Introduction
Liver cancer is one of the most common malignancies
worldwide, with an increasing incidence, especially in China
(1–3). Hepatocellular carcinoma (HCC) is the
most common type of primary liver cancer; it is estimated to be the
second leading cause of cancer-related death in developing
countries and the sixth leading cause in developed countries
(4–6). Despite recent advances in HCC
management, including surgical resection, radiofrequency ablation,
liver transplantation, and transcatheter arterial chemoembolization
(TACE), the prognosis of most HCC patients remains poor. In
addition, the molecular pathogenesis of HCC is not well understood.
Therefore, it is crucial to clarify the effector molecules and
signaling pathways underlying HCC tumor progression and metastasis
to develop novel therapeutic strategies and more effective
treatments.
MicroRNAs (miRNAs/miRs) are endogenous small (19–25
nucleotide) non-coding RNAs. By binding to the 3′-untranslated
regions (3′-UTRs) of targeted messenger RNAs (mRNAs) (7–9),
miRNAs play important roles in post-transcriptional regulation and
numerous biological processes such as proliferation, survival, and
apoptosis (10,11). Recently, several studies have found
that miRNAs are also critical for cancer invasion and metastasis.
Dysregulated expression of miRNAs is correlated with many human
cancers, including HCC. In addition, miRNAs are emerging as
promising diagnostic and prognostic markers for human cancers
(12,13).
miR-34a reportedly acts as a tumor suppressor in
many cancers, including pancreatic cancer, prostate cancer,
glioblastoma, colon cancer, and breast cancer (14–17).
In pancreatic cancer, miR-34a inhibits stem cell self-renewal by
downregulating Bcl-2 and Notch (18). In glioblastoma, miR-34a inhibits
cell proliferation due to its regulation of the TGF-β signaling
network (19). Several recent
studies have shown that the expression of miR-34a is dramatically
decreased in clinical HCC specimens, suggesting that miR-34a
represents a potential target for HCC treatment (20–22).
However, the biological effect and underlying mechanism of miR-34a
in HCC tumorigenesis and metastasis remains to be elucidated. Thus,
further exploration of miR-34a is of utmost significance.
Lactate dehydrogenase A (LDHA) plays an important
role in tumor cell metabolism (23–25).
It has recently been reported that LDHA expression is correlated
with progression and survival outcomes in multiple cancers,
including renal cancer, gastric cancer, esophageal squamous cell
carcinoma, and pancreatic cancer. Moreover, several oncogenes and
deacetylases, including HIF-1α, SIRT2, and MYC, contribute to the
regulation of LDHA expression and post-transcriptional modification
(26,27). Together, these studies indicate
that LDHA could be a novel therapeutic target for multiple human
cancers, including HCC.
In this study, we evaluated the expression of
miR-34a in HCC tissues and cell lines. Furthermore, functional
studies of the effects of miR-34a and LDHA on cell proliferation,
invasion, and glycolysis in HCC cells were performed to explore the
underlying connection between miR-34a and HCC.
Materials and methods
Cell lines and culture
The HCC cell lines Huh7, HCCLM3, Hep3B, Mahlavu,
SNU475, and human hepatocyte line L02 were obtained from the Cell
Bank of the Chinese Academy of Sciences (Shanghai, China) and
cultured in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with
10% fetal bovine serum and 1% penicillin/streptomycin (Gibco;
Thermo Fisher Scientific, Inc.) at 37°C in a humidified atmosphere
with 5% CO2.
Clinical samples
The study protocols were approved by the Ethics
Committee of Qingdao No. 6 People's Hospital following the ethical
standards outlined in the Declaration of Helsinki. Written informed
consent was obtained from all patients. Tissue samples from 22
pairs of HCC tissues (HC) and their corresponding adjacent tissues
(normal, located ~2 cm apart) were collected in the Department of
Hepatology, Qingdao No. 6 People's Hospital. The clinical profiles
of the 22 participants in the study, such as gender, age, BMI,
tumor size, and Child-Pugh class were described in Table I. Tissue samples were obtained and
immediately stored in liquid nitrogen for further reverse
transcription-quantitative polymerase chain reaction (RT-qPCR
analysis.
 | Table I.The clinical profiles of the 22
patients with hepatocellular carcinoma. |
Table I.
