Introduction
Living organisms on the earth exhibit circadian
rhythms with a 24-h alternating pattern, allowing them to adapt to
the daily cycle of light and dark and change of seasons (1). At the molecular level, circadian
rhythms are generated by a set of clock genes and proteins, which
include Aryl hydrocarbon receptor nuclear translocator-like protein
1, human circadian locomoter output cycles protein kaput (hCLOCK)
and period circadian protein homolog (PER)1, 2 and 3. These clock
genes work in accurate feedback loops and keep the levels of
certain mRNAs and proteins oscillating throughout the 24 h period
(2). It has been demonstrated that
>10% of all genes in the mammalian genome are under the
regulation of circadian genes (3).
As core circadian genes regulate gene expression associated with
apoptosis, cell cycle and other signaling pathways in cells, in
turn, they also regulate many physiological functions such as body
temperature, blood pressure, immune responses and hormone
biosynthesis (4). Therefore, these
genes may be important in maintaining the regular biological status
of the organism. There is increasing evidence indicating that
dysfunction of hCLOCK links to the pathogenesis of cancer. For
example, attenuation of all three PER genes may promote the
tumorigenesis of breast cancer (5). In contrast, the expression of hCLOCK
is upregulated in breast cancer tissues when compared to adjacent
normal tissues or healthy ones (1). There is evidence that ablation of
PER2 leads to the development of malignant lymphomas (6). There is also evidence demonstrating
that hCLOCK is involved in pathogenesis of colorectal carcinomas
(CRCs). In our previous studies, it was demonstrated that the
expression level of human (h)PER2 was significantly reduced in
human CRC tissues than in paired surrounding non-cancerous tissues.
Deregulation of hPER2 associated with patient age, histological
grade and TNM stage (7). In
contrast, higher levels of hCLOCK expression were observed in human
CRC tissues when compared with paired non-cancerous ones. hCLOCK
expression was significantly higher in late-stage Dukes' grade
tumors, or poorly differentiated, and in 64.3% of tumor cases with
lymph node metastasis (8).
Although mammalian circadian rhythms have been
extensively studied in recent years, most of them were based on
post-translational modification or negative regulation at the
transcriptional level (9–11). Several groups have focused research
attention on microRNAs (miRNAs/miRs), a class of small noncoding
RNAs that serve essential roles in post-transcriptional regulation
of gene expression (12–13). As they target tumor suppressor
genes or oncogenes, alterations of miRNAs have also been reported
in the initiation and progression of CRCs. For example, miR-124 is
frequently reported to be dysregulated in CRCs and serves a role of
suppressing CRCs by targeting specific genes such as signal
transducer and activator of transcription 3 (STAT3), structural
maintenance of chromosomes protein 4 and RelA-associated inhibitor
(14–17). However, the mechanisms of
cross-talk between miR-124 and circadian genes in regulation of
CRCs are poorly understood.
The present study demonstrated that the expression
level of hCLOCK, a core circadian gene, is significantly increased
in high-grade human CRCs tissues. In contrast, miR-124 is similarly
attenuated in these tissues samples. In the LOVO CRC cell line,
upregulation of miR-124 significantly inhibited expression of
hCLOCK, and consequently inhibited invasion and proliferation,
while at the same time promoting apoptosis of LOVO cells.
Furthermore, it was revealed that miR-124 deregulated hCLOCK by
directly targeting its 3′-untranslated region (3′-UTR). In
conclusion, the present study demonstrated that the attenuation of
miR-124 contributed elevated hCLOCK protein expression in high
grade CRCs tissue, and promoted progression of human CRCs.
Materials and methods
Clinical samples
Resected CRC samples and paired non-cancerous
tissues (males, 28; females, 22; n=50) were obtained from patients
undergoing surgery for colorectal cancer in Huashan Hospital
(Shanghai, China). All patients had a definitive pathological
diagnosis and received neither chemotherapy nor radiotherapy prior
to surgery. Following surgical removal, the samples were preserved
in liquid nitrogen immediately and then moved to −80°C for long
term storage. The age of the patients, tumor site, tumor type,
histological grade and TNM stage according to American Joint
Committee on Cancer classification were recorded (18). Written informed consent was
obtained from all patients used in the present study.
Immunohistochemical staining
Tissues were fixed in formalin, embedded in paraffin
and cut into 4-µm sections. Following deparaffinization, sections
were rehydrated and subjected to antigen retrieval by microwaving
in 0.01 M sodium citrate (pH 6) for 10 min. Sections were incubated
at 4°C overnight with monoclonal antibodies against hCLOCK (#5157;
1:100; Cell signaling Technology, Inc., Danvers, MA, USA). After a
brief wash with PBS, horseradish peroxidase-conjugated secondary
antibody (#A31460, 1:500; Thermo Fisher Scientific, Inc., Waltham,
MA, USA) was added for 60 min at 37°C. After several washes with
PBS, staining was achieved using 3,3′-diaminobenzidine for 5~10
min. Finally, slides were counterstained with Mayer's hemalum and
mounted. Protein staining was evaluated under a light microscope at
×400 magnification. Staining intensity was scored manually by two
independent experienced pathologists as follows: 0=no staining,
1=weak staining, 2=moderate staining and 3=strong staining. Tumor
cells in five fields were randomly selected and scored based on the
percentage of positively stained cells (0–100%). The final score
was calculated by multiplying the intensity score with the
percentage of positive cells.
Cell culture
The SW620, SW480, LOVO and HT-29 CRC cell lines were
purchased from the Chinese Academy of Sciences Institute (Shanghai,
China). The cell lines were cultured in Dulbecco's modified Eagle's
medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with
10% fetal bovine serum (Invitrogen; Thermo Fisher Scientific,
Inc.).
miRNA target prediction
TargetScan (19)
(http://www.targetscan.org), PicTar
(20) (http://www.pictar.org/) and miRNA.org
(21) (http://www.microrna.org) were used to predict the
interaction between miRNAs and the hCLOCK 3′-UTR.
