Introduction
Liver cancer is one of the most common types of
cancer in the world, with an estimated 782,500 new liver cancer
cases and 745,500 mortalities occurring worldwide during 2012; ~50%
of the total number of cases and mortalities were in China
(1). The majority (70–90%) of
primary liver cancer cases worldwide are hepatocellular carcinoma
(HCC) (2). The development of HCC
is the result of a complex, multi-step process associated with
genetic and epigenetic alterations (3,4).
Therefore, studies are required to elucidate the mechanism of HCC,
and novel strategies for early diagnosis, prediction of the
prognosis, and treatment of patients.
More than 2,000 microRNAs (miRNAs/miRs) have been
discovered in humans and they serve an important role in biological
processes by post-transcriptionally regulating protein-coding genes
(5). A number of studies have
associated miRNAs with numerous functions in tumorigenesis,
including cell proliferation, differentiation, apoptosis, invasion,
metastasis, autophagy (6),
epithelial-mesenchymal transition (EMT) (7), lipogenesis (8) and epigenetics (9). The involvement of miRNAs in HCC has
been demonstrated; aberrant miRNA expression may affect a number of
cancer-associated pathways, including cellular tumor antigen p53,
Ras/mitogen-activated protein kinase, phosphatidylinositol
3-kinase/RAC-α serine/threonine protein kinase/protein kinase mTOR,
Wnt/β-catenin, and transforming growth factor-β (10). Therefore, analysis of the
dysregulation of miRNAs in HCC, and the interactions between
targets proteins and miRNAs is of importance.
miR-340 has been identified to be a tumor suppressor
in many types of tumor, including colorectal cancer (11), ovarian cancer (12) and breast cancer (13). According to previous studies,
miR-340 exerts tumor inhibitory functions by suppressing
proliferation, migration, invasion and inducing apoptosis (13,14).
Notably, it was reported that miR-340 impaired the growth of
colorectal cancer by counteracting the Warburg effect (15); similarly, miR-340 may upregulate
the expression of glucose transporter-1 and result in an increase
in lactate secretion and glucose uptake in oral squamous cell
carcinoma (16). In the present
study, the expression of miR-340 in HCC cell lines and clinically
resected human HCC tissues was evaluated, in addition to
investigating the biological function of miR-340 in HCC
development. The roles of miR-340 in HCC development and the
underlying mechanism were investigated. The results of the present
study demonstrated that miR-340 inhibited HCC cell proliferation,
migration and invasion, and induced apoptosis, in addition to
downregulating the expression of S-phase kinase-associated protein
2 (SKP2), which may be a potential therapeutic application of
miR-340 in patients with HCC.
Materials and methods
Ethics statement
All patients and their families agreed to
participate in the present study and gave written informed consent.
The present study was approved by the institutional ethics board of
The First Affiliated Hospital of Chongqing Medical University
(Chongqing, China) and complied with the Declaration of
Helsinki.
Cell lines and cell culture
The human HCC cell lines SMMC-7721, Hep3B and
Bel-7402, in addition to normal human hepatocyte HL-7702 cells,
were purchased from the China Center for Type Culture Collection
(Wuhan, China). Hep3B cells were cultured in Minimum Essential
Medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA),
and the other cells were cultured in RPMI-1640 medium (1X; Gibco;
Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum
(FBS; PAN-Biotech GmbH, Aidenbach, Germany) at 37°C in a humidified
chamber supplemented with 5% CO2.
Hepatocellular carcinoma tissues
A total of 45 frozen primary tumor samples and
corresponding non-cancerous samples (located >3 cm away from the
tumor) were obtained from patients undergoing hepatectomy at The
First Affiliated Hospital of Chongqing Medical University between
September 2015 and March 2016. Patients (9 female and 36 male) were
aged between 22 and 78 years and were at the following tumor
stages: 4 cases of stage I; 10 cases of stage II; and 31 cases of
stage III. Tissue samples were snap frozen in liquid nitrogen at
the time of surgery and stored at −80°C. The tumor, node,
metastasis (TNM) classification of the Union for International
Cancer Control was used. None of the patients received radiotherapy
or chemotherapy prior to surgery.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
miRNA and mRNA was extracted from cell lines or
tissue using Hipure Universal miRNA kit (Magen, Guangzhou, China;
http://www.magentec.com.cn), according
to the manufacturer's protocol. The primers for miRNA and mRNA were
produced using All-in-One™ miRNA qPCR Primer and All-in-One™ mRNA
qPCR Primer (GeneCopoeia, Inc., Rockville, MD, USA), and the
sequences were: miR-340 forward, 5-'GCG GTT ATA AAG CAA TGA GA-3′
and reverse, 5′-GTGCGTGTCGTGGAGTCG-3′; U6 forward,
5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′;
SKP2 forward, 5′-AGTCTCTATGGCAGACCTTAGACC-3′ and reverse,
5′-TTTCTGGAGATTCTTTCTGTAGCC-3′; and GAPDH forward,
5′-CAGTCAGCCGCATCTTCTTTT-3′ and reverse, 5′-GTGACCAGGCGCCCAATAC-3′.
