two
sets.
|
|
| Discovery | Validation |
|---|
|
|
|
|
|
|---|
| Category | Metabolite | VIP | FC | P-value | VIP | FC | P-value |
|---|
| Porphyrin | Coproporphyrin
III | 2.8375 | 0.241717 | 0.000683 | 3.26 | 0.270882 | 0.007657 |
| Hormone | Pregnenolone
sulfate | 2.0109 | 0.308604 | 0.001741 | 2.9401 | 0.299152 | 0.000017 |
|
| Hydrocortisone
(cortisol) | 1.2543 | 1.283218 | 0.085341 | 1.9657 | 1.716536 | 0.002482 |
| Bile acid |
Taurochenodeoxycholate | 2.3718 | 0.465954 | 0.000993 | 3.5658 | 0.388247 | 0.001494 |
|
| Glycocholic
acid | 1.0539 | 0.58811 | 0.066354 | 1.2245 | 0.613199 | 0.048233 |
|
| Taurocholate | 1.0283 | 0.403143 | 0.073469 | 2.1432 | 0.419715 | 0.000974 |
| Amino acid | L-Arginine | 1.5972 | 0.74232 | 0.029519 | 1.7646 | 0.697481 | 0.031024 |
|
| L-Histidine | 1.2185 | 0.73082 | 0.093209 | 1.441 | 0.706818 | 0.041167 |
|
| L-Glutamate | 2.2569 | 0.391223 | 0.000213 | 2.4137 | 0.509103 | 0.001985 |
|
| L-Glutamine | 1.4845 | 1.885866 | 0.067402 | 1.5919 | 1.58102 | 0.035146 |
 | Table IV.Common Kyoto Encyclopedia of Genes
and Genomes pathways involved with genes in chromosome 21 and amino
fluid metabolites from Down syndrome fetuses. |
Table IV.
Common Kyoto Encyclopedia of Genes
and Genomes pathways involved with genes in chromosome 21 and amino
fluid metabolites from Down syndrome fetuses.
| KEGG pathways | Metabolites | Genes in chromosome
21 |
|---|
| Galactose
metabolism | α-D-glucose,
D-mannose, Myo-inositol, stachyose | PFKL |
| Purine
metabolism | L-glutamine,
hypoxanthine, adenosime | PDE9A, GART |
| Histidine
metabolism | L-histidine,
L-methyl-histidine, L-glutamate | FTCD |
| ABC
transporters | Arginine,
histidine, glutamate, glutamine, mannitol, mannose,
myo-inositol | ABCG1 |
| Neuroactive
ligand-receptor interaction | Adenosine,
metabotropic glutamate, dopamine, cortisol, glutamate | GIRK1 |
| Parkinson's
disease | Dopamine,
adenosine | ATP5J, ATP5O,
NDUFV3, UBE2G2 |
| Amyotrophic lateral
sclerosis | Glutamate,
arginine | SOD1 |
| Huntington's
disease | Glutamate | ATP5J, ATP5O,
NDUFV3, SOD1 |
Discussion
Although metabolomics has been applied to numerous
biomedical studies, few have been conducted using this approach for
the monitoring of fetal malformations. One of the reasons for this
is the variability that exists in metabolomic studies in different
experimental paradigms and laboratories. For example, using 1H-NMR,
Graca et al (13,17,18)
identified alterations in metabolites including methionine and
succinate with fetal malformations, however using UPLC-MS, they
identified another group of metabolites that were not identical to
those previously identified. In addition, Bock (12) studied the AF metabolome in fetal DS
specimens, however the metabolites exhibited no particular pattern
due to the small number collected. These researchers indicated that
the results of a metabolomics study may largely depend on the study
population, sampling process and detection approach when applied to
the study of fetal disease.
To explore the repeatability and stability of AF
metabolite identification, the discovery and validation sets were
carefully designed, both of which included 10–15 pairs of controls
and cases of DS fetuses (Table I).
The results indicated that although ~80 and ~60 metabolites,
respectively, were indentified in the two sets (which were
significantly different between cases and controls), there were few
metabolites that were common between the sets. The results
demonstrated that metabolite identification may exhibit large
variability among different samples, even when using the same
approach and protocol. This is suggested to be due to the dynamic
nature and inherent variability of metabolites in AF, and the
limited number of techniques available to accurately identify them.
However, when metabolomic pathways were investigated, the majority
of the altered metabolites identified could be clustered into
similar pathways in the two experiments (Table II). The major pathway alterations
in DS fetuses included amino acids, bile secretion, neuroactive
ligand-receptor interaction and galactose metabolism, all of which
could be associated with the pathophysiology of DS fetuses,
including growth and mental retardation (Table II). Therefore, although metabolite
identification is not consistent in AF, the alterations in
metabolism pathways may be more likely to be invariable over
different experiments.