Total RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Preparation of total RNA from CRC tissue samples and
cultured cell lines was performed using TRIzol reagent (Invitrogen;
Thermo Fisher Scientific, Inc.). The amount of total RNA was
determined by ultraviolet spectrophotometry. Total RNA was reverse
transcribed using a MicroRNA Reverse Transcription kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.) and a miR-124-specific
stem-loop primer purchased from GeneChem Inc. (Daejon, Korea). The
primer sequence for miR-124 was as follows:
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGGCATTC-3′. The PCR
reactions containing SYBR-green (Toyobo Co., Ltd., Osaka, Japan)
for mature miR-124 were amplified on a Corbett Real Time PCR
machine (Bio-Rad, USA). The reaction mixtures for PCR contained 1
µl cDNA, 4 µl 10 mmol/l primers (2 µl forward primers, 2 µl reverse
primers), 10 µl real-time PCR Master Mix (Toyobo Co., Ltd., Osaka,
Japan), and 5 µl H2O, into a final volume of 20 µl, and
run under the following conditions: One cycle at 95°C for 5 min,
and 40 cycles at 95°C for 5 sec, 60°C for 30 sec and 72°C for 5
sec. The sequences for qPCR primers were as follows: Forward,
5′-GCTAAGGCACGCGGTG-3′ and reverse, 5′-CAGTGCAGGGTCCGAGGT-3′. The
relative amounts of miR-124-3p were measured with the
2−ΔΔCq (22)
method.
Western blot analysis
The cells were lysed with radioimmunoprecipitation
assay buffer (Cell Signaling Technology, Inc.) and 1 mM
phenylmethanesulfonyl fluoride. The lysates were incubated at 4°C
for 10 min and centrifuged at 13,400 × g at 4°C for 15 min. Equal
amounts of lysate (25 µg protein per lane) were separated by 12%
SDS-PAGE and transferred to a polyvinylidene fluoride membrane (EMD
Millipore, Billerica, MA, USA). The membranes were blocked in 5%
non-fat skim milk/PBS with Tween-20 at room temperature for 2 h,
and probed with a primary antibody against hCLOCK (Cell signaling
Technology, Inc.; #5157; 1:1,000) at room temperature for 2 h. The
membranes were then incubated for 2 h at room temperature with a
horseradish peroxidase-conjugated secondary antibody (Thermo Fisher
Scientific, Inc.; #A31460; 1:10,000), followed by incubation with
enhanced chemiluminescence western blot detection reagent (GE
Healthcare, Chicago, IL, USA). The hCLOCK primary antibody was
purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX,
USA).
Lentiviral vector construction and
infection
A GV258 lentiviral vector, GV214 vector containing
miR-124 precursor sequences and a negative control (NC) were
purchased from GeneChem, Inc. The human miR-124 precursor gene was
PCR amplified from normal genomic DNA, and cloned into the GV258
lentiviral vector or GV214 for ectopic expression of miR-124.
Primers used for amplification were
5′-AGCTGTACAAGTAAGTTCTTATTCCATCTTCTACC-3′ (forward) and
5′-AGCTGTACAAGTAAGTTCTTATTCCATCTTCTACC-3′ (reverse: with
NheI site). LOVO cells were infected with miR-NC or mir-124
overexpression viruses and selected by puromycin, and were designed
as miR-124 LOVO and miR-NC LOVO, respectively.
Dual-luciferase reporter
construction
GV306 vectors with a fragment of the 3′-UTR of
hCLOCK gene (NM_004898) containing the recognition site for
miR-124-3p at nucleotide position (from nt 930 to nt1501, with
XbaI sites), the corresponding site-directed mutant
containing the mutated seed sequence and the NC were purchased from
GeneChem, Inc. The fragment was constructed via a generate gene
synthesis service (Shanghai GeneChem Co., Ltd., Shanghai, China).
The corresponding site-directed mutant containing the mutated seed
sequence (5′-GTGCCTT-3′ to 5′-CGTAGAG-3′) was also generated. The
wild type and mutated hCLOCK 3′-UTR fragments were then cloned into
pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega
Corporation, Madison, WI, USA) at XbaI/XbaI sites and termed
Luc-hCLOCK-wt and Luc-hCLOCK-mt, respectively. A luciferase vector
without hCLOCK 3′-UTR (Luc-hCLOCK-ctl) was used as experimental
control. All constructs were confirmed by DNA sequencing.
Luciferase assay
293T cells were seeded overnight in antibiotic-free
complete medium at 5×104 cells/well on a 24-well plate.
The cells were transfected with 2 µg miR-124-3p or miR-NC and 400
ng Luc-hCLOCK-NC, Luc-hCLOCK-wt or Luc-hCLOCK-mt for 48 h. The
co-transfection procedure for miRNAs and luciferase reporter
constructs was performed using Lipofectamine 2000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). A luciferase activity
assay was performed using the Dual-Luciferase Reporter Assay system
(Biotime, Shanghai, China) according to the manufacturer's
instructions. The levels of firefly luciferase activities were
obtained by normalizing to Renilla luciferase activities and
relative to a control.
In vitro cell proliferation
assays
Cell proliferation was determined by Cell Counting
kit-8 (Dojindo Molecular Technologies, Inc., Kumamoto, Japan)
assay. In brief, miR-124-LOVO, miR-NC-LOVO and LOVO cells were
seeded into 96-well plates at an density of 1×104 cells
per well. After 12, 24, 36, 48 and 72 h, 10 µl kit reagent was
added to each well, and 2 h later all plates were scanned by a
microplate reader (Thermo Fisher Scientific, Inc.) at 450 nm. Cell
proliferation was calculated on the basis of absorbency.