The clinical profiles of the 22
patients with hepatocellular carcinoma.
| Clinical
variable | No. of patients
(n=22) |
|---|
| Gender |
|
|
Female | 8 |
|
Male | 14 |
| Age |
|
|
Median |
58.7 |
|
Range | 40–79 |
| HBV |
|
|
Positive | 22 |
|
Negative | 0 |
| Tumor size |
|
| ≤5
cm | 13 |
| >5
cm | 9 |
| Child-Pugh
class |
|
| A | 22 |
| B | 0 |
| BMI |
|
|
Median |
21.2 |
|
Range | 16–26.8 |
RNA extraction and RT-qPCR
Total RNA was extracted from the tissues and cell
lines using TRIzol reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). RT was performed using a Prime-Script RT Reagent kit (Takara
Biotechnology Co., Ltd., Dalian, China) and RT-qPCR was performed
using SYBR Premix Ex Taq (Takara Biotechnology Co., Ltd.). MicroRNA
extraction was conducted using a MicroRNA Extraction kit (Tiangen
Biotech Co., Ltd., Beijing, China) and RT-qPCR was performed with
SYBR Premix Ex Taq (Takara Biotechnology Co., Ltd.) according to
the manufacturer's protocol. GAPDH and RNU6B were used as
normalizing controls for mRNA and miRNA quantification,
respectively. The primers were as follows: miR-34a forward,
5′-UGGCAGUGUCUUAGCUGGUUGU-3′ and reverse,
5′-AACCAGCUAAGACACUGCCAUU-3′; LDHA, forward,
5′-TTGGTCCAGCGTAACGTGAAC-3′ and reverse,
5′-CCAGGATGTGTAGCCTTTGAG-3′. The 2−ΔΔCq method was used
to determine relative expression levels.
Cell proliferation assay
HuH7 and HCCLM3 cells were seeded in 96-well plates
and incubated for 24 h before being transfected with miR-34a or
scrambled mimics. After the cells were incubated for another 48 h,
an MTT assay was performed according to the manufacturer's
instructions (Molecular Probes; Thermo Fisher Scientific, Inc.).
The absorbance at 570 nm was determined using a Spectra Max 250
spectrophotometer (Molecular Devices, Sunnyvale, CA, USA).
Foci formation assay
HuH7 and HCCLM3 cells were seeded in 6-well plates
at a density of 2,000 cells per well and transfected with miR-34a
or scrambled mimics. When the number of clones exceeded 50, the
cells were stained with 0.06% crystal violet and the foci numbers
were counted.
Cell invasion assay
A Transwell invasion assay was used to determine the
invasion capacity of the tumor cells. Briefly, HuH7 and HCCLM3
cells were transfected with miR-34a or scrambled mimics, cultivated
for 24 h, and seeded onto the Matrigel-coated chambers (24-well; BD
Biosciences, San Jose, CA, USA) in serum-free DMEM. DMEM containing
10% FBS was added to the lower chamber. The matrix and non-invaded
cells were removed after 48 h of incubation, while the invaded
cells were fixed, stained, and counted.
Wound-healing assay
After transfection, HuH7 cells were seeded in 6-well
plates and grown to 90% confluence. After 24 h, linear scratch
wounds were created using pipette tips and the cells were washed
three times with PBS. The cells were then incubated in DMEM
containing 5% FBS. Cell movement at the wound site was monitored
and photographed at 0 and 24 h. The percentage of wound closure was
analyzed and compared as described previously (24).
Measurement of glucose uptake and
lactate production
To assess glucose uptake and lactate production,
HuH7 and HCCLM3 cells were transfected with miR-34a or scrambled
mimics, and the cell culture medium was collected 48 h after
transfection. An Amplex® Red Glucose/Glucose Oxidase
Assay kit (Invitrogen; Thermo Fisher Scientific, Inc.) was then
used to measure glucose uptake, and a lactate assay kit
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was used to
determine lactate production.
Western blotting
HuH7 and HCCLM3 cells were transfected with either
miR-34a or scrambled mimics. After 48 h, protein was extracted
using RIPA lysis buffer. Protein concentrations were quantified
using a Protein BCA Assay kit (Pierce; Thermo Fisher Scientific,
Inc.). The protein samples were separated by 10% SDS-PAGE then
transferred to polyvinylidene difluoride membranes (EMD Millipore,
Billerica, MA, USA). After blocking in 5% skim milk for 1 h at room
temperature, the membranes were incubated with antibodies against
LDHA and β-actin (Affinity Biosciences, Columbus, OH, USA) at 4°C
overnight. A peroxidase-conjugated secondary antibody (dilution
1:1,500) was applied for 1 h at room temperature to visualize the
target proteins. The target proteins were visualized using Western
blotting detection reagents (Thermo Fisher Scientific, Inc.) and
then exposed to X-ray film (Kodak, Inc., Rochester, NY, USA). The
optical density (OD) (target proteins)/OD (β-actin) was used to
quantify protein expression.
