Introduction
Skin aging is a biological process that, as for
aging of other organs, is mediated by intrinsic and extrinsic
factors. Features of skin aging include wrinkling, reduced
elasticity, sagging, laxity, dullness, roughness and discoloration.
Deep wrinkles, in particular, are a primary consequence of
ultraviolet (UV)-mediated skin aging (1–3).
Histological studies have revealed that a reduction in collagen is
one of the primary features of UV-mediated skin aging (1–5).
This occurs via degradation of collagen and the collagen matrix, in
addition to factors that interfere with their synthesis (3,6).
Underlying this process are molecular mechanisms that modulate
expression of collagen, matrix metalloproteinases and associated
proteins (7).
The sirtuin (SIRT) family is composed of
nicotinamide adenine dinucleotide-dependent protein deacetylases,
which regulate aging in yeast, worms, flies and mammals (8,9). In
humans, seven SIRT genes (SIRT1-7) have been
identified, and these serve roles in numerous age-associated
diseases, including cancer, neurodegenerative diseases, diabetes
and cardiovascular disease (9–12).
In mice, SIRT1 deficiency induces destruction of the skin barrier
and enhances UV-induced injury and sensitivity (10–12).
Additionally, SIRT1 may promote collagen expression in human smooth
muscle cells by the deacetylation of regulatory factor X5 and
alleviation of α-1-type 2 collagen (COL1A2) repression, and
has been implicated in the inhibition of the transforming growth
factor-β/mothers against decapentaplegic signaling pathway in
fibroblasts during fibrosis (13–15).
In mice, a deficiency in SIRT6 led to a skin aging phenotype
(16), and the expression levels
of collagen 1 and 3 are reduced by knockdown of SIRT6 in
dermal fibroblasts (16).
Therefore, regulation of SIRT1 and SIRT6 is associated with skin
aging.
MicroRNAs (miRNAs) are small non-coding RNAs that
are primarily generated by RNA polymerases II and III, and are
processed and matured by the proteins Drosha and Dicer (17,18).
These miRNAs consist of 20-24 nucleotides and associate with the
miRNA-induced silencing complex. This complex targets the 3′
untranslated region (3′UTR) of specific target genes, which
interferes with translation and therefore down-regulates protein
expression (19,20). miRNAs serve a role in numerous
biological processes, including development, differentiation, the
cell cycle, apoptosis, stemness and tumorigenesis (21–25).
In the present study, a novel miRNA that regulates
SIRT6 was identified that may regulate collagen expression.
Materials and methods
Cell culture
Human dermal fibroblasts (HDFs) were purchased from
Lonza (Basel, Switzerland) and cultured in Dulbecco's modified
Eagle's medium (HyClone; GE Healthcare Life Sciences, Logan, UT,
USA) supplemented with 10% fetal bovine serum (Hyclone; GE
Healthcare Life Sciences) and 1% penicillin and streptomycin. Cells
were transfected with 100 nM miRNA (miR)-378b mimic
(5′-ACUGGACUUGGAGGCAGAA-3′; Bioneer Corporation, Dajeon, Korea),
100 nM anti-miR-378b (5′-UUCUGCCUCCAAUCCUGU-3′; Bioneer
Corporation) or 100 nM scrambled control (AccuTarget™ Negative
Control siRNA; Bioneer Corporation) using Lipofectamine®
RNAiMAX Transfection reagent (Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA), according to the manufacturer's protocol.
After transfection for 4 h, cells were washed with PBS and
incubated for 24 h. Transfected HDFs were subsequently exposed to
UVB using a Super light-VI UV illuminator (Boteck, Gunpo, Korea).
After incubation for 24 h following UVB exposure, reverse
transcription-quantitative polymerase chain reaction (RT-qPCR),
western blotting and luciferase assay were performed.
