Introduction
For a long time, microbial fermentation was applied
in milk, soybean, fruit and grain as source of enzymes as a means
to provide palatability, nutritional value, preservative and
medicinal properties (1). Protein
fermentation that mainly occurs through hydrolysis by digestive,
microbial, and plant proteolytic enzymes can release the bioactive
peptides corresponding to cryptic sequences from native proteins
(2–4). Also, the degree of proteolysis
depends on the microbial strain used for fermentation and is
reported to be directly related to the biological activity of the
released peptides (5).
Marine microalgae have been of interest as a biofuel
source with their large biomass in the marine environment and
recently reported in various studies in the field of
pharmaceuticals and nutraceuticals as unconventional protein source
(6). We have previously suggested
that the fermented of Pavlova lutheri (P. lutheri;
microalgae) by Hansenula polymorpha or Candida
rugopelliculosa as protein source has potential antioxidant
activity via reduction of hydroxyl radical or the increment of
antioxidant related enzymes at protein levels in hydroxyl
radical-induced oxidative stress (7).
Bone structure is maintained by upholding a balance
between bone formation by osteoblasts and bone resorption by
osteoclasts (8–10). Osteoblasts originating from
mesenchymal stem cells (MSCs) can translate mechanical signals into
biological responses to regulate bone remodelling in intact bone in
the process of bone repair (11).
The mechanical signal in the cells involves the sequential
activation (via phosphorylation cascade) of various intracellular
signalling molecules, including MAPKs and phosphoinositide 3-kinase
(PI3k)/Akt, resulting in the activation of transcription factors
such as activator protein-1 (AP-1) and nuclear factor (NF)-κB and
subsequent modulation of the expression of genes that regulate
osteoblast maturation and mineralisation (12–17).
The cellular responses in osteoblasts have influence on the
interaction between various systemic factors, such as alkaline
phosphatase (ALP), bone morphogenetic proteins (BMPs), osteocalcin
(OCN), collagen type I (Col I) that are generally considered
stimulants for osteoblast adhesion and differentiation (9,18).
In the present study, the effect of the P.
lutheri fermentation on osteoblastic differentiation in MG-63
osteoblastic cells was compared with the non-ferment. The active
peptide [peptide of P. lutheri fermentation (PPLF)]
responsible for the effect was subsequently purified and the
mechanism by which PPLF is involved in the regulation of
osteoblastic differentiation of MG-63 cells was explored.
Materials and methods
General
In the previous study, we fermented microalgae
Pavlova lutheri by H. polymorpha as below (7). The cellulose-degraded microalga P.
lutheri was autoclaved at 121°C for 30 min in a buffer (pH 7)
solution at a ratio of 1:15 (w/v). Subsequently, H.
polymorpha was inoculated with the autoclaved solution at a
concentration of 1% (v/v) and incubated at 37°C for 12 days.
Fermented microalgae was lyophilized and stored at 80°C until used.
Human osteoblastic (MG-63) cells were obtained from American Type
Culture Collection (Manassas, VA, USA). Dulbecco's Modified Eagle's
Medium (DMEM), Trypsin-ethylenediaminetetraacetic acid (EDTA),
penicillin/streptomycin, fetal bovine serum (FBS) and other
materials required for culturing cells were purchased from Gibco
BRL, Life Technologies (Grand Island, NY, USA). The p38 inhibitor
(SB203580), NF-κB inhibitor [pyrrolidine dithiocarbamate (PDTC)]
and 3-(4,5-dimethyl-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
reagent were purchased from Sigma-Aldrich (St. Louis, MO,
USA). All other chemicals and reagents used in this study were of
analytical grade.
Cell culture and viability assay
Human osteoblastic MG-63 cells were cultured in DMEM
containing 5% FBS in a humidified atmosphere with 5% CO2
at 37°C. Cells were sub-cultured at 3-day intervals using
trypsin-EDTA and were seeded in 96-well plates at a density of
5×103 cells per well. 24 h post plating, media was
replaced with DMEM without serum and cells were subjected to sample
treatment for another 3 days. Subsequently, 100 µl of 1 mg/ml MTT
reagent was added to each well, and incubated for 3 h. The
supernatant was removed and formazan was dissolved in DMSO and its
formation was observed by monitoring the signal at 540 nm using a
microplate reader.
ALP activity assay
Cells were seeded into 12-well plates at a density
of 5×105 cells per well for 24 h and then treated with
samples in DMEM media without serum for 3 days. Cells were then
washed three times with PBS, and were lysed with a lysis buffer
containing 20 mM Tris-HCl (pH 7.5), 150 mM NaCl and 1% Triton
X-100. Briefly, the lysate was mixed with p-NPP substrate solution
which consists of 1.5 mM MgCl2 and 15 mM p-nitrophenyl
phosphate (p-NPP). The activity of phosphatases to catalyze the
hydrolysis of p-NPP to p-nitrophenol was evaluated by measuring the
absorbance at 405 nm (19). ALP
activity was normalized according to the cellular total protein
content of cell lysate determined by BCA protein assay
(Sigma-Aldrich).
Preparation of fermented microalgae
peptide
The lyophilized fermented microalgae was dissolved
in distilled water and passed through disposable Silica-based
bonded-phase cartridges (Sep-Pak Vac C18 20 cc/5 g;
Waters, Milford, MA, USA). The cartridges were activated with 100%
methanol in 0.1% trifluoroacetic acid (TFA) and subsequently the
cleaned product was eluted with 10% methanol in 0.1% TFA. The
eluent was lyophilized then further purified as described
below.