In vitro invasion assays
Cell invasion was analyzed by a Transwell Permeable
Supports system with 12-µm pores (Corning Incorporated, Corning,
NY, USA). Cells were observed under an inverted phase microscope.
Cells (2–3×105) were seeded onto upper inserts with a
Matrigel-coated membrane (BD Biosciences, Franklin Lakes, NJ, USA).
Cells were seeded in serum-free medium and transferred to 10% serum
media for 48 h. After removal of the non-invading cells, the
remaining cells were fixed, stained, eluted by 30% acetic acid,
scanned by a microplate reader (Thermo Fisher Scientific, Inc.) at
450 nm and finally calculated on the basis of absorbency.
Evaluation of cell apoptosis
Annexin V-fluorescein isothiocyanate
(FITC)/propidium iodide (PI) staining was performed to quantify
cell apoptosis. Briefly, cells were collected, washed in cold PBS
twice and suspended in binding buffer at 1×106 cells/ml.
Cells were then stained with Annexin V-FITC and PI according to the
manufacturer's protocol. Cell apoptosis was detected by a FACSVerse
flow cytometer, equipped with FACSuite software version 1.0 (both
from BD Biosciences, Franklin Lakes, NJ, USA).
Statistical analysis
All values are expressed as the mean ± standard
error. Results were analyzed by Student's t-test or one-way
analysis of variance, followed by Fisher's least significant
difference test, as appropriate. Pearson's correlation test was
used to assess the correlation between two variables. Statistical
analysis was performed using SPSS 11.0 (SPSS, Inc., Chicago, IL,
USA). P<0.05 was considered to indicate a statistically
significant difference.
Results
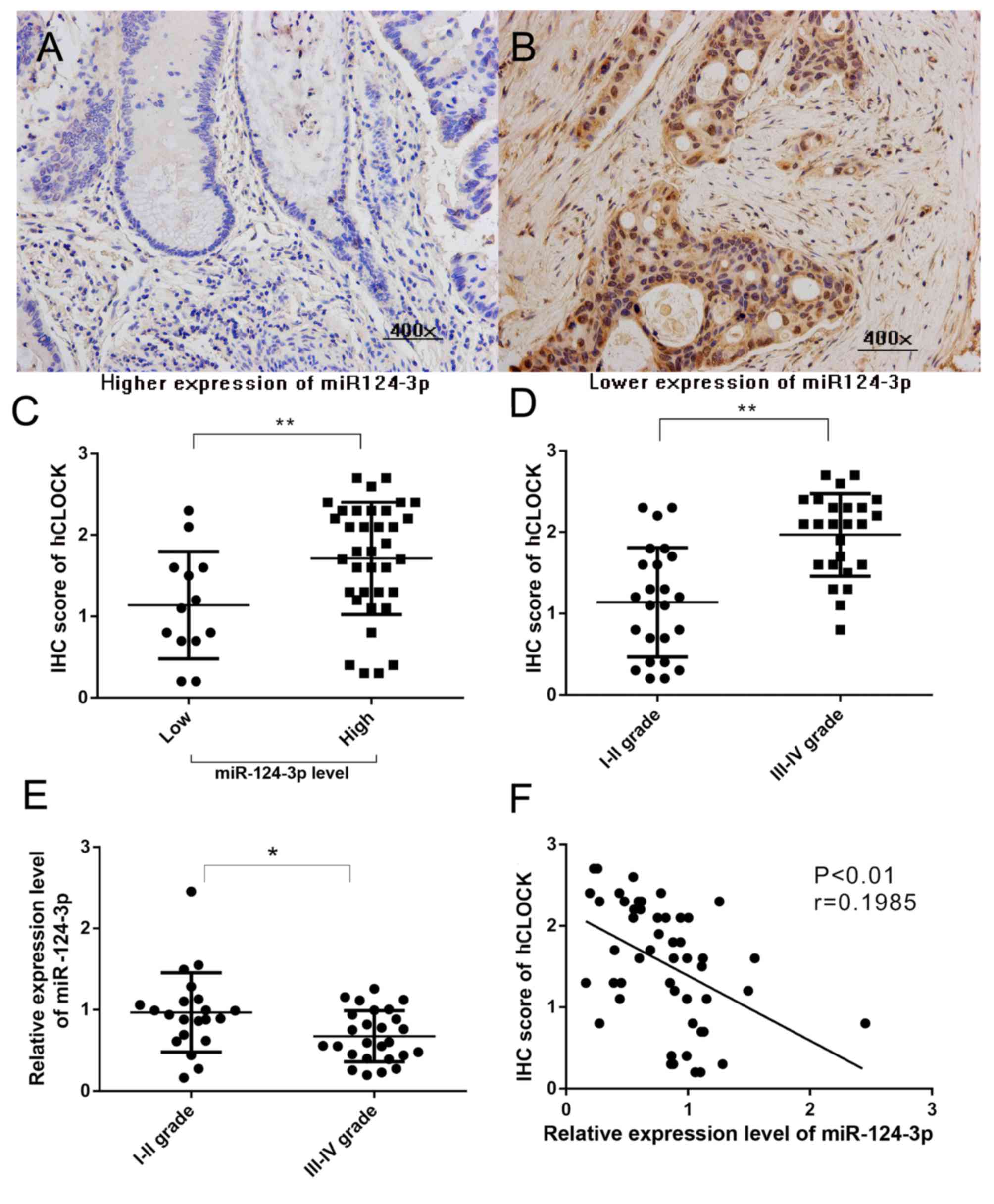
Attenuation of miR-124-3p leads to
overexpression of hCLOCK in CRCs tissues of clinical patients, and
is correlated with patients of the late stage CRC
Our previous study reported that increased hCLOCK
expression is observed in human CRC tissues compared with paired
non-cancerous tissues (8). To
evaluate the association between miR-124-3p level and hCLOCK
expression in CRCs, hCLOCK expression in CRC tissues with different
miR-124-3p expression levels was analyzed. miR-124-3p expression
was defined as low if the relative level in the CRC tissue was
lower than the paired non-cancerous tissue, and high if the
relative miR-124-3p level was higher than the paired non-cancerous
tissue. The hCLOCK level in CRC tissues with high miR-124-3p were
significantly decreased compared with those CRCs with low
miR-124-3p expression (P<0.01; Fig.