The synthesis of cDNA from miRNA and mRNA used All-in-One™ miRNA
First-Strand cDNA Synthesis and All-in-One™ First-Strand cDNA
Synthesis kits (GeneCopoeia, Inc.). The reaction conditions for
miRNA were as follows: 37°C for 60 min; 85°C for 5 min; 4°C
holding, and the reaction system had a volume of 20 µl. The
reaction conditions for mRNA were as follows: 60°C for 5 min, with
a reaction system of 13 µl; 37°C for 60 min; 85°C for 5 min; 4°C
holding, and the reaction system had a volume of 25 µl. All steps
were performed according to the manufacturer's protocol. qPCR was
performed in triplicate for each case using All-in-One™ miRNA kit
and All-in-One™ qPCR Mix (GeneCopoeia, Inc.), respectively, on a
CFX96™ Real-Time System (Bio-Rad Laboratories, Inc., Hercules, CA,
USA). The temperature protocol for the reaction was as follows:
95°C for 10 min; 40 cycles at 95°C for 10 sec and 60°C for 20 sec,
and 72°C for 15 sec. miRNA expression was measured using Cq
(threshold cycle). The 2−ΔΔCq method for relative
quantitation of gene expression was used to determine miRNA
expression levels (17). The ΔCq
was calculated by subtracting the Cq of U6 RNA from the Cq of the
miRNA of interest. Fold changes were generated using the equation
2−ΔΔCq. The expression level of U6 and GAPDH was used as
the internal control for miRNA and mRNA expression,
respectively.
Oligonucleotide transfection
The miR-340 mimics (pLVX-ZsGreen-Puro-miR-340),
miR-340 inhibitor (pLVX-tdTomato-Puro-miR-340 inhibitor) and their
controls (pLVX-ZsGreen and pLVX-tdTomato) were synthesized by
GeneCopoeia, Inc. and transfected into the cells at a final
oligonucleotide concentration of 1 µg/ml. All cell transfections
were performed using EndoFection™-Max (GeneCopoeia, Inc.),
according to the manufacturer's protocol. The density of cells was
3×107 per 6-well plate and subsequent experimentation
was performed 48 h after transfected.
Cell proliferation and apoptosis
For the analysis of cell proliferation, cells were
seeded into 96-well plates at 1×105 cells/well. Cells
were incubated in 10% Cell Counting kit-8 (CCK-8; Dojindo Molecular
Technologies, Inc., Kumamoto, Japan) solution and × diluted in
normal culture medium at 37°C until visual color conversion was
apparent. The SMMC-7721 proliferation rate was determined at 24, 48
and 72 h following transfection. The absorbance in each well was
measured with a VARIOSKAN FLASH (Thermo Fisher Scientific, Inc.) at
450 nm. For the analysis of cellular apoptosis, cells were
transfected with miR-340 mimic, miR-340 inhibitor and their
corresponding control for 24 h. Following transfection, cells were
harvested and washed twice in PBS; miR-340 mimic and its control
were stained with an Annexin V-phycoerythrin/7-aminoactinomycin D
apoptosis kit [Multi Sciences (Lianke) Biotech. Co., Ltd.,
Hangzhou, China], and miR-340 inhibitor and its control was stained
with Annexin V-fluorescein isothiocyanate/propidium iodide
apoptosis kit [Multi Sciences (Lianke) Biotech. Co., Ltd.],
following the manufacturer's protocol, using a BD Influx Flow
Cytometer & Cell Sorter and the results were analyzed using BD
FACS software version 1.0 (BD Biosciences, Franklin Lake, NJ,
USA).
Transwell invasion assay
For analysis of cell migration and invasion,
Transwell chambers (8-µm pore size; EMD Millipore, Billerica, MA,
USA) and Matrigel (diluted 1:9) (Corning Incorporated, Corning, NY,
USA) was used. Cells were incubated at 37°C on 6-well plates for 8
h A total of 12 h subsequent to transfection, cells were starved
for 12 h. For migration assay, 2×105 cells were
suspended in 200 µl serum-free RPMI-1640 and seeded in the top
chamber, and 350 µl RPMI-1640 containing 10% FBS was added to the
lower chamber. For invasion assay, following the above process the
Matrigel on the top of chamber was allowed to solidify (~5 h at
37°C). Following migration or invasion assay was 24 h of incubation
at 37°C in a 5% CO2 atmosphere, after which the cells on
the upper surface of the membrane were removed and fixed in 4%
paraformaldehyde ~30 min, followed by staining with 0.5% crystal
violet ~60 min, all at 25°C. Cells were counted using an upright
microscope (Ci-L; Nikon Corporation, Tokyo, Japan; magnification,
×200).
Western blotting
Prior to extracting the protein, SMMC-7721 cells
were incubated at 37°C on 6-well plates for 48 h following
transfection. Cells were washed twice in cold PBS and lysed in
radioimmunoprecipitation assay lysis buffer (Beyotime Institute of
Biotechnology, Haimen, China) with protease inhibitor cocktail
(Beyotime Institute of Biotechnology). The protein concentration of
cell lysates was quantified using a bicinchoninic acid kit
(Beyotime Institute of Biotechnology), and equal quantities (30 µg)
of protein were separated using SDS-PAGE on 10% gels, followed by
transfer to a polyvinylidene fluoride membrane (EMD Millipore). The
membranes were blocked in 8% skimmed milk diluted with TBS with
Tween-20 [Tris-HCl 24.23 g/l, NaCl 80.06 g/l (pH 7.5), and 0.1%
Tween (1 ml)] at room temperature for 1 h and incubated overnight
at 4°C with primary anti-SKP2 antibody (RLT-4311) or primary
anti-GAPDH antibody (RLM-3215; both 1:500; Ruiyingbio, Suzhou,
China). The membranes were subsequently incubated with goat
anti-rabbit IgG secondary antibody (RS0002) conjugated to
horseradish peroxidase (HRP; 1:5,000; Ruiyingbio) for 3 h at 25°C.