Purification procedures
The cleaned eluent from previous step was loaded
onto HiPrep 16/10 DEAE ion exchange column equilibrated with 20 mM
sodium acetate buffer (pH 4.0) and eluted with a linear gradient of
NaCl (0–2 M) at a flow rate of 1.0 ml/min by 280 nm absorbance
using FPLC (Amersham Bioscience Co., Uppsala, Sweden). Separated
fractions were pooled and desalted by an electrodialyzer (Micro
Acilyzer model G3; Asahi Chemical Industry Co., Tokyo, Japan) with
a 100 Da molecular mass cutoff membrane (Asahi Chemical Industry
Co.). The desalted fractions were lyophilized and investigated for
their ALP activity. The fraction which had the highest ALP activity
was further purified on a Primesphere 10 C18 (20×250 mm)
column at reversed-phase HPLC (Dionex Corp., Sunnyvale, CA, USA)
with a linear gradient of acetonitrile (0–30% in 30 min) containing
0.1% TFA at a flow rate 1.0 ml/min by 215 nm absorbance. Elution
peaks were collected and tested for ALP activity. The highest
active fraction was re-applied to a Acclaim 120 C18
analytical column with 15% acetonitrile (20% v/v, in 15 min)
containing 0.1% TFA at flow rate of 1 ml/min. The purified peptide
was subjected to amino acid sequence analysis.
Amino acid sequence of the purified
peptide
The accurate molecular mass and amino acid sequence
of the purified peptide was determined by quadrupole time-of-flight
mass spectroscopy (Q-TOF MS; Micromass UK Ltd., Altrincham, UK)
coupled to electrospray ionization (ESI) source. The molecular
weight of the purified peptide dissolved in methanol/water (1:1,
vol/vol) was detected by a doubly charged (M+2H)+2 state
in the mass spectrum and the amino acid sequence was identified by
tandem MS analysis. Spray voltage was 4,000 V and nitrogen was
maintained at 40 psi for nebulization. Mass spectra were acquired
over the range from 50 to 1,500 m/z.
Mineralization assay
The levels of mineralization were determined in the
24-well plates using Alizarin Red staining (Sigma-Aldrich) after 7
days treatment with the peptide. Briefly, cells were fixed with 70%
(v/v) ethanol for 1 h and then stained with 40 mM Alizarin Red S
(pH 4.2) for 15 min at room temperature. After removing Alizarin
Red S solution by aspiration, cells were incubated in PBS for 15
min at room temperature on an orbital rotator. Cells were then
rinsed once with fresh PBS and subsequently distained for 15 min
with 10% cetylpyridinium chloride in 10 mM sodium phosphate (pH
7.0). The extracted stain was then transferred to a 96-well plate
and the absorbance at 562 nm was measured using a microplate reader
(Tecan Austria GmbH, Grödje, Austria).
Western blot analysis
Cells were treated with p38 inhibitor (SB203580, 20
µM) or NF-κB inhibitor (PDTC, 10 µM) for 1 h prior to treatment
with the peptide (in the presence of inhibitor) for 72 h. Cells
were then harvested in lysis buffer (RIPA; Sigma-Aldrich). Cleared
total cell lysates (20 µg) were separated by 10% SDS-polyacrylamide
gel electrophoresis and transferred onto a polyvinylidene fluoride
membrane (Amersham Pharmacia Biotech, Amersham, UK). Membrane was
blocked with 5% skim milk and probed with the primary antibody
(diluted 1:1,000) followed by incubation with a secondary antibody
conjugated with horseradish peroxidase (diluted 1:5,000) at room
temperature. The proteins of interest were detected using a
chemiluminescent ECL assay kit (Amersham Pharmacia Biotech)
according to the manufacturer's instructions. Primary and secondary
antibodies including ALP (sc-373737), OCN (sc-365797), β-actin
(sc-47778), p-p38 (sc-7973), p-pERK (sc-7383), p-Jun (sc-822),
p-p65 (sc-166748) and anti-mouse IgG-HRP (7076) purchased from
Santa Cruz Biotechnology Inc. (Santa Cruz, CA, USA) and Cell
Signaling Technology, Inc. (Danvers, MA, USA). Western blots were
visualized using an LAS3000® Luminescent image analyzer
and protein expression was quantified by Multi Gauge v3.0 software
(Fujifilm Life Science, Tokyo, Japan).
Statistical analysis
All experiments were repeated at least three times
and expressed as the means ± SD. Differences between the means of
individual groups were assessed by two-way ANOVA with Tukey's
multiple comparisons test using the statistical software package
GraphPad Prism 6 software (San Diego, CA, USA).