LDHA-expressing vector
Full-length LDHA cDNA was purchased from GeneCopeia
(Rockville, MD, USA) and sub-cloned into the expression vector
pcDNA3.1(+) (GeneCopeia). The vector pcDNA3.1(+) was used as a
negative control.
Statistical analysis
All in vitro experiments were performed in
triplicate. The results are presented as means ± standard
deviation. Statistical comparisons between two groups were analyzed
using t-tests and χ2 tests. Statistical comparisons
between multiple groups were analyzed using one-way ANOVA followed
by Newman-Keuls post-hoc comparison test. P<0.05 was considered
to indicate a statistically significant difference (SPSS 16.0;
SPPS, Inc., Chicago, IL, USA).
Results
miR-34a was significantly
downregulated in HCC cell lines and clinical specimens
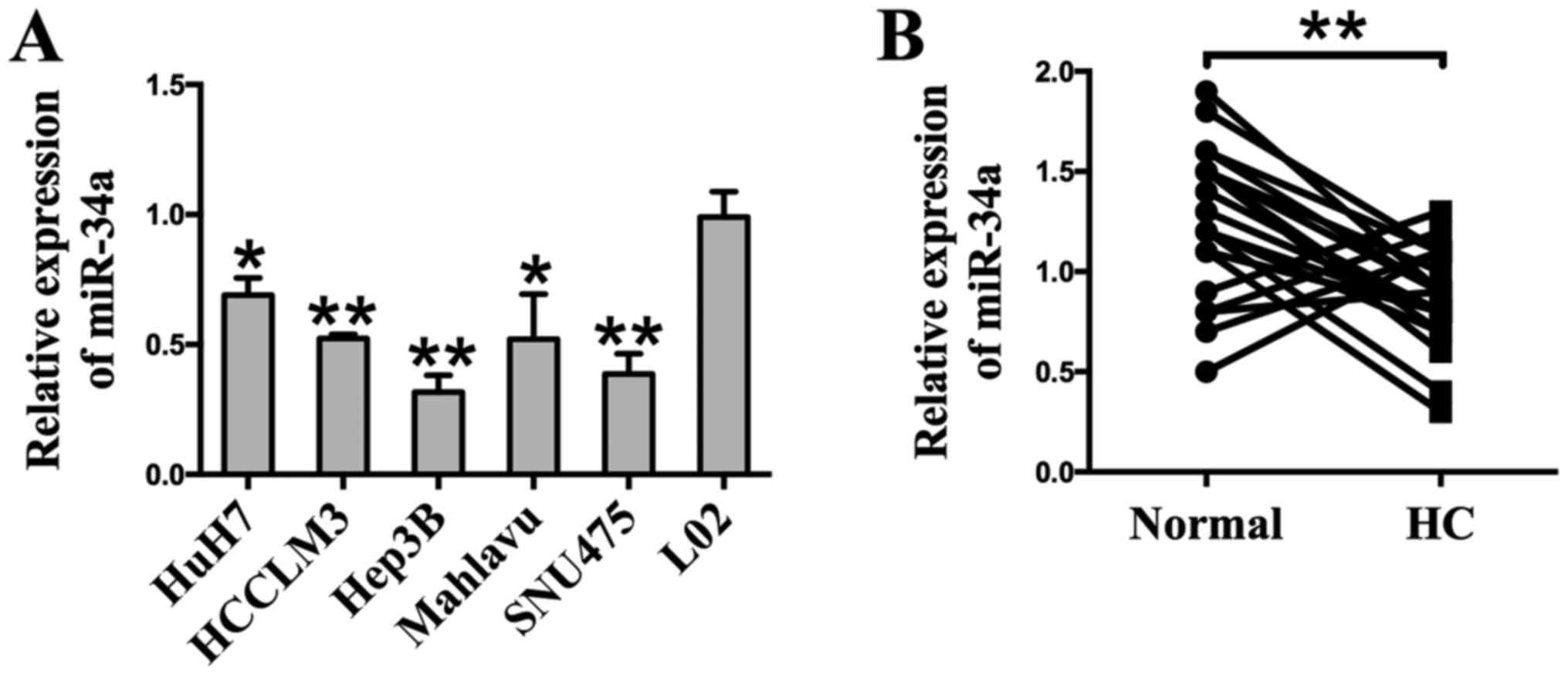
A RT-qPCR analysis was employed to detect the
expression of miR-34a. The results show that the expression of
miR-34a was markedly downregulated in six different HCC cell lines
(Huh7, HCCLM3, Hep3B, Mahlavu, and SNU475) compared to the human
hepatocyte cell line L02 (Fig.