RNA extraction and RT-qPCR
Total RNA was isolated from cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). cDNA was synthesized using 2 µg RNA with the miScript II RT
kit (Qiagen GmbH, Hilden, Germany), according to the manufacturer's
protocol. Following cDNA synthesis, the mRNA expression levels of
COL1A1 and the gene encoding β-actin were detected using the
StepOnePlus Real-Time PCR system (Applied Biosystems; Thermo Fisher
Scientific, Inc.), using EvaGreen™ premix (Solis BioDyne, Tartu,
Estonia) and the following specific primers: Forward,
5′-AGGGCCAAGACATC-3′ and reverse, 5′-AGATCACGTCATCGCACAACA-3′ for
human COL1A1; and forward, 5′-GGATTCCTATGTGGGCGACGA-3′ and
reverse, 5′-CGCTCGGTGAGGATCTTCATG-3′ for human β-actin. Expression
levels of miR-378b were detected with miR-378b specific primers
(Qiagen GmbH) using the hsa-mir-378b miScript Primer assay (cat.
no. MI0014154) and Hs_RNU6-2_11 miScript Primer assay (cat. no.
MS00033740) from Qiagen GmbH, and the miScript SYBR®
Green PCR kit (Qiagen GmbH) with the StepOnePlus Real-Time PCR
system. Forward primers were included in the miScript Primer assay
kits and reverse primers were included in the miScript
SYBR® Green PCR kit. The expression levels of
COL1A1 and miR-378b were normalized to β-actin and U6
respectively, using the 2−ΔΔCq method (26). All RT-qPCRs were performed as
follows: Initialization step at 94°C for 5 min, followed by 40
cycles (denaturing, 94°C for 30 sec; annealing, 60°C for 30 sec;
polymerization, 72°C for 30 sec) and a final elongation step at
72°C, for 5 min. All experiments were repeated three times. Data
was analyzed with Excel 2016 (Microsoft Corporation, Redmond, WA,
USA) and presented as the mean value of viable cells ± standard
deviation.
Target prediction and identification
of miR-378b
Predicted targets of miR-378b were identified using
the bioinformatic analysis tool, microRNA.org
(www.microrna.org). A luciferase reporter
construct containing the predicted target sequence of miR-378b in
the SIRT6 3′UTR was generated by ligating a region (+1,312
to +1,329) of the human SIRT6 gene into the XbaI
restriction site, downstream of the luciferase gene in the pGL3
vector (Promega Corporation, Madison, WI, USA). HDFs were
subsequently transfected with the 1 µg reporter assay vector and
the 0.2 µg pSV-β-galactosidase control plasmid (Promega
Corporation), with or without an miR-378b mimic or anti-miR-378b,
using Lipofectamine RNAiMAX Transfection reagent (Invitrogen;
Thermo Fisher Scientific) for 4 h, and subsequently incubated at
37°C for 24 h. After incubation for 24 h, luciferase and
β-galactosidase assays were performed using Luciferase Assay
Reagent (Promega Corporation) and the β-galactosidase Detection kit
II (Clontech Laboratories, Inc., Mountainview, CA, USA), according
to the manufacturer's protocol. Luciferase results were normalized
using β-galactosidase activity.
Western blot analysis
Cells were lysed in radioimmunoprecipitation assay
buffer, containing 1% NP-40, 150 mM NaCl, 10 mM Tris-HCl (pH 8.0),
1 mM EDTA and complete protease inhibitor cocktail (Roche
Diagnostics, Basel, Switzerland). Extracted proteins (20 µg) were
loaded onto 12% gels and separated by electrophoresis. Proteins
were subsequently transferred onto nitrocellulose membranes (EMD
Millipore, Billerica, MA, USA) and blocked with blocking buffer
[(5% skim milk in TBS-Tween-20 buffer (50 mM Tris, 150 mM NaCl,
0.1% Tween 20)] at 25°C for 1 h. Protein expression levels of SIRT6
and β-actin were detected using rabbit anti-SIRT6 (1:2,000; D8D12;
cat. no. 12486; Cell Signaling Technology, Inc., Danvers, MA, USA)
anti-β-actin (1:10,000; N-21; cat. no. sc-130656; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) primary antibodies, followed
by anti-mouse IgG horseradish peroxidase (HRP)-conjugated (1:5,000;
cat. no. 7076; Cell Signaling Technology, Inc.) and anti-rabbit IgG
HRP-conjugated antibody (1;3,000; cat. no. 7074; Cell Signaling
Technology, Inc.) secondary antibodies. Membranes were incubated
with primary antibody at 25°C for 4 h, followed by incubation with
secondary antibody at 25°C for 1 h. Proteins were visualized using
SuperSignal™ West Pico Chemiluminescent Substrate (Thermo Fisher
Scientific, Inc.). The intensity of each band was measured using
ImageJ software Version 1.50 (National Institutes of Health,
Bethesda, MD, USA).