Results
Comparison of the ALP activities of
fermented and non-fermented P
lutheri
We previously investigated the antioxidant
activities of the fermented P. lutheri microalgae by H.
polymorpha or C. rugopelliculosa and suggested the potential
of fermented P. lutheri as protein source possessing
antioxidant activity (7). In this
study, we investigated the difference between the effect of
fermented and non-fermented P. lutheri on osteoblastic
differentiation in MG-63 cells. The lyophilized P. lutheri
was treated with cellulose (1% dry powder weight of the raw
substance) to break its cell wall and was subsequently fermented by
inoculation with H. polymorpha for 12 days (7). Different concentrations (10, 50, 100
µg/ml) of fermented and non-fermented P. lutheri (with
degraded cell-wall) were added to the cells for 3 days followed by
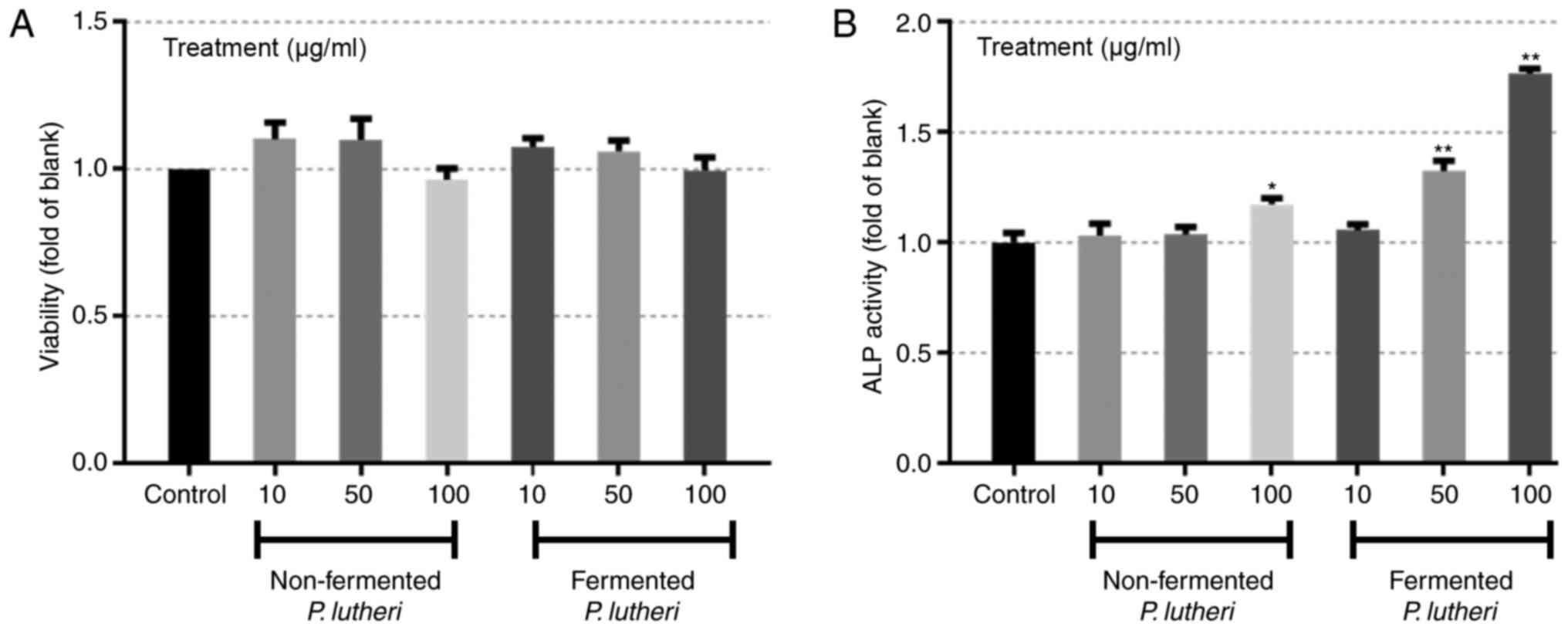
measuring their toxicity and ALP activity. The cells treated with
either fermented or non-fermented P. lutheri did not show
any significant difference in viability compared to control which
is non-treatment group (Fig. 1A),
however a significant increase in the ALP activity of the cells
were observed when treated with 100 µg/ml non-fermented P.
lutheri with P<0.05 and 50 and 100 µg/ml fermented P.
lutheri with P<0.01 (Fig.
1B).