1A-C). The present study confirmed that a higher level of
hCLOCK expression was significantly correlated with patients with
late stage CRC (P<0.001; Fig.
1D). Furthermore, the results demonstrated that downregulated
expression of miR-124-3p was significantly correlated with late TNM
stage in patients (Fig. 1E).
Significant negative correlations were observed between miR-124-3p
level and hCLOCK expression (r=0.1985, P<0.01) in CRC tissues
(Fig. 1F).
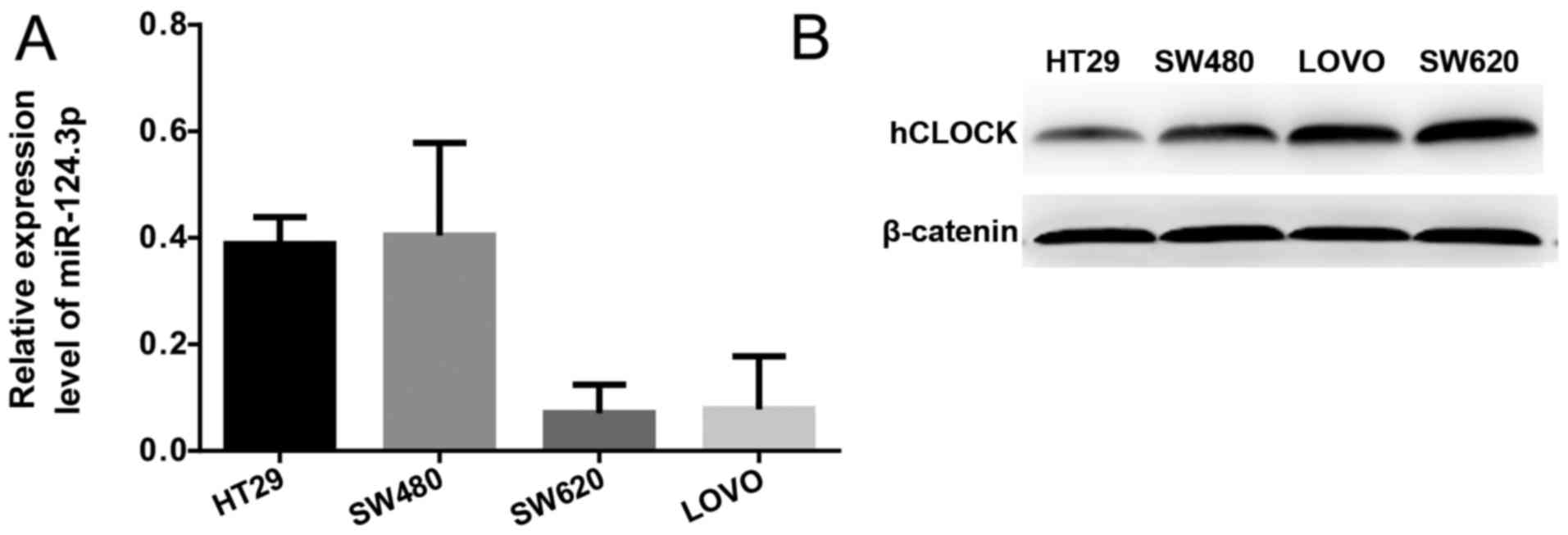
Expression level of miR-124-3p is
negatively correlated with the protein expression level of hCLOCK
in human CRC cells
Protein expression levels of hCLOCK and the mRNA
expression levels of miR-124-3p in the SW620, SW480, HT29 and LOVO
human CRC cell lines were examined by western blotting and RT-qPCR,
respectively. It was demonstrated that miR-124-3p expression was
significantly increased in more invasive CRC cell lines (SW620 and
LOVO) compared with less invasive CRC cell lines (SW480 and HT29;
Fig. 2A). In contrast, the protein
expression levels of hCLOCK was increased in less invasive CRC cell
lines compared with more invasive ones (Fig. 2B).