Statistical analysis
Statistical significance was calculated using
one-way analysis of variance with Tukey's post-hoc test. P<0.05
was considered to indicate a statistically significant difference
using Excel 2016 (Microsoft Corporation, Redmond, WA, USA). Data
are presented as the mean ± standard error.
Results and discussion
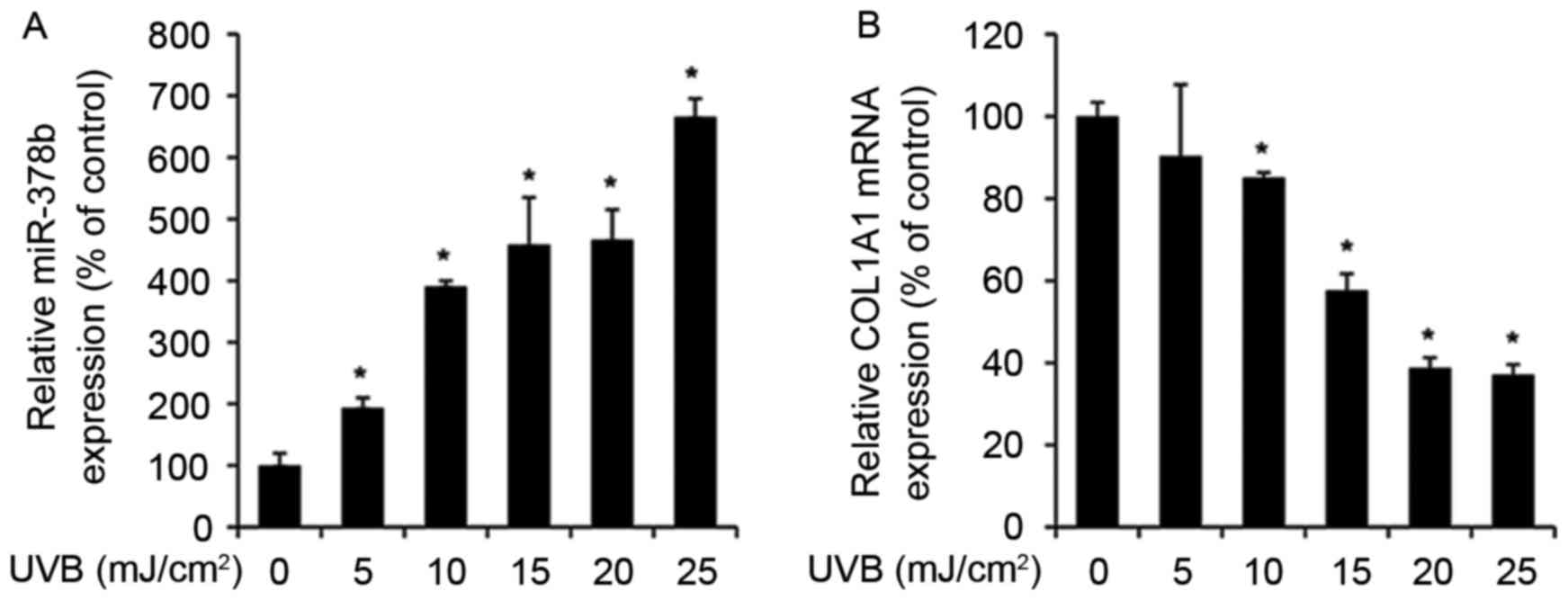
UVB exposure enhances miR-378b and
reduces COL1A1 expression levels in HDFs
The expression levels of various miRNAs alter during
skin aging and in UVB-exposed cells (27). Therefore, to determine whether
miR-378b is modulated by UVB, miR-378b expression was measured by
RT-qPCR in UVB-exposed HDFs. miR-378b expression levels were
significantly enhanced in cells exposed to 5–25 mJ/cm2
UVB in a dose-dependent manner, compared to untreated cells (0
mJ/cm2 UVB; P<0.05; Fig.