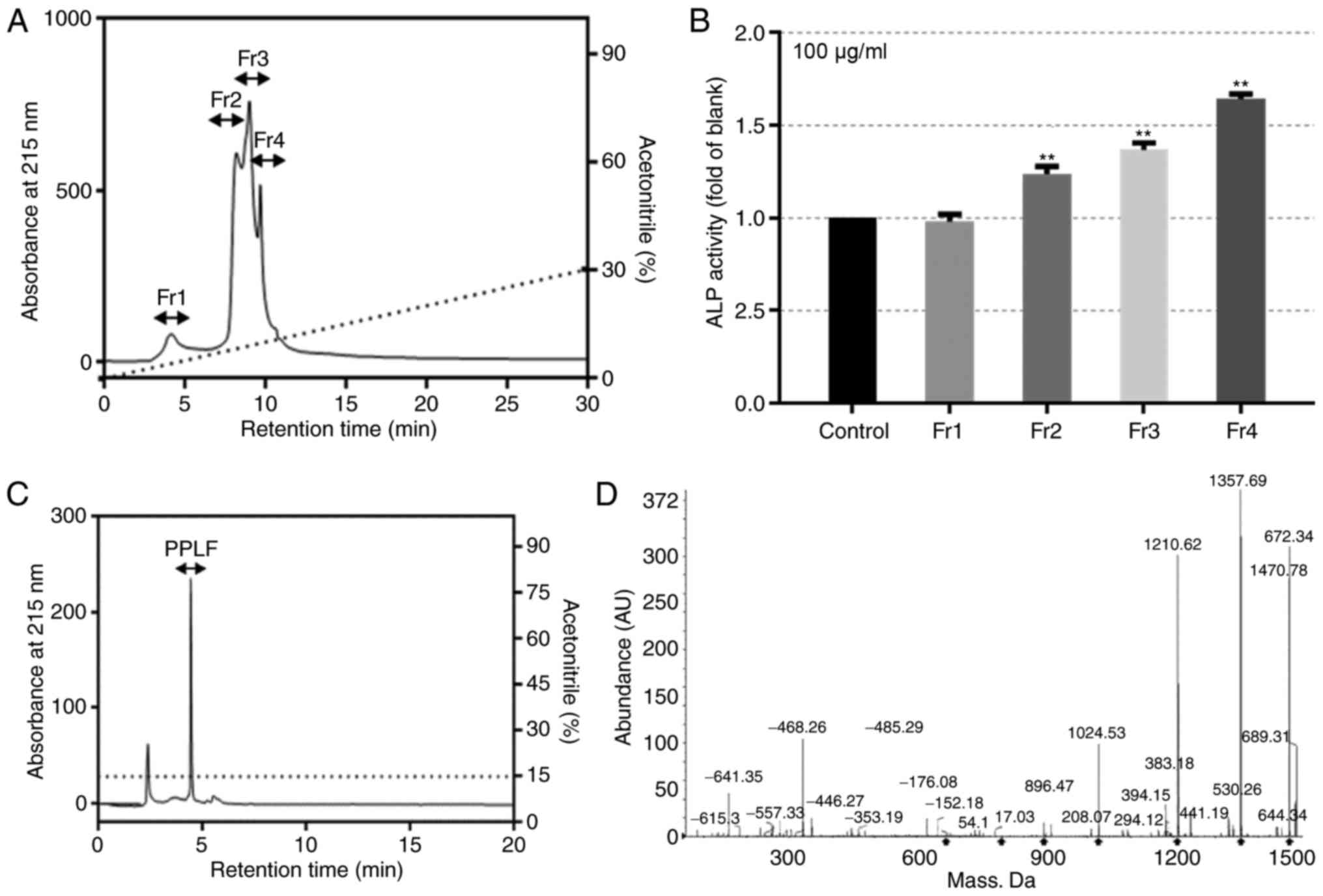
We then further traced the purification profile of
peptide from fermented P. lutheri and characterized the
effect on osteoblastic differentiation in MG-63 cells.
Fractionation profile of fermented P.
lutheri and the effect on ALP activity
The lyophilized fermented P. lutheri was
dissolved in distilled water and passed through disposable Sep-Pak
Vac C18 cartridges to salt out. The cleaned product was
then concentrated and further separated using a weak anion exchange
chromatographic method on a FPLC at 280 nm. The monitored peaks
were desalted and analyzed for their ALP activity (data not shown).
The selected fraction with highest ALP activity was split to four
portions through reversed-phase chromatographic method on a HPLC at
215 nm (Fig. 2A). Each portion was
measured for its ALP activity at 100 µg/ml and Fraction 4 (Fr.4)
showed the highest activity with of 1.642 fold compared to the
control (P<0.01; Fig. 2B). The
peptide PPLF was purified from the additional analysis using an
Acclaim 120 C18 analytical column and was determined as
Glu-Pro-Gln-Trp-Phe-Leu (MW 908.9 Da) by ESI/MS spectroscopy
(Fig. 2C and D).