When the significantly altered metabolites were
identified from both experiments, it was noted that they belonged
to 4 categories: 4 were amino acids, 3 were bile acids, 2 were
hormones and 1 was porphyrin (Table
II, Fig. 3). Alterations in
these metabolites have been associated with several negative
pregnancy outcomes including abortion, fetal growth abnormality,
prematurity and low birth weight (19). For example, in a rat intrauterine
growth restriction model, elevated maternal and fetal
corticosterone levels were reported in serum and AF (20). In humans, analysis of total urinary
steroids was effective in detecting fetuses with Smith-Lemli-Opotz
syndrome (21), and progesterone
levels were altered in 87% of maternal urine in the presence of a
DS fetus (22). Concomitantly,
increased levels of cortisol and decreased levels of pregnenolone
sulfate were detected in DS fetuses, which may be associated with
abnormal bone and brain development. Similarly, bile acids
including taurochenodeoxycholic acid, glycocholic acid and
taurocholic acid all increased in AF from women with preterm
deliveries (23). Due to the fact
that the accumulation of bile acids can trigger an inflammatory
response in maternal and fetal lungs (24), high circulating levels in the
preterm birth fetus may reflect uterine stress due to preterm
birth. Considering that mothers who carry DS fetuses did not
exhibit a higher incidence of pregnancy complications, the decrease
in bile acids that was observed may reflect an underdeveloped liver
or impaired bile acid metabolism in the DS fetus.
In addition to changes in hormones and bile salts,
altered amino acid concentrations are widely reported for fetuses
in adverse pregnancies (23,25).
A previous study demonstrated that amino acids can be actively
taken up by the fetus, and that metabolic factors such as
insulin-like growth factor-1 treatment may promote gut utilization
of amino acids from the AF pool (26). Among amino acids, arginine is the
donor for NO in vivo, and its concentration is associated
with decreased NO levels. Molecular mechanistic studies suggest
that arginine is involved in diverse functions including placental
angiogenesis, antioxidant stress, the immune system and placental
apoptosis (27–30). In the present study, the decrease
in L-arginine concentration suggested that the impaired growth of
DS fetuses may also be involved in downregulating the NO signaling
pathway, however the details warrant further validation. As an
essential amino acid for fetuses, a decrease in histidine levels
has been reported in several types of abnormal fetal growth
including preterm fetuses and fetuses with skeletal dysplasia
(23,25). However, in the case of
chorioamnionitis, histidine increased significantly, suggesting an
association with uterine inflammation and infection (31). The majority of cases of DS fetuses
are not accompanied by uterine infection, and therefore, similar to
the situation for preterm fetuses and fetuses with skeletal
dysplasia, the reduction in histidine in AF may be due to altered
histidine metabolism.
As a relatively recent type of ‘omics’ technology,
one of the most important goals of metabolomics is to assist in
uncovering the etiology of diseases at functional levels. It is
notable that in the present study, although only certain
significantly altered metabolites were identified, some were
directly associated with the DS phenotype at a molecular level. It
was identified that metabolites involved in erythropoiesis
(coproporphyrin III) were significantly altered in the DS fetus.
Coproporphyrin III is a byproduct of heme biosynthesis, and is
excreted normally in feces as a decomposition product of bilirubin;
it has been demonstrated that coproporphyrin increases in human
urine due to lead poisoning, which is associated with decomposition
products of erythropoiesis (32).
A significant decrease in coproporphyrin III was observed in AF
taken from fetuses with DS, which may reflect an aberration in
fetal erythropoiesis. Correspondingly, patients with DS frequently
exhibit blood cell abnormalities, including abnormal blood counts,
transient myeloproliferative disorders and acute megakaryoblastic
leukemia. Using a stem cell model, DS cells were demonstrated to
exhibit enhanced erythropoiesis and reduced myelopoiesis (33). In vitro and mouse
transplantation assays have indicated that trisomy 21 progenitors
manifested enhanced production of erythroid and megakaryocytic
cells that proliferated excessively (34). Therefore, the decrease in
coproporphyrin III in DS fetuses may be potentially coupled at a
molecular level to their abnormal erythropoiesis.