1A). In addition, in HDFs exposed to the same doses of UVB,
expression levels of COL1A1 were reduced in a dose-dependent
manner compared with untreated cells, and were inversely associated
with miR-378b expression levels (Fig.
1B).
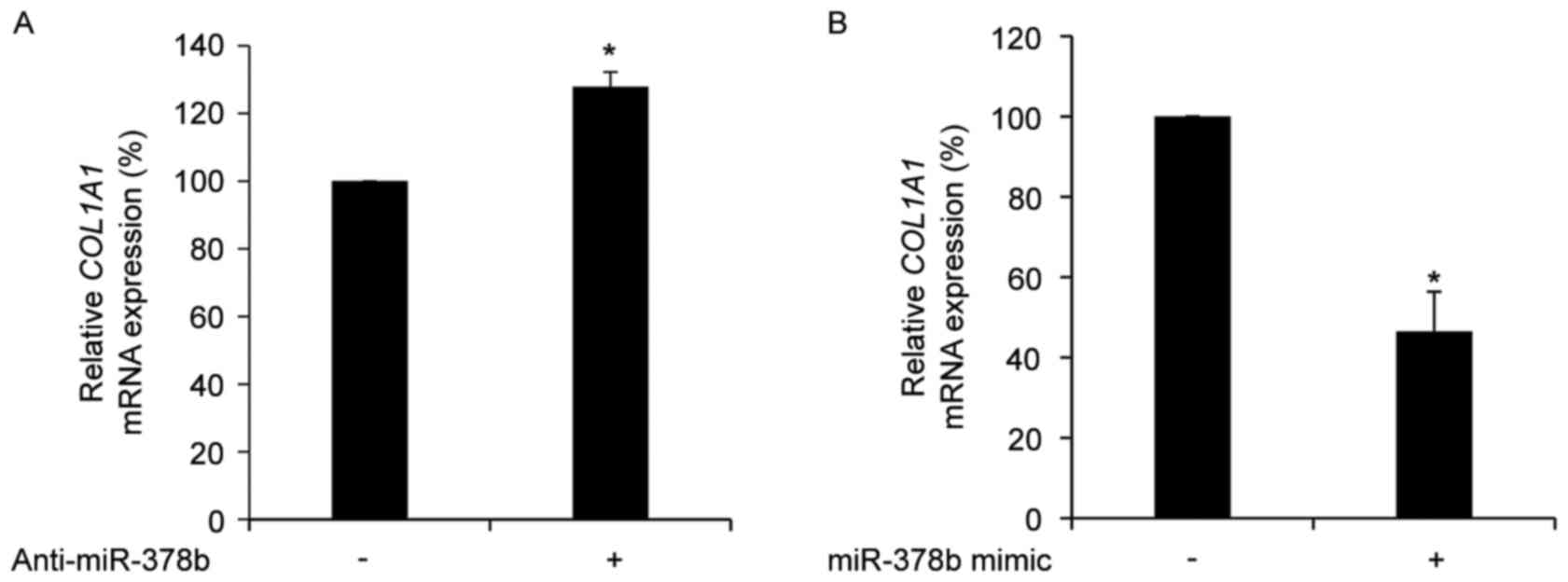
COL1A1 is negatively regulated by
miR-378b in HDFs
To determine whether the expression of COL1A1
is directly affected by miR-378b, the mRNA expression levels of
COL1A1 in HDFs treated with anti-miR-378b or an miR-387b
mimic were measured. Transfection with anti-miR-378b significantly
up-regulated COL1A1 mRNA expression levels (P<0.05;
Fig. 2A), whereas transfection
with an miR-378b mimic resulted in a significant reduction in
COL1A1 mRNA expression levels compared with cells
transfected with a scramble control (P<0.05; Fig. 2B). However, the 3′UTR of
COL1A1 does not contain a predicted binding site for
miR-378b. In addition, the miR-378b mimic did not directly modulate
the luciferase activity of a luciferase-COL1A1 3′UTR fusion
construct in HDFs (data not shown). Therefore, a target prediction
for miR-378b was performed using microRNA.org,
which revealed that SIRT6 contained a binding site for the
miR-378b seed sequence in its 3′UTR. SIRT6 has been identified as
an anti-aging protein due to its ability to regulate the mRNA
expression levels of COL1A1 and COL3A1 in HDFs
(16). Therefore, it was
hypothesized that miR-378b may directly regulate SIRT6
expression.
miR-378b negatively regulates SIRT6 by
binding to its 3′UTR
The majority of miRNAs regulate target genes by
binding to their 3′UTRs (28,29).