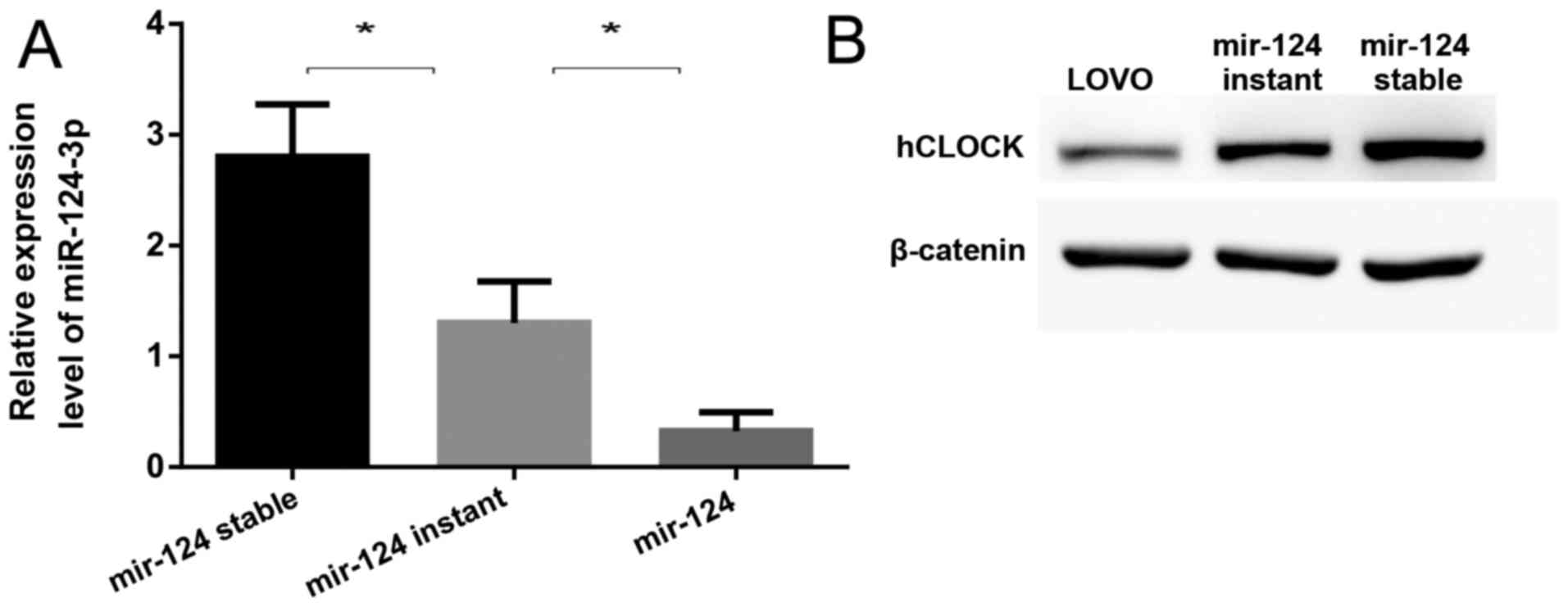
Upregulation of miR-124 contributes to
suppression of hCLOCK expression in human CRC cells
Using three widely used microRNA target prediction
databases (TargetScan, PicTar and miRNA.org),
miR-124-3p was predicted to putatively bind to the 3′-UTR of
hCLOCK. To confirm whether miR-124-3p suppresses the expression of
hCLOCK, the miR-124 precursor was transfected into LOVO cells by
lentiviral vectors or plasmids, representing mir-124 stable
expression (miR-124-stable) and mir-124 instant expression
(miR-124-instant), respectively. It was demonstrated that the
expression of miR-124-3p was significantly increased in
miR-124-stable and miR-124-instant cells compared to LOVO cells
(Fig. 3A), while hCLOCK expression
was significantly suppressed in both cell lines (Fig. 3B). Notably, it was revealed that
the higher the miR-124-3p expression was, the lower the hCLOCK
protein level was in these cells (Fig.
3).
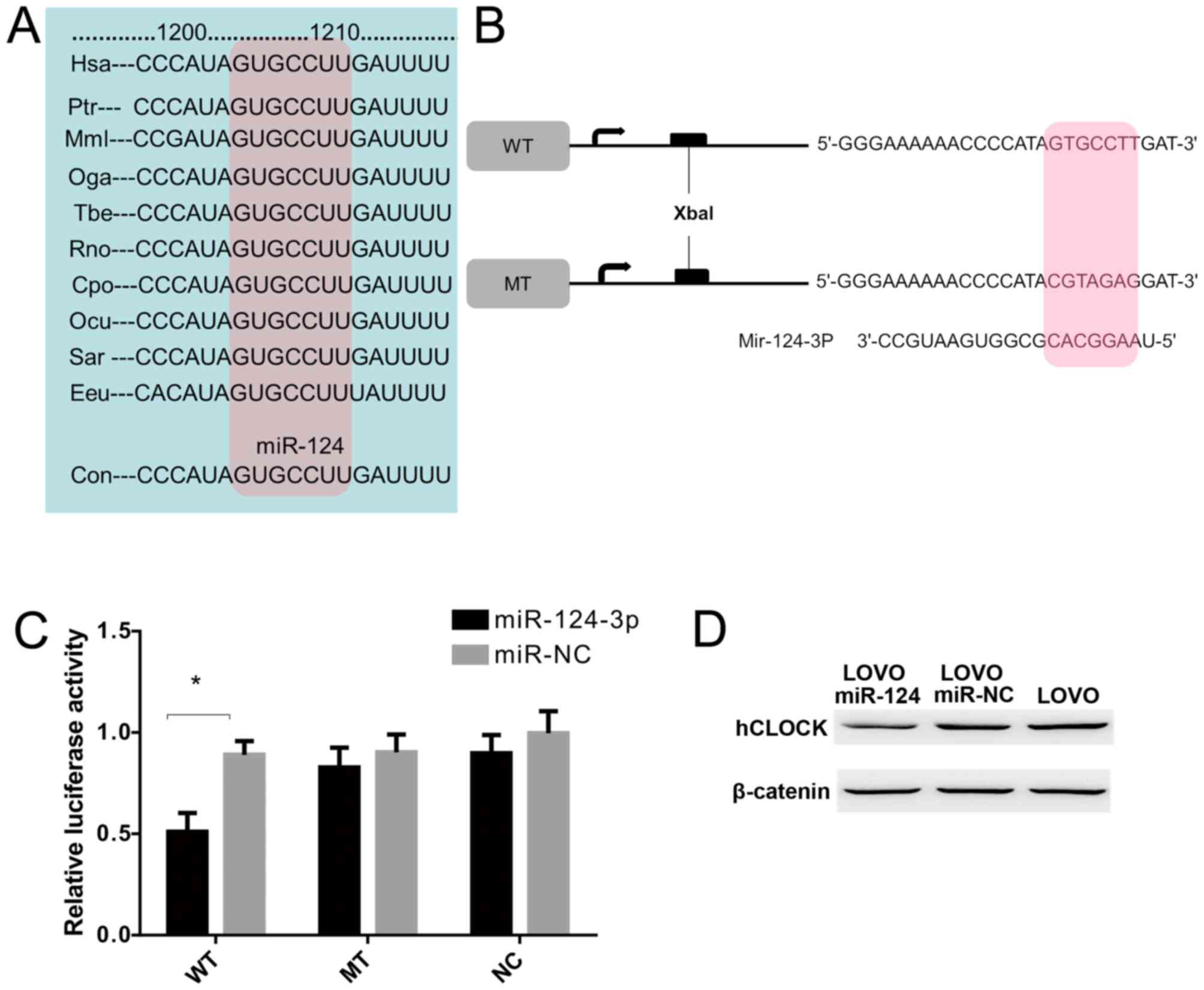
hCLOCK is a direct target of
miR-124-3p
To examine whether hCLOCK is a direct target of
miR-124-3p, a miR-124-3p based luciferase assay was performed to
observe whether miR-124-3p binds to the 3′-UTR of hCLOCK mRNA. The
miR-124 targeting sequence in 3′-UTR of hCLOCK was identified to
conserved among different species (Fig. 4A). Wild-type and mutant hCLOCK
3′-UTR were cloned downstream of the luciferase reporter gene
(Fig. 4B). The results
demonstrated that miR-124-3p directly bound to the hCLOCK 3′-UTR
and significantly reduced the luciferase activities, while cells
with mutant hCLOCK 3′-UTR exhibited higher luciferase activities
(Fig. 4C). Furthermore, western
blot analysis demonstrated that exogenous miR-124-3p expression
suppressed endogenous hCLOCK protein levels in LOVO cells (Fig. 4D). Taken together, these results
indicated that hCLOCK is a direct target for miR-124-3p in CRC
cells.
miR-124-3p induces apoptosis and
inhibits invasion and proliferation of CRC cells
As presented in Fig.
5A, transfection of the miR-124 lentiviruses caused a
significant increase in the expression of miR-124-3p in LOVO cells
relative to LOVO cells transfected with mir-NC viruses (LOVO
miR-NC) and non-transfected LOVO cells (LOVO). Compared with
LOVO-miR-NC and LOVO cells, the invasion ability was significantly
inhibited in LOVO cells transfected with miR-124 (LOVO miR-124;