The proteins were visualized using Chemiluminescent HRP Substrate
(EMD Millipore). The density was measured using Image Lab™ Software
version 4.1 (Bio-Rad Laboratories, Inc.), and values were
normalized to the densitometric values of GAPDH in each sample.
Online prediction for miRNA target
genes
Using online prediction software for miRNA target
genes, including PicTar (http://pictar.mdc-berlin.de/), miRanda (http://www.microrna.org/microrna/home.do) and
Targetscan (http://www.targetscan.org/vert_71/).
Statistical analysis
Each experiment was repeated at least three times.
Data are presented as the mean ± standard deviation. Statistical
analyses were performed using Student's t-test, differences among
groups were analyzed using one-way analysis of variance followed by
least significant difference post hoc analysis, and the
associations between miR-340 expression and clinical pathological
factors were analyzed using the χ2 test. P<0.05 was
considered to indicate a statistically significant difference. All
statistical analyses were performed using SPSS 19.0 software (IBM
Corp., Armonk, NY, USA).
Results
Expression of miR-340 in clinical
patients with HCC and HCC cell lines
In order to examine the expression of miR-340 in
HCC, the expression of miR-340 in the normal hepatocyte line
HL-7702 and hepatocellular carcinoma cell lines Hep3B, SMMC-7721
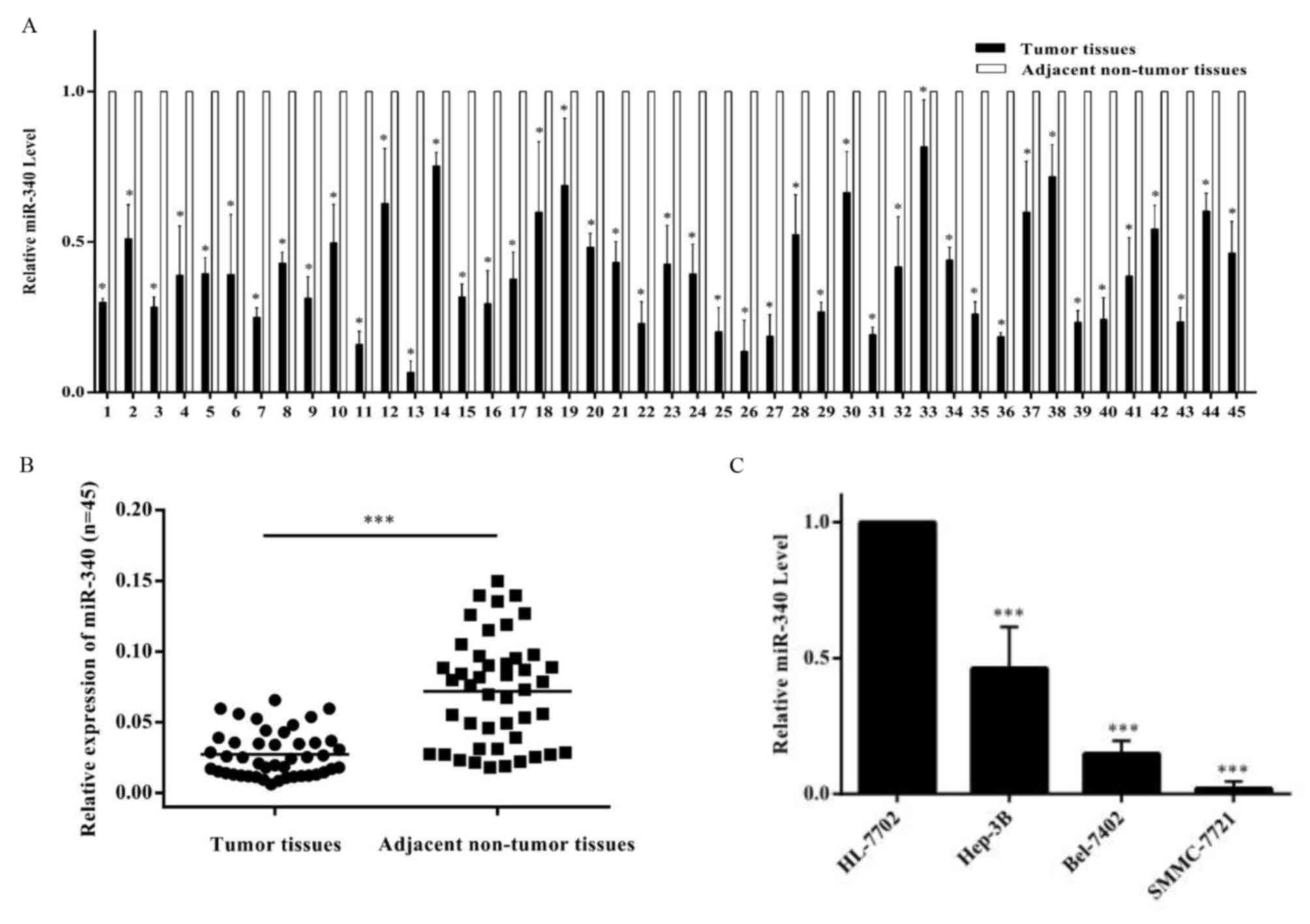
and Bel-7402 was determined (Fig.