It was therefore determined whether the miR-378b mimic directly
interacted with the SIRT6 3′UTR, by measuring the luciferase
activity from a luciferase-SIRT6 3′UTR fusion construct. It
was determined, using microRNA.org,
that the seed sequence from miR-378b matched a region between
+1,312 and +1,324 bp of the SIRT6 3′UTR (Fig. 3A). The SIRT6 3′UTR was
subsequently cloned into a luciferase plasmid and transfected into
HDFs. Co-transfection of an miR-378b mimic significantly reduced
luciferase activity compared with pGL3-SIRT6-3′UTR only
(P<0.05; Fig. 3B), whereas
co-transfection with an miR-378b mimic and anti-miR-378b
significantly enhanced luciferase activity compared with cells
transfected with SIRT6 3′UTR and an miR-378b mimic
(P<0.05; Fig. 3B). Thus, these
results suggested that miR-378b directly binds to, and interferes
with, SIRT6 mRNA.
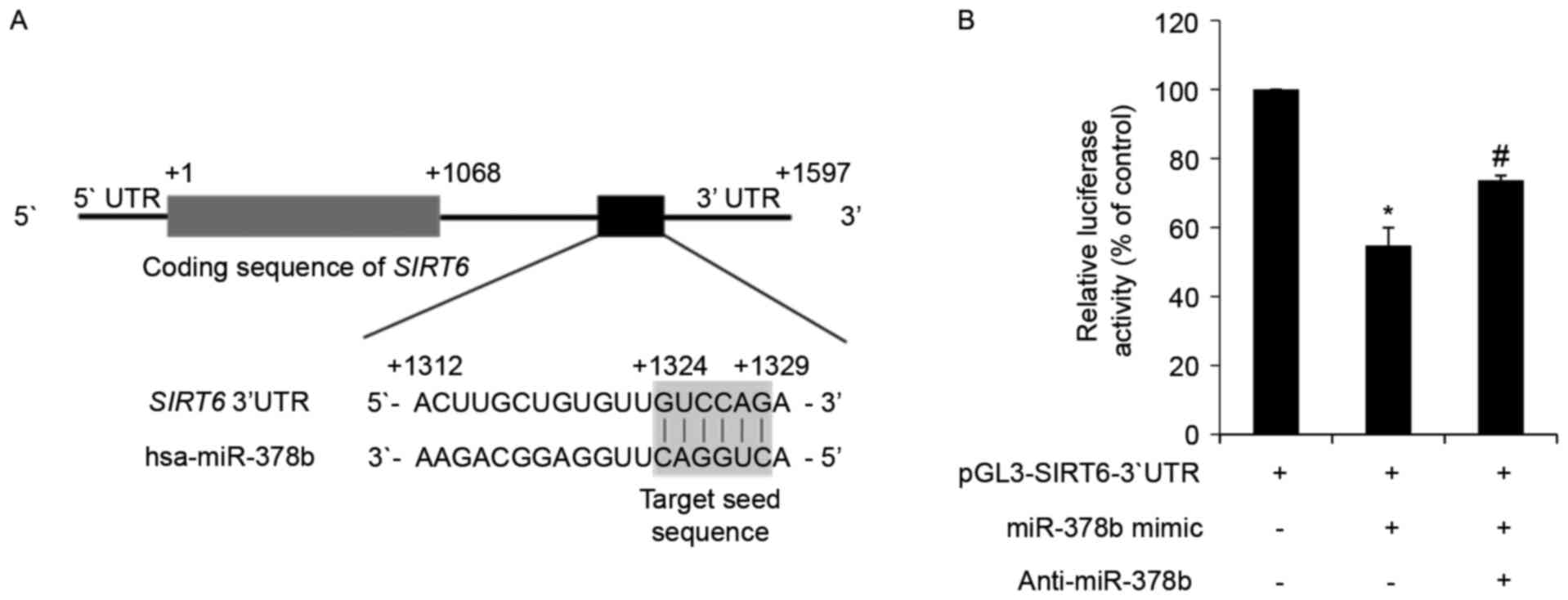 | Figure 3.miR-378b directly targets SIRT6
mRNA and represses its translation. (A) In silico analysis
of SIRT6 mRNA and the predicted target sequence of miR-378b.