1A). To determine the expression of miR-34a in clinical
specimens, HCC tissues (HC) and their matched adjacent normal
tissues (Normal) were examined through RT-qPCR analysis. Compared
with adjacent normal tissues, we found that 77.3% (17 of 22
patients, P<0.01) of tumor tissues showed decreased miR-34a
levels (Fig. 1B). Taken together,
these results indicate that miR-34a is downregulated at a high
frequency in HCC, and may be related to HCC carcinogenesis.
miR-34a inhibits cell proliferation
and invasion
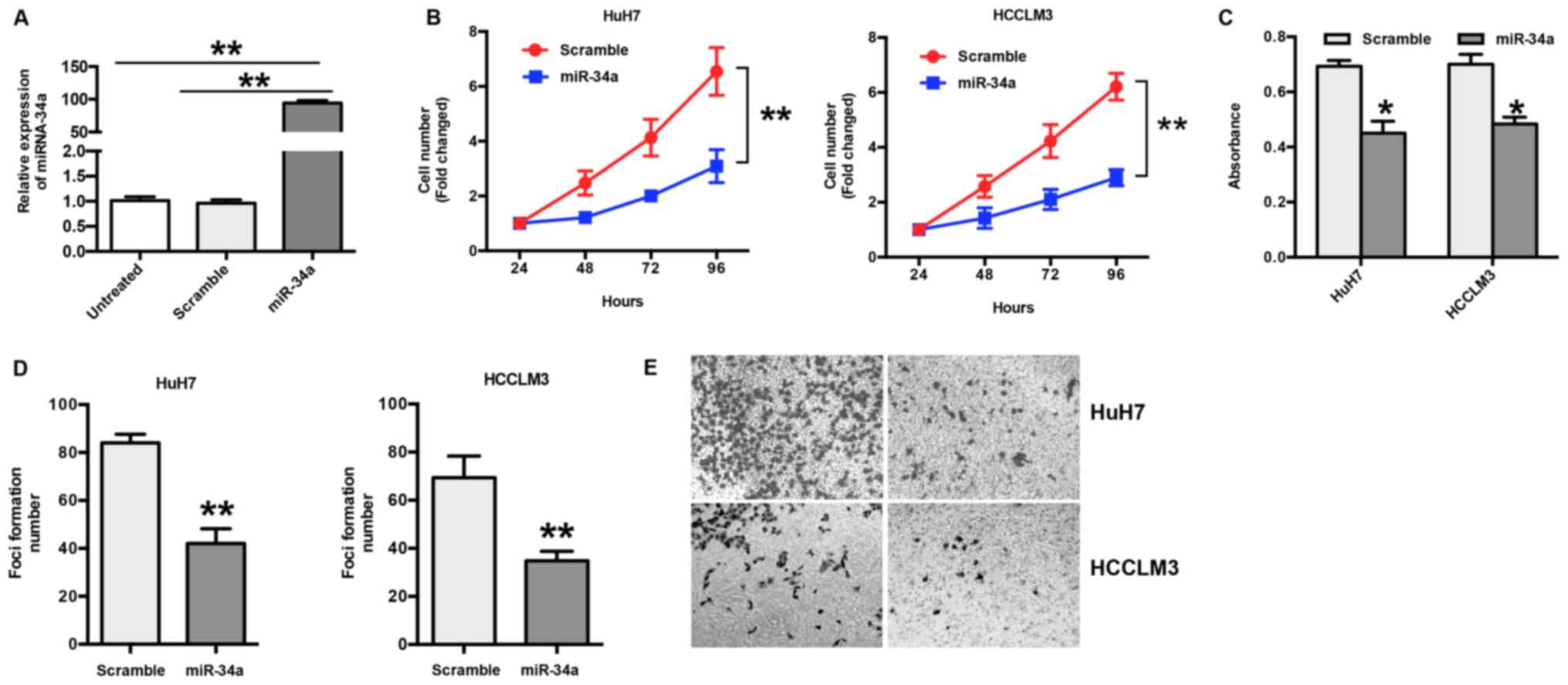
The expression of miR-34a was examined in HuH7 and
HCCLM3 cells following transfection with miR-34a or scramble
mimics. The RT-qPCR results show a significant increase in miR-34a
(~94 fold) in transfected cells compared to scramble or untreated
cells (P<0.001) (Fig. 2A). To
explore the biological effects of miR-34a in HCC, HuH7 and HCCLM3
cells were transfected with miR-34a or scramble mimics, and the
number of cells was counted. The results show that ectopic
expression of miR-34a significantly suppressed the proliferation of
HuH7 and HCCLM3 cells in a time-dependent manner (P<0.05)
(Fig. 2B); this was further
confirmed by an MTT assay (Fig.