Fig. 5B-E). miR-124-3p
transfection significantly increased the apoptosis rate (LOVO
miR-124, 15.54±3.81% vs. LOVO miR-NC, 6.91±1.63% vs. LOVO.
5.27±1.30%; Fig. 5F-I).
Furthermore, the proliferation of LOVO cells was significantly
inhibited 36 and 48 h after transfection with miR-124-3p (Fig. 5J).
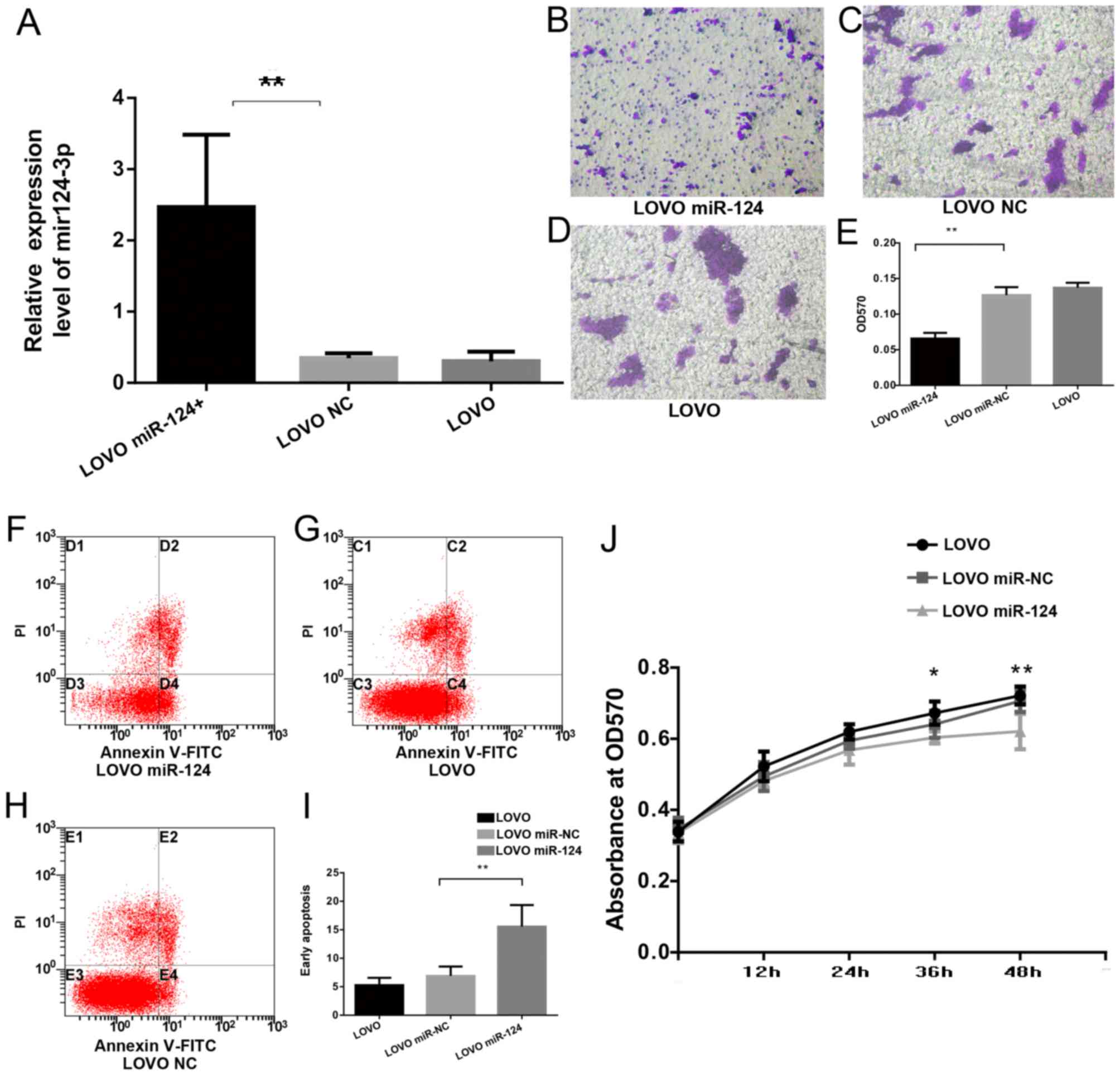 | Figure 5.miR-124-3p induces apoptosis and
inhibits invasion and proliferation of CRC cells. (A) Reverse
transcription-quantitative polymerase chain reaction was used to
detect the expression of miR-124-3p. (B-D) Cell invasion was
analyzed by a Transwell Permeable Supports system in (B) LOVO
miR-142, (C) LOVO NC and (D) LOVO cells, and (E) the results were
quantified. Annexin V/PI staining of (F) LOVO miR-142, (G) LOVO NC
and (H) LOVO cells, and (I) quantification of apoptosis. (J) Cell
proliferation assay examining proliferation rate of LOVO cells
transfected with miR-124 or miR-NC, and LOVO cells without
transfection. Data are presented as the mean ± standard deviation
(n=3). *P<0.05 and **P<0.01, LOVO miR-142 vs. LOVO NC or LOVO
cells, or as indicated. miR, microRNA; hCLOCK, human circadian
locomoter output cycles protein kaput; NC, negative control; FITC,
fluorescein isothiocyanate; PI, propidium iodide; OD, optical
density. |
Discussion
CRC is the third most common cancer in men and the
second in women worldwide (23).
Despite improvement in therapeutic strategies for CRCs, including
surgical technique and chemo-radiotherapy, the prognosis remains
extremely poor (24). Further
studies on the molecular mechanisms underlying CRC initiation and
progression are crucial for developing effective and specific
treatment strategies (25–26).
In our previous study, it was demonstrated that
disruption of circadian rhythms, which are regulated by circadian
genes, was one of the endogenous factors that contribute to CRC
tumor initiation and progression (7,8). As
a core circadian gene, hCLOCK is highly involved in this process.
However, the regulation mechanism of hCLOCK in CRC initiation and
progression are poorly understood. The present study hypothesized
that attenuation of miRNAs targeting hCLOCK would lead to decreased
expression of hCLOCK in CRCs, thereby inhibiting CRC initiation and
progression. After searching for miRNAs targeting hCLOCK through
miRNA predicting databases, miR-124, which is one of the microRNAs