The ATG start codon is indicated by +1, and the 3′ end of the
SIRT6 coding region is indicated by +1,068. The region of
the 3′UTR containing the miR-378b recognition sequences is located
from +1,312 to +1,329 in the SIRT6 transcript. (B) HDFs were
transfected with a reporter construct containing the miR-378b
recognition sequences from SIRT6 fused to luciferase, and
pSV-β-galactosidase, which was the control vector for
normalization. Additional groups were co-transfected with an
miR-378b mimic and anti-miR-378b. Following 24 h transfection,
cells were harvested and luciferase assays were performed. Data are
expressed as the mean ± standard error of three independent
experiments, as a percentage of the
pGL3-SIRT6-3′UTR-transfected group. *P<0.05 vs.
pGL3-SIRT6-3′UTR only-transfected group.
#P<0.05 vs. pGL3-SIRT6-3′UTR and miR-378b
mimic co-transfected group. miR-378b, microRNA-378b; SIRT6,
sirtuin 6; UTR, untranslated region; HDFs, human dermal
fibroblasts. |
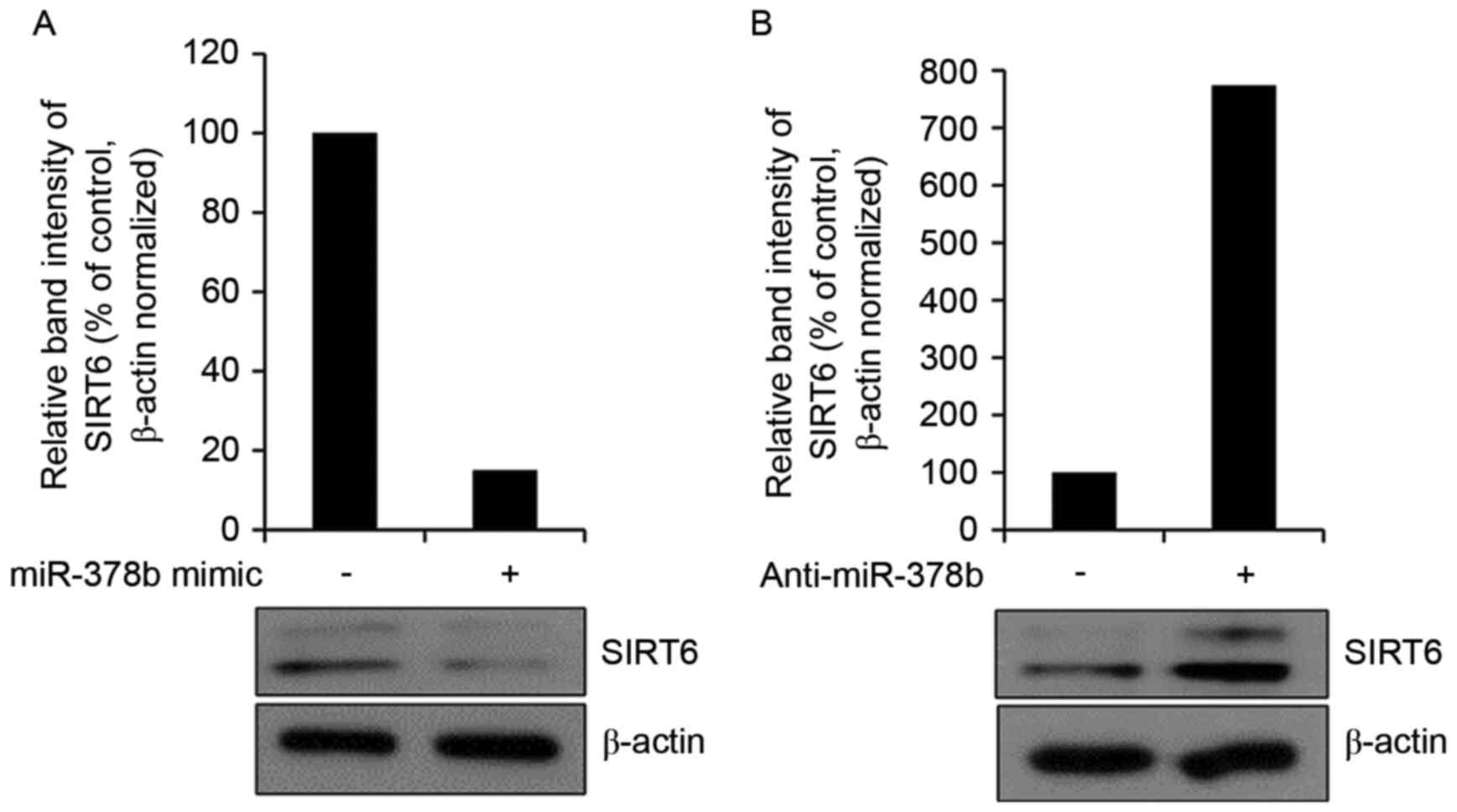
miR-378b represses endogenous SIRT6
expression in HDFs
It was subsequently determined whether the miR-378b
mimic affected endogenous SIRT6 protein expression by western blot
analysis. Overexpression of miR-378b reduced the expression levels
of SIRT6 (Fig. 4A). Transfection
with anti-miR-378b had the reverse effect and enhanced SIRT6
protein expression levels (Fig.
4B). These results suggested that miR-378b may inhibit
COL1A1 mRNA expression via effects on SIRT6. In
addition, a previous study demonstrated that SIRT6 regulates the
expression of genes associated with stress and aging (30). In HDFs from younger individuals,
SIRT6 was highly activated compared with HDFs from older
individuals (31). Therefore,
miR-378b, which was upregulated by UVB in the present study
(Fig. 1A), may regulate aging and
collagen expression via interference of translation of SIRT6
mRNA. Investigation of the association between miR-378b and skin