1). Compared with HL-7702, RT-qPCR analysis revealed that the
expression of miR-340 was decreased in the three hepatocellular
carcinoma cell lines (Fig. 1C). In
tissues, it was observed that the miR-340 level in tumor tissues
was consistently decreased compared with adjacent non-tumor tissues
(Fig. 1A). It was observed that
the expression of miR-340 in tumor tissues was significantly
decreased compared with adjacent non-tumor tissues (Fig. 1B). Further analysis demonstrated
that patients with HCC who were hepatitis B virus (HBV)+
or exhibited copy numbers of HBV DNA >1.0×103 had
decreased expression of miR-340. In addition, the miR-340 level was
decreased in patients who with larger tumor sizes (≥5 cm) and
higher TNM grades (Table I). The
results of the present study demonstrated that miR-340 may be a
tumor suppressor which is associated with HCC pathogenesis, and
miR-340 may correlate with disease severity.
 | Table I.Association between miR-340 expression
and clinicopathological features. |
Table I.
Association between miR-340 expression
and clinicopathological features.
|
|
| miR-340
expression |
|
|---|
|
|
|
|
|
|---|
| Feature | No. patients
(n=45) | Low | High | P-value |
|---|
| Age, years |
|
|
|
|
|
<40 | 5 | 3 | 2 | 0.751 |
| ≥40 | 40 | 21 | 19 |
|
| Sex |
|
|
|
|
| Male | 36 | 21 | 15 | 0.179 |
|
Female | 9 | 3 | 6 |
|
| HBsAg |
|
|
|
|
| + | 32 | 21 | 11 | 0.010 |
| − | 13 | 3 | 10 |
|
| HBV DNA |
|
|
|
|
|
<1.0×103 | 23 | 7 | 16 | 0.002 |
|
≥1.0×103 | 22 | 17 | 5 |
|
| AFP, µg/l |
|
|
|
|
|
<400 | 33 | 15 | 18 | 0.079 |
| ≥400 | 12 | 9 | 3 |
|
| ALT, U/l |
|
|
|
|
|
<40 | 19 | 11 | 8 | 0.600 |
|
≥40 | 26 | 13 | 13 |
|
| AST, U/l |
|
|
|
|
|
<40 | 15 | 8 | 7 | 1.000 |
|
≥40 | 30 | 16 | 14 |
|
| Tumor size, cm |
|
|
|
|
|
<5 | 13 | 2 | 11 | 0.001 |
| ≥5 | 32 | 22 | 10 |
|
| Cirrhosis |
|
|
|
|
|
Yes | 32 | 20 | 12 | 0.053 |
| No | 13 | 4 | 9 |
|
| TNM staging |
|
|
|
|
| I | 4 | 0 | 4 | 0.002 |
| II | 10 | 2 | 8 |
|
|
III | 31 | 22 | 9 |
|
miR-340 inhibits HCC cell
proliferation and induces apoptosis
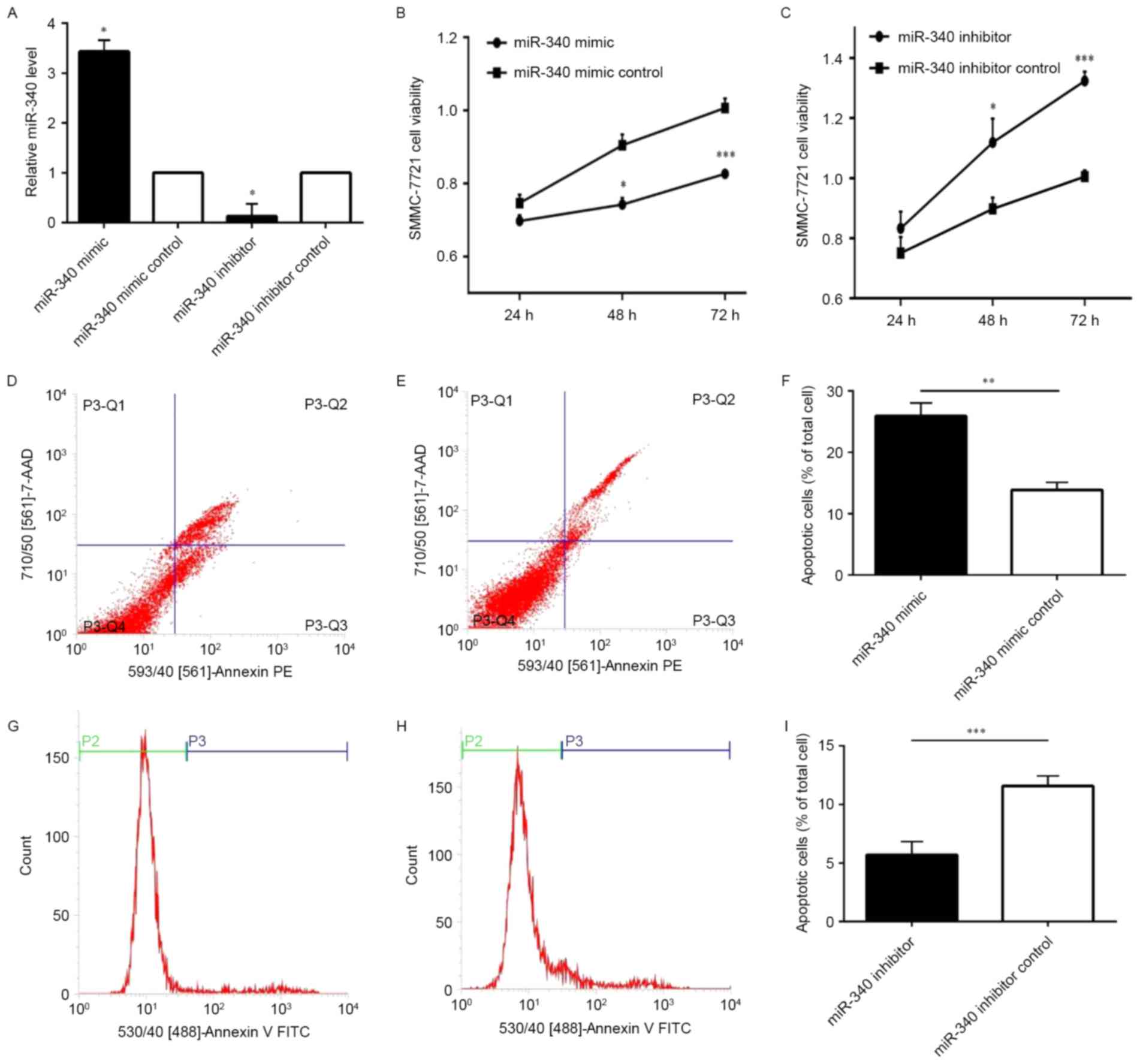
Using RT-qPCR analysis, the expression of miR-340 in
SMMC-7721 following transfection was confirmed. The results of the
present study demonstrated that the expression of miR-340 in
SMMC-7721 cells transfected with the miR-340 mimic was increased
compared with the miR-340 mimic control. Following transfection of
the miR-340 inhibitor and miR-340 inhibitor control, the expression
of the latter was increased compared with the former (Fig. 2A). The CCK-8 assay demonstrated
that the proliferation of SMMC-7721 cells transfected with miR-340
mimic was significantly decreased compared with the miR-340 mimic
control group (Fig. 2B).