There have been few metabolomics studies of fetal
malformations that make intrinsic associations between metabolites
and fetal disease genotypes, likely due to the fact that the
majority of experiments used a mixture of fetal abnormality samples
that included abnormalities in the central nervous, cardiac and
urogenital systems (5,35). Therefore, whether the significantly
altered metabolites could directly reflect the etiology of fetal
diseases or whether they are by-products of affected signaling
pathways in malformed fetuses remains unclear. Due to the fact that
DS is a fetal disease with a clear genetic background, and further
analyses were conducted combining metabolomics and genomics. The
results suggested that metabolite alterations in DS fetuses are
partially associated with its genetic etiology; namely, the extra
copy of chromosome 21. The identified pathways for histidine
metabolism, neuroactive ligand-receptor interaction, and neural
diseases are also located on chromosome 21 (Table III).
Among metabolites that exhibit functional overlap
between metabolic alterations and extra copies of chromosome 21 in
the DS fetus, it is notable that alterations in glutamine-glutamate
metabolism are involved in nearly every overlapping signaling
pathway (Table IV). In previous
studies of fetal malformations, alterations in glutamine-glutamate
have been reported (13,17,18),
suggesting a change in a pivotal element of this signaling pathway
in fetal development. In DS, aberrations in glutamatergic
transmission constituted a major cause of behavioral deficits, and
glutamate is an important hippocampal neuron survival factor
(36). Clinically, glutamate
uptake is significantly decreased in platelets and fibroblasts from
DS patients, and a significant deficit in glutamate has been
observed in the hippocampus with DS (37,38).
In the present study, it was identified that L-glutamate is the
most significantly reduced metabolite in fetal AF (P<0.005 for
both experimental sets; Table
III). Due to the fact that glutamine is synthesized from
glutamate and the glutamate-glutamine cycle serves key roles in
neuronal activation, the increased glutamine and decreased
glutamate may reflect an imbalance in this cycle. Genetic analysis
also indicated that several genes identified on chromosome 21 were
involved in glutamaterigic transmission. For instance, the mouse
Glur-5 gene maps to chromosome 16 (39), and the homologous human GLUR5 gene
maps to the corresponding region of human chromosome 21; the dosage
imbalance regarding GLUR5 may thereby have a role in the DS
phenotype in both mice and humans (39). Therefore, it is hypothesized that
the observed glutamine-glutamate alterations are tightly associated
with the apparent molecular mechanisms of DS etiology.
In conclusion, data from the present study suggest
that metabolic identification was variable when taken from
different samples in separate experiments. In the DS fetus,
alterations in the four metabolic pathways of porphyrin, bile
acids, amino acids and hormones were validated, and significant
changes in metabolites of coproporphyrin III, pregnenolone sulfate,
taurochenodeoxycholate, L-arginine, taurocholate, hydrocortisone,
L-histidine, glycocholic acid, L-glutamate and L-glutamine were
identified. Analysis of these metabolic alterations associated them
with aberrant gene expression on chromosome 21 of DS fetuses,
particularly with respect to intellectual impairment and abnormal
erythropoiesis. Therefore, alterations in AF metabolites may
provide important information in the understanding of fetal disease
pathophysiology beyond that of genomics, epigenomics, and
proteomics. This may aid in the development of tests for the
diagnosis of fetal diseases, providing an additional tool for
exploring the etiology of fetal disease.
Acknowledgements
The authors would like to thank LetPub for its
linguistic assistance during the preparation of this manuscript.
The present study was supported by the National Key R&D Program
of China (grant no. 2016YFC1000405), the Natural Science Foundation
of China (grant no. 81370751) and the Guangdong Natural Science
Foundation (grant no. 2014A030313502).
References
|
1
|
Carmichael SL: Birth defects epidemiology.
Eur J Med Genet. 57:355–358. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hui L and Bianchi DW: Cell-free fetal
nucleic acids in amniotic fluid. Hum Reprod Update. 17:362–371.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Underwood MA, Gilbert WM and Sherman MP:
Amniotic fluid: Not just fetal urine anymore. J Perinatol.
25:341–348. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Da Sacco S, Sedrakyan S, Boldrin F,
Giuliani S, Parnigotto P, Habibian R, Warburton D, De Filippo RE
and Perin L: Human amniotic fluid as a potential new source of
organ specific precursor cells for future regenerative medicine
applications. J Urol. 183:1193–1200. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kamath-Rayne BD, Smith HC, Muglia LJ and
Morrow AL: Amniotic fluid: The use of high-dimensional biology to
understand fetal well-being. Reprod Sci. 21:6–19. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Larrabee PB, Johnson KL, Lai C, Ordovas J,
Cowan JM, Tantravahi U and Bianchi DW: Global gene expression
analysis of the living human fetus using cell-free messenger RNA in
amniotic fluid. JAMA. 293:836–842. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hui L, Slonim DK, Wick HC, Johnson KL and
Bianchi DW: The amniotic fluid transcriptome: A source of novel
information about human fetal development. Obstet Gynecol.