extensively studied in CRCs, was identified to directly target the
hCLOCK 3′-UTR. Increasing evidence has demonstrated that miR-124 is
dysregulated in CRCs and serves a suppressive role for CRC by
targeting specific genes (15–18).
miR-124 is frequently silenced in CRCs, possibly due to
hypermethylation of the miR-124 promoter region (27). mir-124 promoter hypermethylation
may be one of the factors affecting the expression level of
miR-124, and may be responsible for high expression of hCLOCK in
CRCs.
Therefore, the present study aimed to examine the
expression level of miR-124-3p and hCLOCK in human CRC tissues. The
results indicated that the expression level of hCLOCK is
significantly upregulated in the III/IV grade TNM stage CRCs
tissues compared with I/II grade ones. In contrast, miR-124 was
attenuated in the same tissue samples. The hCLOCK expression level
in CRC tissues with high levels of mir-124-3p was significantly
reduced, compared with CRC tissues with low levels of mir-124-3p.
Significant negative correlations were demonstrated between
miR-124-3p and hCLOCK expression levels in CRCs. The same
experiments performed using human CRC cell lines also revealed the
same phenomenon. The expression level of hCLOCK was significantly
upregulated in the more invasive CRC cell lines SW620 and LOVO. In
contrast, miR-124 was attenuated in less invasive CRC cell lines
SW480 and HT29. Upregulation of miR-124 significantly inhibited the
expression level of hCLOCK in LOVO cells. Furthermore, the impact
of miR-124 on proliferation, invasion and apoptosis in CRC cells
was examined. Upregulation of miR-124 significantly inhibited
proliferation and invasion and induced apoptosis of LOVO cells.
Furthermore, dual-luciferase assays indicated that hCLOCK is a
direct target of miR-124.
In conclusion, the present study demonstrated that
hCLOCK has an enhancing role miR-124 has a suppressive role in the
progression of human CRCs. Furthermore, hCLOCK mRNA was a direct
target of miR-124. Attenuation of miR-124 in CRCs contributed to
the high hCLOCK expression, and finally promoted the progression of
human CRCs. These findings expand the understanding of the
underlying mechanisms of circadian rhythms in regulating human
CRCs, and may provide a novel perspective for future clinical
therapy of this type of cancer.