Simultaneously, the proliferation ratio of SMMC-7721 cells
transfected with miR-340 inhibitor was significantly increased
compared with the miR-340 inhibitor control group (Fig. 2C). Using flow cytometry, it was
observed that the apoptosis ratio of SMMC-7721 cells transfected
with miR-340 mimic was significantly increased than miR-340 mimic
control group (Fig. 2D-F);
however, the apoptosis ratio of SMMC-7721 cells transfected with
miR-340 inhibitor was significantly decreased compared with the
miR-340 inhibitor control (Fig.
2G-I). The results of the present study demonstrated that
miR-340 was able to repress cell proliferation and induce
apoptosis.
miR-340 inhibits HCC cell migration
and invasion
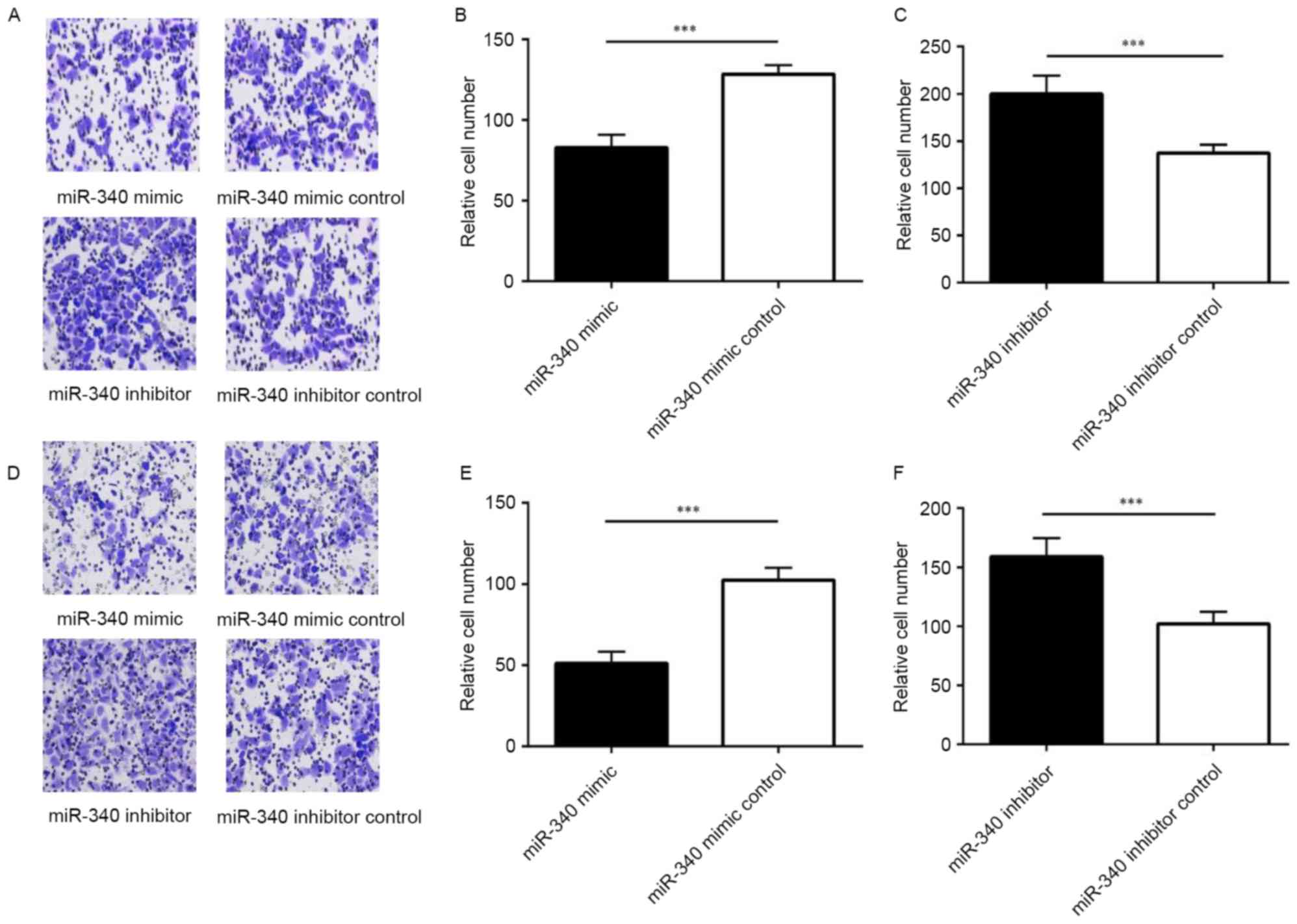
Using a Transwell invasion assay, it was observed
that the migration of SMMC-7721 cells with miR-340 mimics
significantly decreased compared with the miR-340 mimic control
group, while the migration of SMMC-7721 cells transfected with
miR-340 inhibitor increased compared with the miR-340 inhibitor
control group (Fig. 3A-C).