119:111–118. 2011. View Article : Google Scholar
|
|
8
|
Queloz PA, Crettaz D, Thadikkaran L, Sapin
V, Gallot D, Jani J, Deprest J, Lémery D, Barelli S and Tissot JD:
Proteomic analyses of amniotic fluid: Potential applications in
health and diseases. J Chromatogr B Analyt Technol Biomed Life Sci.
850:336–342. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Romero R, Kusanovic JP, Gotsch F, Erez O,
Vaisbuch E, Mazaki-Tovi S, Moser A, Tam S, Leszyk J, Master SR, et
al: Isobaric labeling and tandem mass spectrometry: A novel
approach for profiling and quantifying proteins differentially
expressed in amniotic fluid in preterm labor with and without
intra-amniotic infection/inflammation. J Matern Fetal Neonatal Med.
23:261–280. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nicholson JK, Connelly J, Lindon JC and
Holmes E: Metabonomics: A platform for studying drug toxicity and
gene function. Nat Rev Drug Discov. 1:153–161. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Roux A, Lison D, Junot C and Heilier JF:
Applications of liquid chromatography coupled to mass
spectrometry-based metabolomics in clinical chemistry and
toxicology: A review. Clin Biochem. 44:119–135. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bock JL: Metabolic profiling of amniotic
fluid by proton nuclear magnetic resonance spectroscopy:
Correlation with fetal maturation and other clinical variables.
Clin Chem. 40:56–61. 1994.PubMed/NCBI
|
|
13
|
Graca G, Goodfellow BJ, Barros AS, Diaz S,
Duarte IF, Spagou K, Veselkov K, Want EJ, Lindon JC, Carreira IM,
et al: UPLC-MS metabolic profiling of second trimester amniotic
fluid and maternal urine and comparison with NMR spectral profiling
for the identification of pregnancy disorder biomarkers. Mol
Biosyst. 8:1243–1254. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ivanisevic J, Zhu ZJ, Plate L, Tautenhahn
R, Chen S, O'Brien PJ, Johnson CH, Marletta MA, Patti GJ and
Siuzdak G: Toward ‘omic scale metabolite profiling: A dual
separation-mass spectrometry approach for coverage of lipid and
central carbon metabolism. Anal Chem. 85:6876–6884. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Smith CA, Want EJ, O'Maille G, Abagyan R
and Siuzdak G: XCMS: Processing mass spectrometry data for
metabolite profiling using nonlinear peak alignment, matching, and
identification. Anal Chem. 78:779–787. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xia J, Sinelnikov IV, Han B and Wishart
DS: MetaboAnalyst 3.0-making metabolomics more meaningful. Nucleic
Acids Res. 43:W251–W257. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Graca G, Duarte IF, Barros AS, Goodfellow
BJ, Diaz S, Carreira IM, Couceiro AB, Galhano E and Gil AM: (1)H
NMR based metabonomics of human amniotic fluid for the metabolic
characterization of fetus malformations. J Proteome Res.
8:4144–4150. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Graca G, Duarte IF, Barros AS, Goodfellow
BJ, Diaz SO, Pinto J, Carreira IM, Galhano E, Pita C and Gil AM:
Impact of prenatal disorders on the metabolic profile of second
trimester amniotic fluid: A nuclear magnetic resonance metabonomic
study. J Proteome Res. 9:6016–6024. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Field T and Diego M: Cortisol: The culprit
prenatal stress variable. Int J Neurosci. 118:11812008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Feng JH, Yan YE, Liang G, Liu YS, Li XJ,
Zhang BJ, Chen LB, Yu H, He XH and Wang H: Maternal and fetal
metabonomic alterations in prenatal nicotine exposure-induced rat
intrauterine growth retardation. Mol Cell Endocrinol. 394:59–69.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Marcos J, Craig WY, Palomaki GE, Kloza EM,
Haddow JE, Roberson M, Bradley LA and Shackleton CH: Maternal urine
and serum steroid measurements to identify steroid sulfatase
deficiency (STSD) in second trimester pregnancies. Prenat Diagn.
29:771–780. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Trivedi DK and Iles RK: Shotgun
metabolomic profiles in maternal urine identify potential mass
spectral markers of abnormal fetal biochemistry-dihydrouracil and
progesterone in the metabolism of Down syndrome. Biomed Chromatogr.