aging in human skin samples is required in future studies to
indicate whether miR-378b is a marker of photo-aging.
Acknowledgements
The present study was supported by the Research
Professor Program of Konkuk University (to Professor Hwa Jun Cha)
and the Korean Health Technology R&D Project, Ministry of
Health & Welfare, Republic of Korea (grant no. HN13C0075).
References
|
1
|
Lee YK, Cha HJ, Hong M, Yoon Y, Lee H and
An S: Role of NF-κB-p53 crosstalk in ultraviolet A-induced cell
death and G1 arrest in human dermal fibroblasts. Arch Dermatol Res.
304:73–79. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Varani J, Dame MK, Rittie L, Fligiel SE,
Kang S, Fisher GJ and Voorhees JJ: Decreased collagen production in
chronologically aged skin: Roles of age-dependent alteration in
fibroblast function and defective mechanical stimulation. Am J
Pathol. 168:1861–1868. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wulf HC, Sandby-Møller J, Kobayasi T and
Gniadecki R: Skin aging and natural photoprotection. Micron.
35:185–191. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Quan T and Fisher GJ: Role of
age-associated alterations of the dermal extracellular matrix
microenvironment in human skin aging: A mini-review. Gerontology.
61:427–434. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Darlenski R, Kazandjieva J and Tsankov N:
Skin barrier function: Morphological basis and regulatory
mechanisms. J Clin Med. 4:36–45. 2011.
|
|
6
|
Egbert M, Ruetze M, Sattler M, Wenck H,
Gallinat S, Lucius R and Weise JM: The matricellular protein
periostin contributes to proper collagen function and is
downregulated during skin aging. J Dermatol Sci. 73:40–48. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Newton VL, Mcconnell JC, Hibbert SA,
Graham HK and Watson RE: Skin aging: Molecular pathology, dermal
remodelling and the imaging revolution. G Ital Dermatol Venereol.