Following injection of the Matrigel into the chamber, the
invasiveness of SMMC-7721 cells transfected with miR-340 mimic was
decreased compared with the miR-340 mimic control group. In
addition, the invasiveness of cells transfected with miR-340
inhibitor was increased compared with the miR-340 inhibitor control
group (Fig. 3D-F). Therefore,
miR-340 was able to inhibit migration and invasion in the SMMC-7721
cell line.
miR-340 downregulates the expression
of SKP2
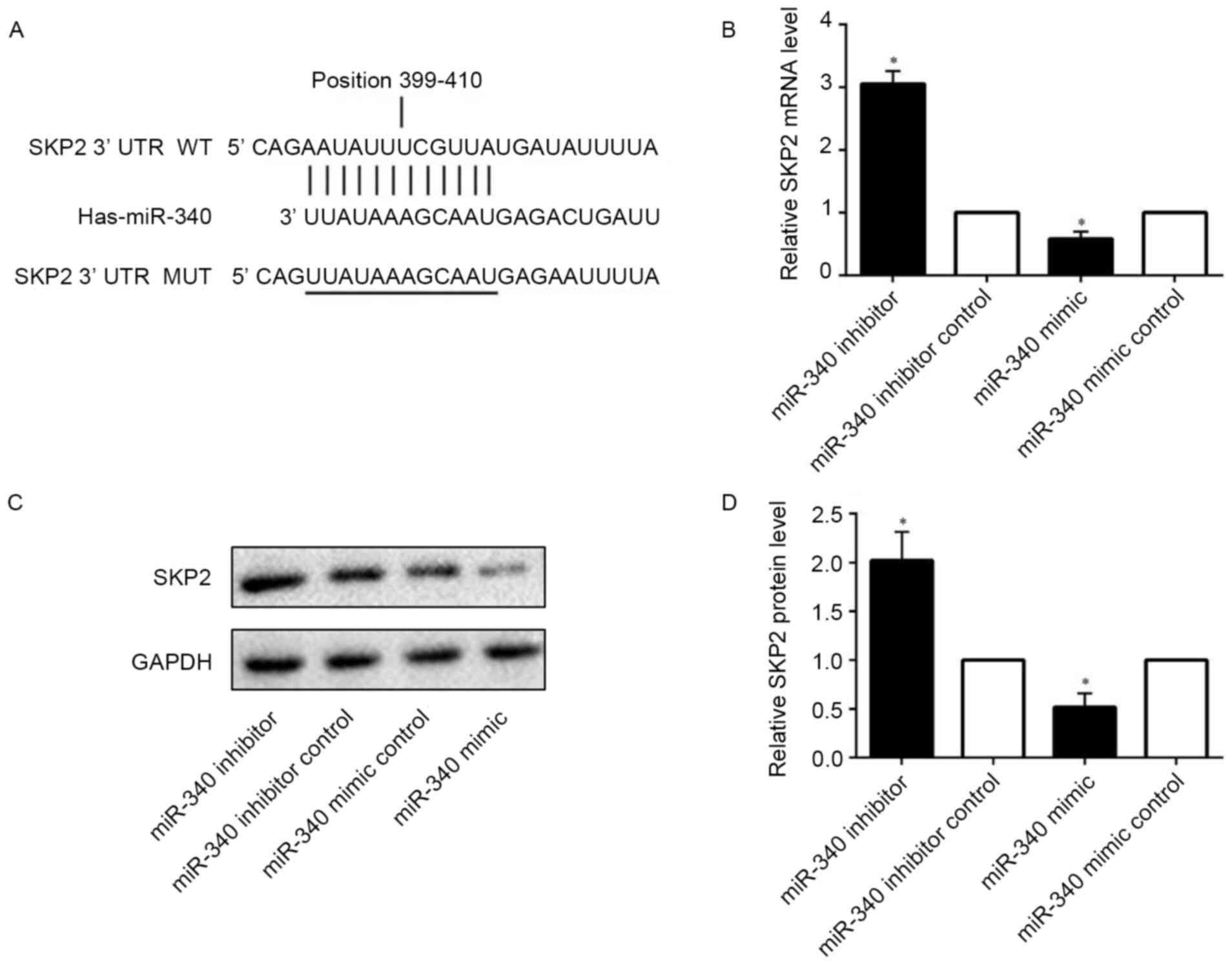
Online prediction software indicated that SKP2,
which is associated with cell proliferation and apoptosis, is a
target gene of miR-340. Bioinformatic analysis demonstrated that
the SKP2 3′ untranslated region (UTR) contains a seed sequence
which complements miR-340. This suggested that miR-340 may bind
strongly to the SKP2 3′UTR (Fig.