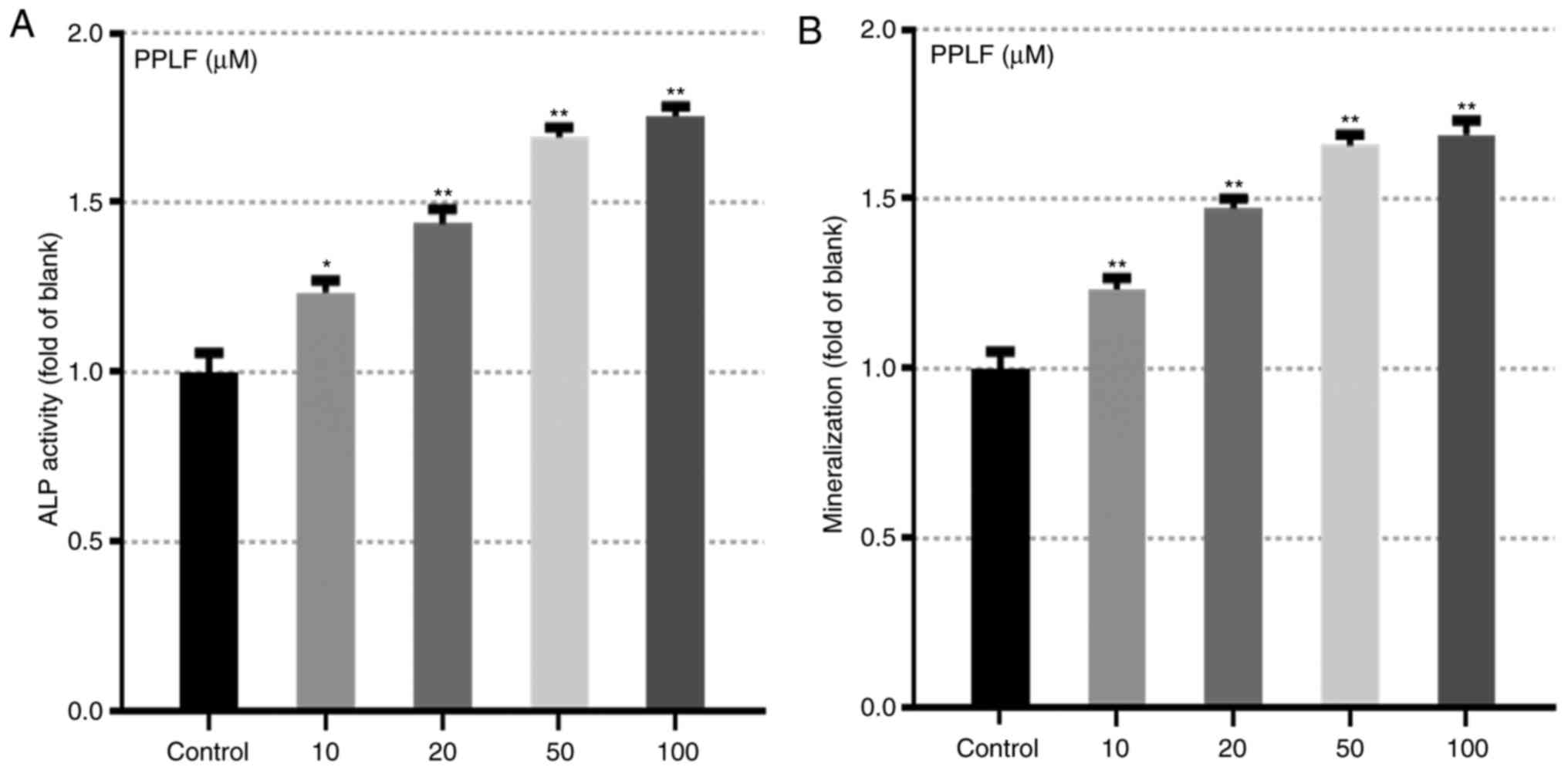
PPLF peptide prompted the markers for
differentiation
The effect of PPLF on ALP activity and
mineralization which are main makers for osteoblastic
differentiation (8,18) was assessed in MG-63 cells. Cells
treated with PPLF showed significantly increased levels of ALP
release (Fig. 3A) and
mineralization (P<0.05 and P<0.01) (Fig. 3B). In order to further assess the
effects of PPLF, we analyzed protein levels of two of the known
early markers of differentiation ALP and OCN in cells treated with
PPLF (10, 20, 50 µM) for 3 days. Both protein levels showed to be
up-regulated by PPLF treatment compared to the non-treated cells
(P<0.05; Fig. 4). These results
suggest that PPLF induces the expression of the early markers of
osteoblastic differentiation in MG-63 cells.
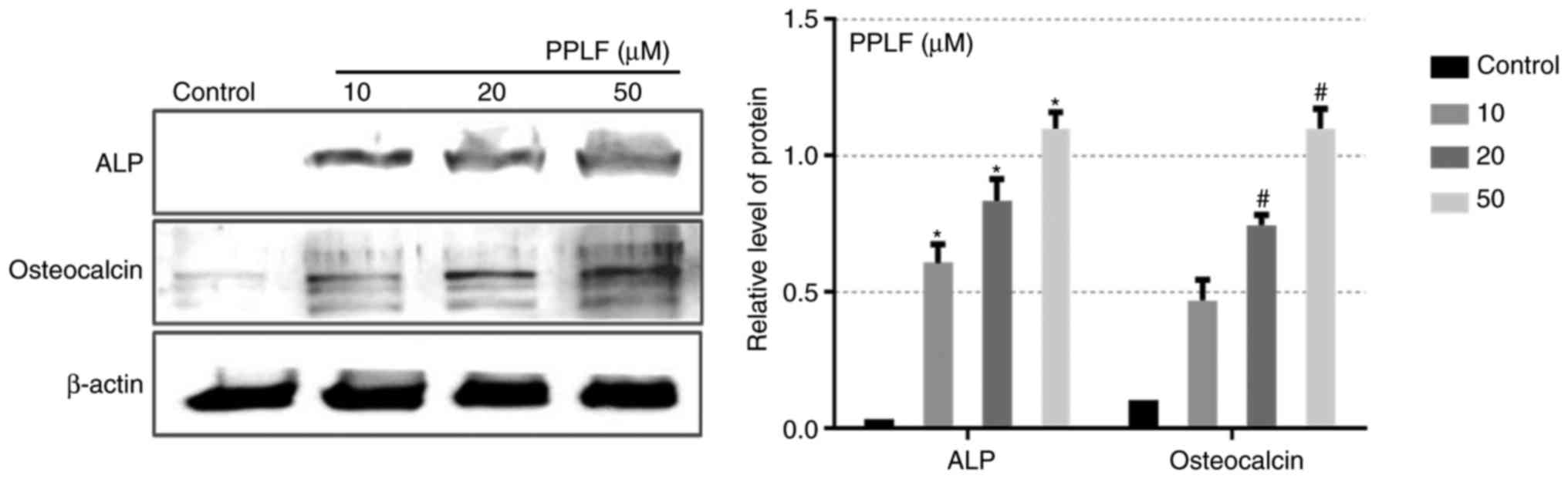
PPLF induces ALP and OCN expression
via p38/p65 pathway
To determine the mechanism of the PPLF effect on
differentiation in MG-63 cells, the phosphorylation levels of MAPK
pathway (p38, ERK1/2, Jun) and NF-κB (p65) which are known to
control ALP and OCN expression and are involved in osteoblastic
differentiation were examined (8,10,18).