Acknowledgements
This work was supported by the Project for National
Basic Science Personnel Training Fund (no. J1310009) and the
National Natural Science Foundation of China (no. 81570771).
References
|
1
|
Hoffman AE, Yi CH, Zheng T, Stevens RG,
Leaderer D, Zhang Y, Holford TR, Hansen J, Paulson J and Zhu Y:
CLOCK in breast tumorigenesis: Genetic, epigenetic, and
transcriptional profiling analyses. Cancer Res. 70:1459–1468. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ko CH and Takahashi JS: Molecular
components of the mammalian circadian Clock. Hum Mol Genet.
15:R271–R277. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Storch KF, Lipan O, Leykin I, Viswanathan
N, Davis FC, Wong WH and Weitz CJ: Extensive and divergent
circadian gene expression in liver and heart. Nature. 417:78–83.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Benca R, Duncan MJ, Frank E, McClung C,
Nelson RJ and Vicentic A: Biological rhythms, higher brain
function, and behavior: Gaps, opportunities, and challenges. Brain
Res Rev. 62:57–70. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen ST, Choo KB, Hou MF, Yeh KT, Kuo SJ
and Chang JG: Deregulated expression of the PER1, PER2 and PER3
genes in breast cancers. Carcinogenesis. 26:1241–1246. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fu L, Pelicano H, Liu J, Huang P and Lee
C: The circadian gene period2 plays an important role in tumor
suppression and DNA damage response in vivo. Cell. 111:41–50. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang Y, Hua L, Lu C and Chen Z: Expression
of circadian Clock gene human period2 (hPer2) in human colorectal
carcinoma. World J Surg Oncol. 9:1662011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang L, Chen B, Wang Y, Sun N, Lu C, Qian
R and Hua L: hClock gene expression in human colorectal carcinoma.
Mol Med Rep. 8:1017–1022. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dibner C, Schibler U and Albrecht U: The
mammalian circadian timing system: Organization and coordination of
central and peripheral clocks. Ann Rev Physiol. 72:517–549. 2010.
View Article : Google Scholar
|
|
10
|
Harms E, Kivimäe S, Young MW and Saez L:
Posttranscriptional and posttranslational regulation of clock
genes. J Biol Rhythms. 19:361–373. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Asher G, Gatfield D, Stratmann M, Reinke
H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW and Schibler U:
SIRT1 regulates circadian clock gene expression through PER2
deacetylation. Cell. 134:317–328. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Long JM and Lahiri DK: Advances in
microRNA experimental approaches to study physiological regulation
of gene products implicated in CNS disorders. Exp Neurol.
235:402–418. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang MJ, Li Y, Wang R, Wang C, Yu YY, Yang
L, Zhang Y, Zhou B, Zhou ZG and Sun XF: Downregulation of
microRNA-124 is an independent prognostic factor in patients with
colorectal cancer. Int J Colorectal Dis. 28:183–189. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang J, Lu Y, Yue X, Li H, Luo X, Wang Y,
Wang K and Wan J: MiR-124 suppresses growth of human colorectal
cancer by inhibiting STAT3. PLoS One. 8:e703002013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jinushi T, Shibayama Y, Kinoshita I,
Oizumi S, Jinushi M, Aota T, Takahashi T, Horita S, Dosaka-Akita H
and Iseki K: Low expression levels of microRNA-124-5p correlated
with poor prognosis in colorectal cancer via targeting of SMC4.
Cancer Med. 3:1544–1552. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu K, Zhao H, Yao H, Lei S, Lei Z, Li T
and Qi H: MicroRNA-124 regulates the proliferation of colorectal
cancer cells by targeting iASPP. Biomed Res Int.
2013:8675372013.PubMed/NCBI
|
|
18
|
Edge SB, Byrd DR, Compton CC, Fritz AG,
Greene FL and Trotti A: AJCC Cancer Staging Manual. 7th edition.
New York: Springer; 2010
|
|
19
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:2015. View Article : Google Scholar
|
|
20
|
Krek A, Grün D, Poy MN, Wolf R, Rosenberg
L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M
and Rajewsky N: Combinatorial microRNA target predictions. Nat
Genet. 37:495–500. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Betel D, Wilson M, Gabow A, Marks DS and
Sander C: The microRNA.org resource: targets and expression.
Nucleic Acids Res. 36(Database Issue): D149–D153. 2008.PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Venook AP, Niedzwiecki D, Lenz HJ, et al:
CALGB/SWOG 80405: Phase III trial of irinotecan/5-FU/leucovorin
(FOLFIRI) or oxaliplatin/5-FU/leucovorin (mFOLFOX6) with
bevacizumab (BV) or cetuximab (CET) for patients with KRAS
wild-type untreated metastatic adenocarcinoma of the colon or
rectum [abstract]; 2014 American Society of Clinical Oncology
Annual Meeting; 2014 May 30-June 3; Chicago, USA. Alexandria (VA).
American Society of Clinical Oncology. 2014;
|
|
25
|
Cancer Genome Atlas Network, .
Comprehensive molecular characterization of human colon and rectal
cancer. Nature. 487:330–337. 2012. View Article : Google Scholar :
|
|
26
|
Fearon ER: Molecular genetics of
colorectal cancer. Annu Rev Pathol. 6:479–507. 2011. View Article : Google Scholar
|
|
27
|
Harada T, Yamamoto E, Yamano HO, Nojima M,
Maruyama R, Kumegawa K, Ashida M, Yoshikawa K, Kimura T, Harada E,
et al: Analysis of DNA methylation in bowel lavage fluid for
detection of colorectal cancer. Cancer Prev Res (Phila).
7:1002–1010. 2014. View Article : Google Scholar
|