2C). In addition, the results of the foci formation assay show
that the overexpression of miR-34a led to decreased foci formation
of HuH7 and HCCLM3 cells (P<0.01) (Fig. 2D). To further explore the function
of miR-34a in HCC, a Transwell invasion assay was performed. The
results show that overexpression of miR-34a significantly inhibited
invasion in HuH7 and HCCLM3 cells compared with the scramble group
(Fig. 2E).
miR-34a inhibits glycolysis in
HCC
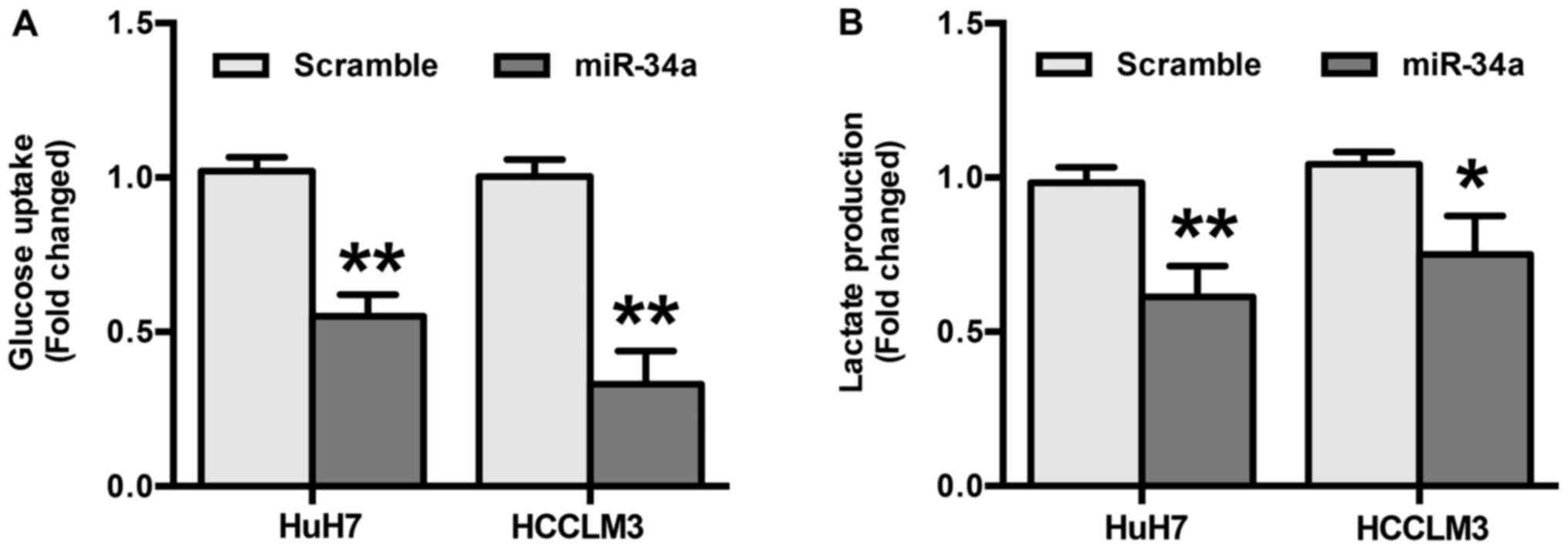
To explore the role miR-34a in glycolysis in HCC,
differences in metabolic parameters were detected after HuH7 and
HCCLM3 cells were transfected with miR-34a or scramble mimics. The
results show that overexpression of miR-34a significantly decreased
glucose uptake (P<0.01) (Fig.