As shown in Fig. 5, p-p38 and
p-p65 exhibited a significant increment by PPLF at 20 and 50 µM
(P<0.05), however p-ERK1/2 and p-Jun did not show any change
with PPLF treatment. The result suggests that PPLF treatment
activates p38/p65 signaling pathway. To examine whether the p38/p65
phosphorylation by PPLF treatment was connected to the expression
of ALP and OCN, we assessed the expression of ALP and OCN in the
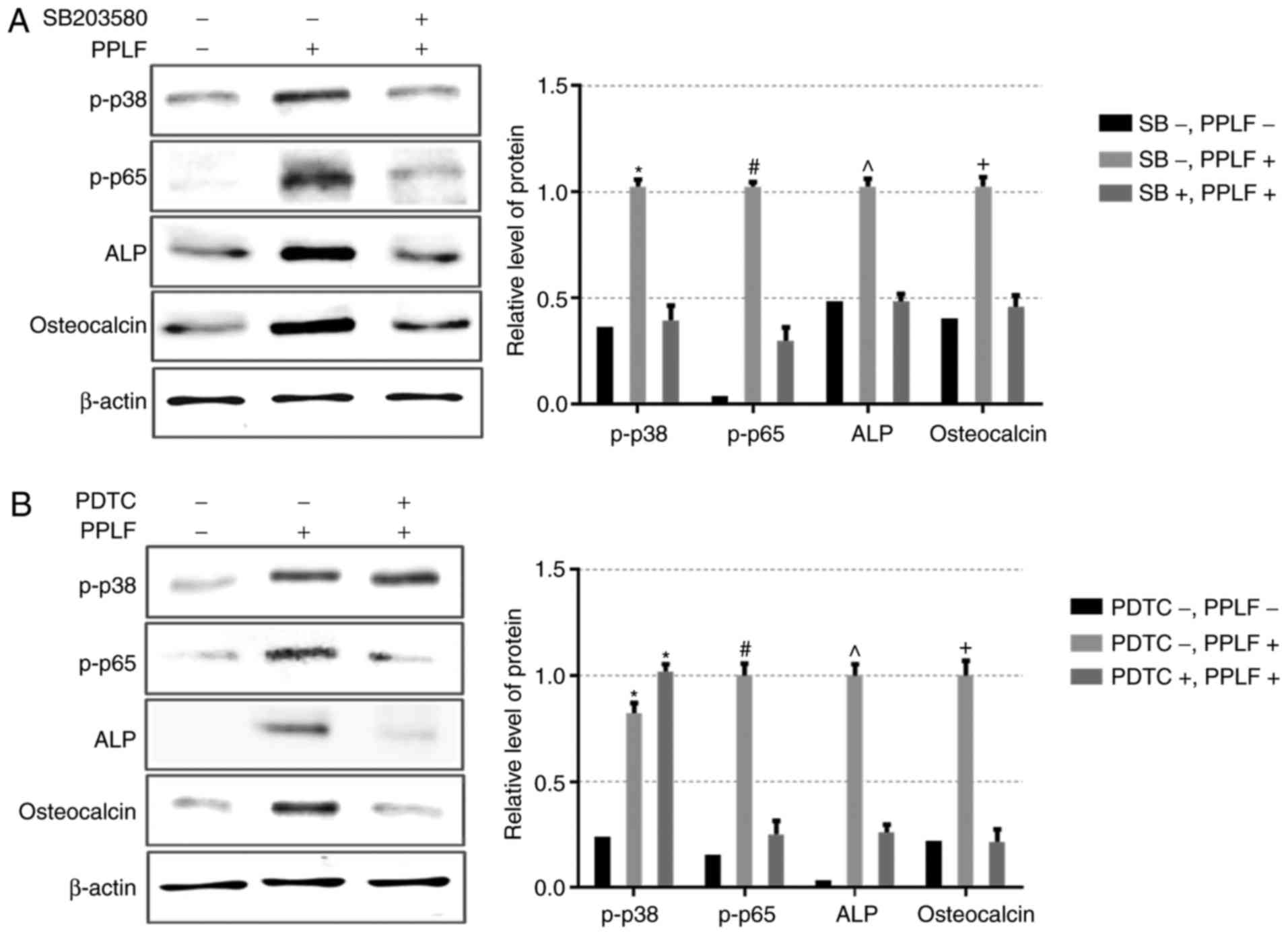
cells treated PPLF (50 µM) when pre-treated with p38 inhibitor
SB203580 (20 µM) or NF-κB inhibitor PDTC (10 µM). Pre-treatment
with SB203580 inhibited both p38 and p65 phosphorylation levels and
resulted in a decrease in the expression of ALP and OCN (Fig. 6A). We also observed the same result
on ALP and OCN expression by PDTC treatment, however p38
phosphorylation showed no-significant change compared to PPLF
treatment suggesting that p38 is upstream of p65 (Fig. 6B). These data suggest that
treatment with PPLF induces ALP and OCN expression via p38/p65
signaling pathway in MG-63 cells.