4A). Using RT-qPCR analysis, it was observed that the level of
SKP2 mRNA was decreased following transfection of miR-340 mimic
into SMMC-7721 cells; by contrast, transfecting miR-340 inhibitor
into SMMC-7721 cells was able to increase the level of SKP2 mRNA
(Fig. 4B). The protein level of
SKP2 was detected by western blotting (Fig. 4C). Compared with the corresponding
control group, miR-340 mimic inhibited the level of SKP2; by
contrast, miR-340 inhibitor increased the level of SKP2 (Fig. 4D). Therefore, miR-340 was able to
influence the expression of SKP2.
Discussion
Considering the importance of miRNAs in the
post-transcriptional regulation gene expression, a number of
studies have demonstrated their role in human carcinogenesis.
Previously, the function of miRNA in tumors has been elucidated,
and it has been demonstrated that miRNAs may be either oncogenic or
anti-oncogenic, acting as therapeutic targets and prognostic tools
(10,18). In the present study, a novel cancer
suppressor, miR-340, was identified in human HCC. It was observed
that the expression of miR-340 was decreased in three human HCC
cell lines compared with normal human hepatocytes. Simultaneously,
miR-340 was detected to be downregulated in 45 pairs of HCC tissues
compared with adjacent normal controls. Statistical analysis
demonstrated that the miR-340 level was decreased in tumor tissues
compared with corresponding adjacent non-tumor tissues. Patients
with HCC who were HBV+ or exhibited copy numbers of HBV
DNA >1.0×103 had decreased expression of miR-340. In
addition, the miR-340 level was decreased in patients with larger
tumor sizes (≥5 cm) and higher TNM grades. The results of the
present study demonstrated that the expression of miR-340 may be
associated with HBV infection. A previous study reported that HBV X
protein may activate nuclear factor-κB, which may combine with the
promoter region or enhancement region of miRNA to induce
transactivation (19). Therefore,
further studies are required to verify the association between
miR-340 and HBV infection. miR-340 significantly suppressed
SMMC-7721 proliferation and induced apoptosis. The results of the
present study indicated that miR-340 suppressed migration and
invasion in HCC cell lines. SKP2 was considered to be a potential
target of miR-340. A previous study detected the overexpression of
SKP2 in late-stage HCC, and overexpression predicted poor survival
outcomes (20). Consequently, the
present study aimed to investigate whether miR-340 may suppress the
expression of SKP2. Using RT-qPCR and western blot analyses, it was
observed that upregulating the expression of miR-340 may inhibit
SKP2 mRNA and protein expression, while downregulating the
expression of miR-340 increased its expression at the mRNA and
protein levels. However, the mechanism underlying the
overexpression of SKP2 regulated by miR-340 requires further
investigation.
The antitumor effect of miR-340 has been previously
identified. As a tumor suppressor, the expression of the miRNA may
be increased in adjacent normal tissues compared with tumor tissue,
in addition to in normal cell lines compared with cancer cell
lines. miR-340 has been detected at a lower expression level in
tumor tissues compared with adjacent normal tissues, in addition to
in cancer cell lines compared with normal cell lines, including
osteosarcoma, glioblastoma and liver metastasis of colorectal
cancer; in addition, the miR-340 expression level in these types of
cancer may predict tumor progression and prognosis (11,21,22).
Notably, in melanoma, miR-340 acts as a tumor suppressor although
its expression in melanoma cell lines is increased compared with
normal melanocyte cell lines (23). This previous result indicated that
the roles of miR-340 may not be consistent with the pattern of its
expression in melanoma cells; consequently, when assessing the
function of a miRNA, the level of its expression may not be the
sole consideration. miR-340 is involved in oncogenesis in various
ways; miR-340 inhibits proliferation, migration and invasion, and
induces apoptosis, in ovarian cancer, prostate cancer, laryngeal
squamous cell carcinoma and glioblastoma (12,22,24–26).
The results of the present study demonstrated the same function of
miR-340 in HCC compared with these previous studies. miR-340 has
been demonstrated to mediate glycometabolism by down-regulating
glucose transporter-1 in oral squamous cell carcinoma (16) and accelerating the transformation
of pyruvate kinase PKM (PKM)2 to PKM1, and the PKM1/PKM2 ratio is
able to repress the rate of glycolysis in colorectal cancer
(15), ultimately resulting in
resistance to the Warburg effect. Whether this resistance presents
in HCC requires further study. Overexpression of miR-340 was
demonstrated to reduce the expression of mesenchymal phenotypic
markers while increasing the expression of epithelial phenotypic
markers, thereby inhibiting EMT in breast cancer cells (27). Autophagy may be observed in a
number of physiological and pathological conditions, and it has
been reported that miR-340 is able to repress E3 ubiquitin-protein
ligase XIAP, which is an important anti-autophagy factor in tumor
cells, and thereby promotes autophagy (22,28).
miR-340 serves important roles in carcinogenesis,
and the results of the present study indicated that miR-340
inhibits HCC cell proliferation, migration and invasion, in
addition to inducing apoptosis. The mechanism underlying the
function of miR-30 in HCC may be accounted for by the
downregulation of SKP2, which requires further study. Due to the
differential expression of miR-340 in HCC tumor tissues compared
with normal adjacent tissues, it may be suggested that miR-340 may
have potential for predicting tumor progression and prognosis.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
McGlynn KA, Petrick JL and London WT:
Global epidemiology of hepatocellular carcinoma: An emphasis on
demographic and regional variability. Clin Liver Dis. 19:223–238.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zucman-Rossi J, Villanueva A, Nault JC and
Llovet JM: Genetic landscape and biomarkers of hepatocellular
carcinoma. Gastroenterology. 149:1226–1239.e4. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Szabo G and Bala S: MicroRNAs in liver
disease. Nat Rev Gastroenterol Hepatol. 10:542–552. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wu SY, Lan SH and Liu HS: Autophagy and
microRNA in hepatitis B virus-related hepatocellular carcinoma.