29:1173–1183. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Menon R, Jones J, Gunst PR, Kacerovsky M,
Fortunato SJ, Saade GR and Basraon S: Amniotic fluid metabolomic
analysis in spontaneous preterm birth. Reprod Sci. 21:791–803.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Herraez E, Lozano E, Poli E, Keitel V, De
Luca D, Williamson C, Marin JJ and Macias RI: Role of macrophages
in bile acid-induced inflammatory response of fetal lung during
maternal cholestasis. J Mol Med (Berl). 92:359–372. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kale E and Kale A: Amniotic fluid amino
acid concentrations in fetal skeletal dysplasia. Clin Exp Obstet
Gynecol. 41:280–282. 2014.PubMed/NCBI
|
|
26
|
Bloomfield FH, van Zijl PL, Bauer MK and
Harding JE: Effects of intrauterine growth restriction and
intraamniotic insulin-like growth factor-I treatment on blood and
amniotic fluid concentrations and on fetal gut uptake of amino
acids in late-gestation ovine fetuses. J Pediatr Gastroenterol
Nutr. 35:287–297. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bednov A, Espinoza J, Betancourt A,
Vedernikov Y, Belfort M and Yallampalli C: L-arginine prevents
hypoxia-induced vasoconstriction in dual-perfused human placental
cotyledons. Placenta. 36:1254–1259. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
da Costa CM, de Freitas MR, Brazão V, dos
Santos CD, Sala MA, do Prado Júnior JC and Abrahão AA: Does
L-arginine availability during the early pregnancy alters the
immune response of Trypanosoma cruzi infected and pregnant Wistar
rats? Exp Parasitol. 142:59–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pimentel AM, Pereira NR, Costa CA, Mann
GE, Cordeiro VS, de Moura RS, Brunini TM, Mendes-Ribeiro AC and
Resende AC: L-arginine-nitric oxide pathway and oxidative stress in
plasma and platelets of patients with pre-eclampsia. Hypertens Res.
36:783–788. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shen SF and Hua CH: Effect of L-arginine
on the expression of Bcl-2 and Bax in the placenta of fetal growth
restriction. J Matern Fetal Neonatal Med. 24:822–826. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dudzik D, Revello R, Barbas C and Bartha
JL: LC-MS-based metabolomics identification of novel biomarkers of
chorioamnionitis and its associated perinatal neurological damage.
J Proteome Res. 14:1432–1444. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sakai T: Biomarkers of lead exposure. Ind
Health. 38:127–142. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chou ST, Byrska-Bishop M, Tober JM, Yao Y,
Vandorn D, Opalinska JB, Mills JA, Choi JK, Speck NA, Gadue P, et
al: Trisomy 21-associated defects in human primitive hematopoiesis
revealed through induced pluripotent stem cells. Proc Natl Acad Sci
USA. 109:17573–17578. 2012; View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chou ST, Opalinska JB, Yao Y, Fernandes
MA, Kalota A, Brooks JS, Choi JK, Gewirtz AM, Danet-Desnoyers GA,
Nemiroff RL and Weiss MJ: Trisomy 21 enhances human fetal
erythro-megakaryocytic development. Blood. 112:4503–4506. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Palmas F, Fattuoni C, Noto A, Barberini L,
Dessi A and Fanos V: The choice of amniotic fluid in metabolomics
for the monitoring of fetus health. Expert Rev Mol Diagn.
16:473–486. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bambrick LL, Yarowsky PJ and Krueger BK:
Glutamate as a hippocampal neuron survival factor: An inherited
defect in the trisomy 16 mouse. Proc Natl Acad Sci USA.
92:9692–9696. 1995; View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Begni B, Brighina L, Fumagalli L, Andreoni
S, Castelli E, Francesconi C, Del Bo R, Bresolin N and Ferrarese C:
Altered glutamate uptake in peripheral tissues from Down syndrome
patients. Neurosci Lett. 343:73–76. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Reynolds GP and Warner CE: Amino acid
neurotransmitter deficits in adult Down's syndrome brain tissue.
Neurosci Lett. 94:224–227. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Gregor P, Reeves RH, Jabs EW, Yang X,
Dackowski W, Rochelle JM, Brown RH Jr, Haines JL, O'Hara BF, Uhl
GR, et al: Chromosomal localization of glutamate receptor genes:
Relationship to familial amyotrophic lateral sclerosis and other
neurological disorders of mice and humans. Proc Natl Acad Sci USA.
90:3053–3057. 1993; View Article : Google Scholar : PubMed/NCBI
|