 | Figure 5.Phosphorylation levels of the three
major types of mitogen-associated protein kinases, p38, ERK1/2 and
Jun, and NF-κB (p65) following PPLF treatment in MG-63 cells. Cells
were treated with PPLF (10, 20 and 50 µM) for 3 days and the levels
of p-p38, p-ERK1/2, p-Jun and p-p65 were assessed by western
blotting. The results were normalized to β-actin. *P<0.05 vs.
p-p38 control; #P<0.05 vs. p-p65 control. PPLF,
peptide of Pavlova lutheri fermentation; ALP, alkaline
phosphatase; ERK, extracellular signal-regulated kinase; NF,
nuclear factor; p-, phosphorylated. |
Discussion
Microbial fermentation by proteolytic action can
produce small molecule peptides from the parent proteins with
various activities. These peptides have been reported to have role
as part of dietary proteins in controlling and influencing health
(3,20). In the present study, we have shown
that the fermented of microalgae P. lutheri (by H.
polymorpha) causes a significant elevation in differentiation
at a concentration of 50 and 100 µg/ml in human osteoblastic MG-63
cells compared to the cells treated with non-fermented P.
lutheri (Fig. 1B). Following
this, we fractionalized the fermented P. lutheri using
chromatography separation and measured the ALP activity of each
separated fraction to purify the active responsible peptide
(Fig. 2). Through ESI/MS
spectroscopy, the purified peptide (PPLF) was sequenced as
Glu-Pro-Gln-Trp-Phe-Leu (MW 908.9) which has rich-hydrophobic amino
acids including one acidic residue (Glu), four basic residues (Pro,
Trp, Phe, Leu), and one aromatic residue (Trp). High levels of
hydrophobic and aromatic amino acids have been shown to rejuvenate
cellular activity, cell differentiation and metabolism (21).
Subsequently, we showed that PPLF significantly
promoted ALP release and mineralization concentration-dependently
and further observed an increase in the protein levels of ALP and
OCN by PPLF treatment (Figs. 3 and
4). ALP, OCN and mineralization
are specific markers of differentiation that are known to be
involved in differentiation of mature osteoblasts and induced by
p38 MAPK and p65 (a subunit of NF-κB) (22). p38/p65 are widely conserved family
of serine threonine protein kinases implicated in several cellular
programs such as cell proliferation, calcification, and apoptosis
(23). A number of studies have
reported that p38 and p65 pathways are required in osteoblast
differentiation and act downstream of BMP receptors and play
important roles in osteoblast differentiation and bone remodeling
(16,24,25).
Indeed, p38 pathway has been found to contribute to bone formation
by phosphorylating the p65 subunit of NF-κB transcription complex
and thus increasing NF-κB transcriptional activity (23). In this study, we showed that PPLF
increased the activation of p38 and p65 whereas did not have any
effect on the ERK1/2 and Jun phosphorylation (Fig. 5). Furthermore, we demonstrated that
selective inhibitors of p38 and p65 significantly inhibited the
expression of differentiation specific markers (ALP and OCN)
indicating that PPLF induces osteoblastic differentiation in MG-63
cells by triggering activation of p38/p65 signaling pathway
(Fig. 6).
Our data shows that fermented P. lutheri
promotes differentiation of MG-63 cells and the peptide PPLF which
was purified from the fermented P. lutheri is responsible
for the increased differentiation in MG-63 cells through p38/p65
activation. Our work shows the potential of the fermented P.
lutheri as an inducer of osteoblast differentiation and further
suggests PPLF peptide as a likely candidate for the treatment of
bone loss and promotion of bone health.
Acknowledgements
The present study was supported by a grant from the
Marine Biotechnology Program (20150220) funded by the Ministry of
Oceans and Fisheries, Republic of Korea.
References
|
1
|
Steinkraus KH: Fermentations in world food
processing. Compr Rev Food Sci Food Saf. 1:23–32. 2002. View Article : Google Scholar
|
|
2
|
Kudoh Y, Matsuda S, Igoshi K and Oki T:
Antioxidative peptide from milk fermented with Lactobacillus
delbrueckii subsp bulgaricus IFO13953. J Jpn Soc Food
Sci. 48:44–50. 2001. View Article : Google Scholar
|
|
3
|
Korhonen H and Pihlanto A: Bioactive
peptides: Production and functionality. Int Dairy J. 16:945–960.
2006. View Article : Google Scholar
|
|
4
|
Korhonen H and Pihlanto A: Bioactive
peptides from food proteinsHandbook of Food Products Manufacturing:
Health, Meat, Milk, Poultry, Seafood and Vegetables. John Wiley
& Sons, Inc.; Hoboken, NJ: pp. 5–37. 2007
|
|
5
|
Gupta A, Mann B, Kumar R and Sangwan RB:
Antioxidant activity of cheddar cheeses at different stages of
ripening. Int J Dairy Technol. 62:339–347. 2009. View Article : Google Scholar
|
|
6
|
Spolaore P, Joannis-Cassan C, Duran E and
Isambert A: Commercial applications of microalgae. J Biosci Bioeng.