World J Gastroenterol. 22:176–187. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang J and Ma L: MicroRNA control of
epithelial-mesenchymal transition and metastasis. Cancer Metastasis
Rev. 31:653–662. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
de Cedrón MG and de Molina AR:
Microtargeting cancer metabolism: Opening new therapeutic windows
based on lipid metabolism. J Lipid Res. 57:193–206. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Peschansky VJ and Wahlestedt C: Non-coding
RNAs as direct and indirect modulators of epigenetic regulation.
Epigenetics. 9:3–12. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Callegari E, Elamin BK, Sabbioni S,
Gramantieri L and Negrini M: Role of microRNAs in hepatocellular
carcinoma: A clinical perspective. Onco Targets Ther. 6:1167–1178.
2013.PubMed/NCBI
|
|
11
|
Takeyama H, Yamamoto H, Yamashita S, Wu X,
Takahashi H, Nishimura J, Haraguchi N, Miyake Y, Suzuki R, Murata
K, et al: Decreased miR-340 expression in bone marrow is associated
with liver metastasis of colorectal cancer. Mol Cancer Ther.
13:976–985. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li P, Sun Y and Liu Q: MicroRNA-340
induces apoptosis and inhibits metastasis of ovarian cancer cells
by inactivation of NF-x03BA;B1. Cell Physiol Biochem. 38:1915–1927.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mohammadi-Yeganeh S, Paryan M, Arefian E,
Vasei M, Ghanbarian H, Mahdian R, Karimipoor M and Soleimani M:
MicroRNA-340 inhibits the migration, invasion, and metastasis of
breast cancer cells by targeting Wnt pathway. Tumour Biol.
37:8993–9000. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fernandez S, Risolino M, Mandia N, Talotta
F, Soini Y, Incoronato M, Condorelli G, Banfi S and Verde P:
miR-340 inhibits tumor cell proliferation and induces apoptosis by
targeting multiple negative regulators of p27 in non-small cell
lung cancer. Oncogene. 34:3240–3250. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sun Y, Zhao X, Zhou Y and Hu Y: miR-124,
miR-137 and miR-340 regulate colorectal cancer growth via
inhibition of the Warburg effect. Oncol Rep. 28:1346–1352. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xu P, Li Y, Zhang H, Li M and Zhu H:
MicroRNA-340 mediates metabolic shift in oral squamous cell
carcinoma by targeting glucose transporter-1. J Oral Maxillofac
Surg. 74:844–850. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Negrini M, Ferracin M, Sabbioni S and
Croce CM: MicroRNAs in human cancer: From research to therapy. J
Cell Sci. 120:1833–1840. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang X, Liu S, Hu T, Liu S, He Y and Sun
S: Up-regulated microRNA-143 transcribed by nuclear factor kappa B
enhances hepatocarcinoma metastasis by repressing fibronectin
expression. Hepatology. 50:490–499. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lee SW, Li CF, Jin G, Cai Z, Han F, Chan
CH, Yang WL, Li BK, Rezaeian AH, Li HY, et al: Skp2-dependent
ubiquitination and activation of LKB1 is essential for cancer cell
survival under energy stress. Mol Cell. 57:1022–1033. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cai H, Lin L, Cai H, Tang M and Wang Z:
Combined microRNA-340 and ROCK1 mRNA profiling predicts tumor
progression and prognosis in pediatric osteosarcoma. Int J Mol Sci.
15:560–573. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huang D, Qiu S, Ge R, He L, Li M, Li Y and
Peng Y: miR-340 suppresses glioblastoma multiforme. Oncotarget.
6:9257–9270. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Strong AM Poenitzsch, Setaluri V and
Spiegelman VS: MicroRNA-340 as a modulator of RAS-RAF-MAPK
signaling in melanoma. Arch Biochem Biophys. 563:118–124. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang K, Tang Y, He L and Dai Y:
MicroRNA-340 inhibits prostate cancer cell proliferation and
metastasis by targeting the MDM2-p53 pathway. Oncol Rep.
35:887–895. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wei P, Qiao B, Li Q, Han X, Zhang H, Huo Q
and Sun J: microRNA-340 suppresses tumorigenic potential of
prostate cancer cells by targeting high-mobility group
nucleosome-binding domain 5. DNA Cell Biol. 35:33–43. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yu W, Zhang G, Lu B, Li J, Wu Z, Ma H,
Wang H and Lian R: MiR-340 impedes the progression of laryngeal
squamous cell carcinoma by targeting EZH2. Gene. 577:193–201. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hou LK, Yu Y, Xie YG, Wang J, Mao JF,
Zhang B, Wang X and Cao XC: miR-340 and ZEB1 negative feedback loop
regulates TGF-β- mediated breast cancer progression. Oncotarget.
7:26016–26026. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Huang X, Wu Z, Mei Y and Wu M: XIAP
inhibits autophagy via XIAP-Mdm2-p53 signalling. EMBO J.
32:2204–2216. 2013. View Article : Google Scholar : PubMed/NCBI
|