3A). In addition, the results indicate that miR-34a mimics
induced a decrease in the production of extracellular lactate
(P<0.05) (Fig. 3B). Taken
together, these results suggest that the inhibition of glycolysis
by miR-34a may be responsible for the suppression of migration and
invasion in HCC cells.
miR-34a inhibits LDHA expression in
HCC
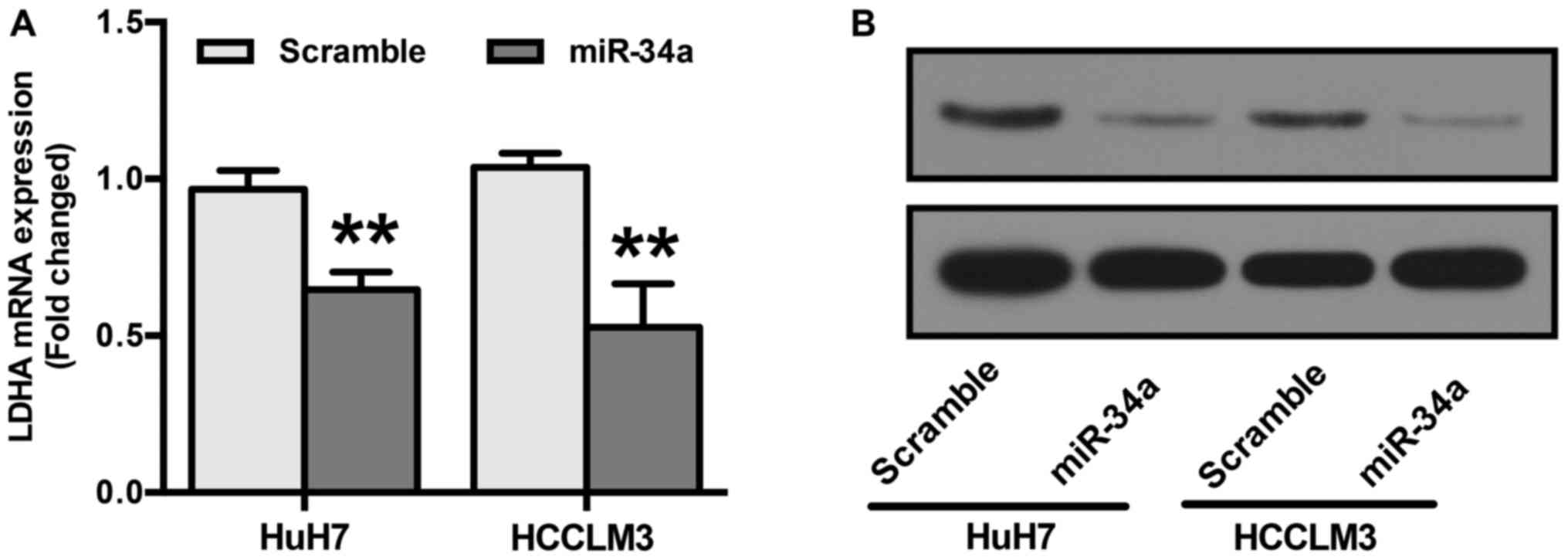
LDHA is reportedly a direct target of miR-34a
(28). To confirm this, RT-qPCR
and Western blot analyses were performed. HuH7 and HCCLM3 cells
were transfected with miR-34a or scramble mimics before examining
LDHA expression. Our results show that cells transfected with
miR-34a mimics showed a significant reduction in both the mRNA and
protein levels of LDHA (Fig. 4A and
B).
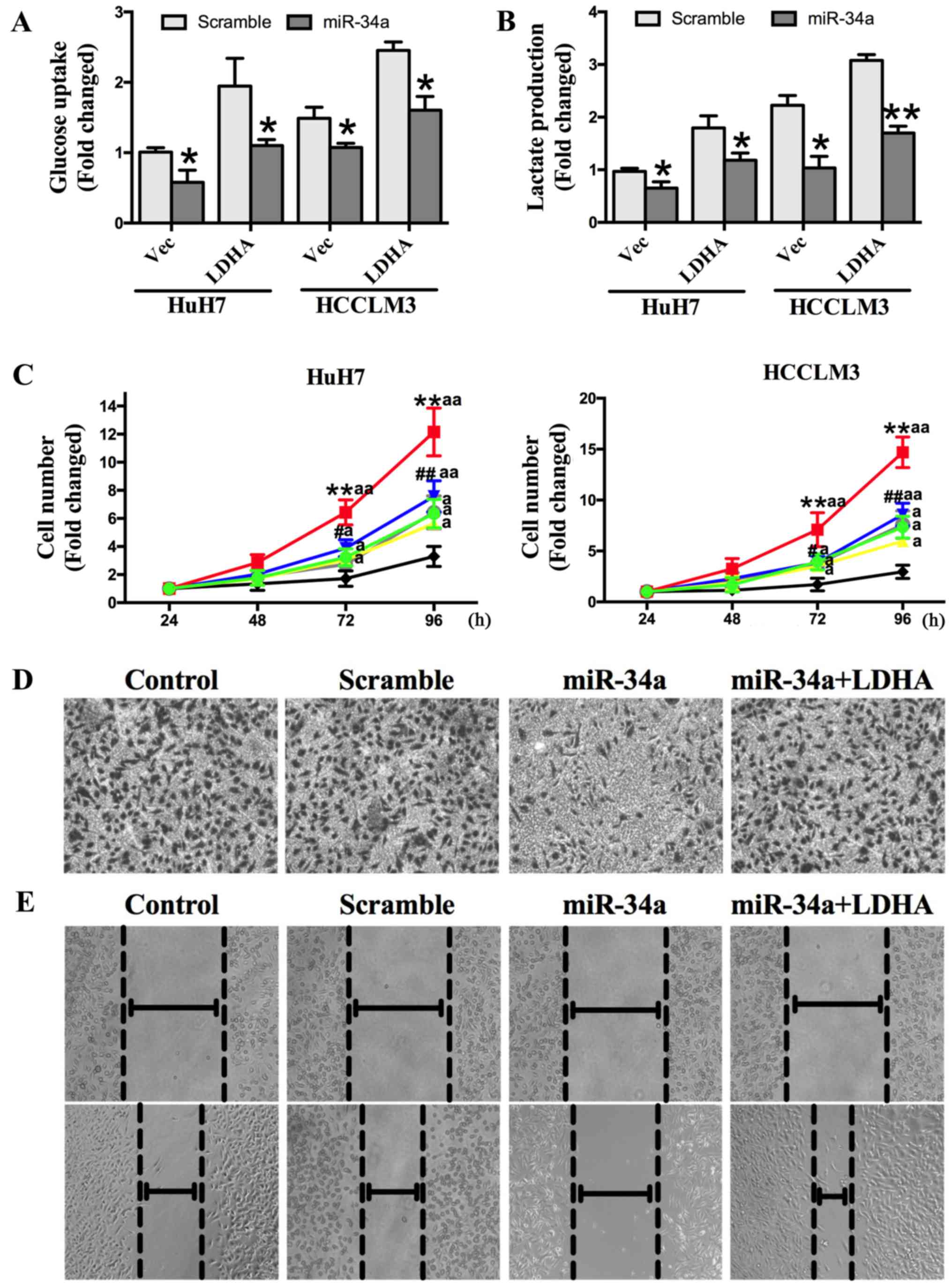
miR-34a inhibits LDHA-induced
glycolysis, cell proliferation, and invasion
To confirm that miR-34a inhibited glycolysis, cell
proliferation, and invasion of HCC cells by targeting LDHA, we
transfected HuH7 and HCCLM3 cells with an LDHA-expressing vector,
control vector, control vector + miR-34a mimics, control vector + a
scrambled oligonucleotide, LDHA expressing vector + miR-34a mimics,
or LDHA expressing vector + a scrambled oligonucleotide. The
results show that overexpression of LDHA could increase HCC cell
glucose uptake and lactate production, and that this effect was
abolished by transfection with miR-34a mimics (Fig. 5A and B). We also found that the
overexpression of LDHA could promote cell proliferation, while
miR-34a inhibited cell proliferation. The increased cell
proliferation induced by LDHA was significantly repressed by
miR-34a (Fig. 5C). Transwell
invasion and wound-healing assays showed that miR-34a mimics