150:665–674. 2015.PubMed/NCBI
|
|
8
|
Borradaile NM and Pickering JG: NAD (+),
sirtuins, and cardiovascular disease. Curr Pharm Des. 15:110–117.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Law IK, Liu L, Xu A, Lam KS, Vanhoutte PM,
Che CM, Leung PT and Wang Y: Identification and characterization of
proteins interacting with SIRT1 and SIRT3: Implications in the
anti-aging and metabolic effects of sirtuins. Proteomics.
9:2444–2456. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Han SH: Potential role of sirtuin as a
therapeutic target for neurodegenerative diseases. J Clin Neurol.
5:120–125. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Roth M and Chen WY: Sorting out functions
of sirtuins in cancer. Oncogene. 33:1609–1620. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Balcerczyk A and Pirola L: Therapeutic
potential of activators and inhibitors of sirtuins. Biofactors.
36:383–393. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chou WW, Chen KC, Wang YS, Wang JY, Liang
CL and Juo SH: The role of SIRT1/AKT/ERK pathway in ultraviolet B
induced damage on human retinal pigment epithelial cells. Toxicol
In Vitro. 27:1728–1736. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fan W and Luo J: SIRT1 regulates
UV-induced DNA repair through deacetylating XPA. Mol Cell.
39:247–258. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xia J, Wu X, Yang Y, Zhao Y, Fang M, Xie
W, Wang H and Xu Y: SIRT1 deacetylates RFX5 and antagonizes
repression of collagen type I (COL1A2) transcription in smooth
muscle cells. Biochem Biophys Res Commun. 428:264–270. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Baohua Y and Li L: Effects of SIRT6
silencing on collagen metabolism in human dermal fibroblasts. Cell
Biol Int. 36:105–108. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
An IS, An S, Park S, Lee SN and Bae S:
Involvement of microRNAs in epigallocatechin gallate-mediated UVB
protection in human dermal fibroblasts. Oncol Rep. 29:253–259.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lee MJ, Cha HJ, Lim KM, Lee OK, Bae S, Kim
CH, Lee KH, Lee YN, Ahn KJ and An S: Analysis of the microRNA
expression profile of normal human dermal papilla cells treated
with 5α-dihydrotestosterone. Mol Med Rep. 12:1205–1212. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ambros V: microRNAs: Tiny regulators with
great potential. Cell. 107:823–826. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fahs F, Bi X, Yu FS, Zhou L and Mi QS: New
insights into microRNAs in skin wound healing. IUBMB Life.
67:889–896. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Virant-Klun I, Ståhlberg A, Kubista M and
Skutella T: MicroRNAs: From female fertility, germ cells, and stem
cells to cancer in humans. Stem Cells Int. 2016:39849372016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nothnick WB: Non-coding RNAs in uterine
development, function and disease. Adv Exp Med Biol. 886:171–189.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Garg M: MicroRNAs, stem cells and cancer
stem cells. World J Stem Cells. 4:62–70. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dumortier O and Van Obberghen E: MicroRNAs
in pancreas development. Diabetes Obes Metab. 14 Suppl 3:S22–S28.
2012. View Article : Google Scholar
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhou BR, Xu Y and Luo D: Effect of UVB
irradiation on microRNA expression in mouse epidermis. Oncol Lett.
3:560–564. 2012.PubMed/NCBI
|
|
28
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chang YM, Juan HF, Lee TY, Chang YY, Yeh
YM, Li WH and Shih AC: Prediction of human miRNAs using
tissue-selective motifs in 3′ UTRs. Proc Natl Acad Sci USA. 105:pp.
17061–17066. 2008; View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kawahara TL, Rapicavoli NA, Wu AR, Qu K,
Quake SR and Chang HY: Dynamic chromatin localization of Sirt6
shapes stress- and aging-related transcriptional networks. PLoS
Genet. 7:e10021532011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sharma A, Diecke S, Zhang WY, Lan F, He C,
Mordwinkin NM, Chua KF and Wu JC: The role of SIRT6 protein in
aging and reprogramming of human induced pluripotent stem cells. J
Biol Chem. 288:18439–18447. 2013. View Article : Google Scholar : PubMed/NCBI
|