101:87–96. 2006. View Article : Google Scholar
|
|
7
|
Qian ZJ, Jung WK, Kang KH, Ryu B, Kim SK,
Je JY, Heo SJ, Oh C, Kang DH, Park WS and Choi IW: In vitro
antioxidant activities of the fermented marine microalga Pavlova
lutheri (haptophyta) with the yeast Hansenula
polymorpha. J Phycol. 48:475–482. 2012. View Article : Google Scholar
|
|
8
|
Xiao Y, Haase H, Young WG and Bartold PM:
Development and transplantation of a mineralized matrix formed by
osteoblasts in vitro for bone regeneration. Cell Transplant.
13:15–25. 2004. View Article : Google Scholar
|
|
9
|
de Crombrugghe B, Lefebvre V and Nakashima
K: Regulatory mechanisms in the pathways of cartilage and bone
formation. Curr Opin Cell Biol. 13:721–727. 2001. View Article : Google Scholar
|
|
10
|
Teitelbaum SL: Bone resorption by
osteoclasts. Science. 289:1504–1508. 2000. View Article : Google Scholar
|
|
11
|
Ducy P, Schinke T and Karsenty G: The
osteoblast: A sophisticated fibroblast under central surveillance.
Science. 289:1501–1504. 2000. View Article : Google Scholar
|
|
12
|
Day TF, Guo X, Garrett-Beal L and Yang Y:
Wnt/beta-catenin signaling in mesenchymal progenitors controls
osteoblast and chondrocyte differentiation during vertebrate
skeletogenesis. Dev Cell. 8:739–750. 2005. View Article : Google Scholar
|
|
13
|
Greenblatt MB, Shim JH and Glimcher LH:
Mitogen-activated protein kinase pathways in osteoblasts. Annu Rev
Cell Dev Biol. 29:63–79. 2013. View Article : Google Scholar
|
|
14
|
Thouverey C and Caverzasio J: The p38α
MAPK positively regulates osteoblast function and postnatal bone
acquisition. Cell Mol Life Sci. 69:3115–3125. 2012. View Article : Google Scholar
|
|
15
|
Danciu TE, Adam RM, Naruse K, Freeman MR
and Hauschka PV: Calcium regulates the PI3K-Akt pathway in
stretched osteoblasts. Febs Lett. 536:193–197. 2003. View Article : Google Scholar
|
|
16
|
Peverali FA, Basdra EK and Papavassiliou
AG: Stretch-mediated activation of selective MAPK subtypes and
potentiation of AP-1 binding in human osteoblastic cells. Mol Med.
7:68–78. 2001.
|
|
17
|
Granet C, Boutahar N, Vico L, Alexandre C
and Lafage-Proust MH: MAPK and SRC-kinases control EGR-1 and
NF-kappa B inductions by changes in mechanical environment in
osteoblasts. Biochem Biophys Res Commun. 284:622–631. 2001.
View Article : Google Scholar
|
|
18
|
Eichner A, Brock J, Heldin CH and
Souchelnytskyi S: Bone morphogenetic protein-7 (OP1) and
transforming growth factor-beta 1 modulate 1,25(OH)2-vitamin
D3-induced differentiation of human osteoblasts. Exp Cell Res.
275:132–142. 2002. View Article : Google Scholar
|
|
19
|
Ryu B, Li Y, Qian ZJ, Kim MM and Kim SK:
Differentiation of human osteosarcoma cells by isolated
phlorotannins is subtly linked to COX-2, iNOS, MMPs and MAPK
signaling: Implication for chronic articular disease. Chem Biol
Interact. 179:192–201. 2009. View Article : Google Scholar
|
|
20
|
Moller NP, Scholz-Ahrens KE, Roos N and
Schrezenmeir J: Bioactive peptides and proteins from foods:
Indication for health effects. Eur J Nutr. 47:171–182. 2008.
View Article : Google Scholar
|
|
21
|
Eriksson LS: Administration of aspartate
to patients with liver-cirrhosis. Clin Nutr. 4:88–96. 1985.
View Article : Google Scholar
|
|
22
|
Ryu B, Qian ZJ and Kim SK: Purification of
a peptide from seahorse, that inhibits TPA-induced MMP, iNOS and
COX-2 expression through MAPK and NF-kappaB activation, and induces
human osteoblastic and chondrocytic differentiation. Chem Biol
Interact. 184:413–422. 2010. View Article : Google Scholar
|
|
23
|
Huang H, Ryu J, Ha J, Chang EJ, Kim HJ,
Kim HM, Kitamura T, Lee ZH and Kim HH: Osteoclast differentiation
requires TAK1 and MKK6 for NFATc1 induction and NF-kappaB
transactivation by RANKL. Cell death Differ. 13:1879–1891. 2006.
View Article : Google Scholar
|
|
24
|
Villa I, Melzi R, Pagani F, Ravasi F,
Rubinacci A and Guidobono F: Effects of calcitonin gene-related
peptide and amylin on human osteoblast-like cells proliferation.
Eur J Pharmacol. 409:273–278. 2000. View Article : Google Scholar
|
|
25
|
Wang L, Li JY, Zhang XZ, Liu L, Wan ZM, Li
RX and Guo Y: Involvement of p38MAPK/NF-κB signaling pathways in
osteoblasts differentiation in response to mechanical stretch. Ann
Biomed Eng. 40:1884–1894. 2012. View Article : Google Scholar
|