decreased invasion capacity, which was increased by the
overexpression of LDHA (Fig. 5D and
E). Together, these findings demonstrate that miR-34a inhibits
HCC glycolysis, and cell proliferation and invasion in vitro
by targeting LDHA.
Discussion
Although outstanding advances in surgical techniques
and radio-chemotherapy regimens have been achieved in recent
decades, the prognosis for most patients with HCC remains poor.
Therefore, novel therapeutic strategies are needed to treat HCC
more effectively (3,4,6).
Increasing evidence has demonstrated that miRNAs
play a vital role in the pathogenesis, clinical metastasis, and
progression of multiple cancers, including HCC (29–31).
Fang et al (32) reported
that miR-383 was downregulated in HCC tissues compared with their
adjacent normal tissues. In addition, overexpression of miR-383
significantly suppressed cell proliferation and invasion,
indicating that miR-383 may act as a tumor suppressor in HCC. Kota
et al (33) demonstrated
that systemic administration of miR-26a resulted in significant
inhibition of HCC cell proliferation, and induced protection
against disease progression without toxicity. Thus, understanding
the crucial role of miRNAs in HCC may provide novel diagnostic,
prognostic, and therapeutic potential.
miR-34a is a member of the highly conserved miR-34
family. It is located at the chromosome lp36 locus, which is
particularly susceptible to molecular events that may disturb the
balance of proliferation and apoptosis (34,35).
miR-34a is also one of multiple recently discovered miRNAs that are
regulated by p53 (17,36). Studies have shown that miR-34a is
an important molecule that inhibits the growth of many cancers.
Decreased expression of miR-34a has been found in pancreatic
cancer, cervical cancer, prostate cancer, glioblastoma, colon
cancer, esophageal squamous cell carcinoma, breast cancer, and lung
cancer (37–39). In cancers like pancreatic cancer
and glioblastoma, miR-34a was reported to inhibit cell
proliferation and induce apoptosis by downregulating Bcl-2 and
Notch or the TGF-β signaling network (18,19,40).
However, the role of miR-34a in the pathogenesis of HCC is
unclear.
Several studies have shown that miR-34a may act as
an oncogene (41). Pineau et
al (42) found that miR-34a
was highly expressed in liver cancer, and was positively related to
HCC progression. In a tamoxifen-induced mouse liver cancer model,
Pogribny et al (43) found
that miR-34 expression was increased. However, more and more
studies have recently shown that the expression of miR-34a is
dramatically decreased in clinical HCC specimens, suggesting that
miR-34a is a potential target for HCC treatment (20–22).
Dang et al (44) detected
miR-34a expression in 60 HCC tissues and adjacent normal tissues
and found that miRNA-34a expression in HCC tissues was
significantly lower than in normal tissues. Moreover, a miR-34a
mimic was reported to inhibit HCC cell growth and induce apoptosis.
However, the mechanism remains unknown. In this study, we found
that miR-34a was downregulated in human liver cancer tissues, and
that the upregulation of miR-34a inhibits liver cancer cell
proliferation, migration, and invasion, consistent with previous
studies (20–22,44).
Moreover, we found that miR-34a negatively regulates the expression
of LDHA in HCC cell lines, which consequently inhibits
LDHA-dependent glucose uptake in cancer cells, leading to reduced
cell proliferation and invasion.
Aberrant metabolism has been shown to play an
important role in the progression and metastasis of multiple
cancers (45–47). Among a series of enzymes involved
in cancer metabolism, LDHA is reported to be of vital importance
and is involved in proliferation and glycolysis in gastric cancer,
breast cancer, and pancreatic cancer (23,25,27,48).
Indeed, some references have already proved that miR-34a negatively
regulates the expression level of LDHA in various cancers (23,25,28,49).
By performing luciferase reporter assays, or cloning of 3′-UTRs,
miR-34a was reported to have direct role on LDHA mRNA stability in
various cell lines (23,28), including HCC cells (50). In a previous study based on HCC
cells, Wang et al reported that miR-34a specifically bind to
the 3′-UTR region of LDHA, acting as a negative regulator (50). In this study, we confirmed that
LDHA is also involved in HCC metabolism, and an LDHA-expressing
vector significantly improved HCC cell glycolysis, proliferation,
migration, and invasion. Furthermore, we also confirmed that LDHA
may be a target gene for miR-34a. The function of miR-34a in HCC
may be partly due to its regulation of LDHA and the subsequent
reprogramming of glucose metabolism. The increased cell glycolysis,
proliferation, and invasion triggered by LDHA was effectively
inhibited by miR-34a in HCC cell lines, which indicates that the
miR-34a-LDHA axis could be a promising therapeutic target for more
effective HCC treatment.
To the best of our knowledge, this is the first
study of the effect of miR-34a on HCC glucose metabolism. However,
some limitations of our study should be noted. First, this was an
in vitro study that used clinical samples, but it lacked an
in vivo evaluation of the potential impact of miR-34a.
Second, the carcinogenesis of HCC is complicated, which means
miR-34a may also target other genes. In addition, although the
results of this study showed that LDHA was downregulated in miR-34a
transfected HuH7 and HCCLM3 cells, further analysis about the
direct role of miR-34a on LDHA mRNA stability was not conducted,
which may decrease the robustness of the conclusion of our study.
Nevertheless, our study provides useful insight into the effect of
miR-34a on cell proliferation, invasion, and glycolysis in HCC cell
lines. Future studies are needed.
In summary, the results of this study suggest that
miR-34a inhibits HCC glycolysis, cell proliferation, and invasion
in vitro by targeting LDHA. miR-34a functions as a negative
regulator of glucose metabolism, which may serve as a novel marker
for liver cancer prognosis.
The results of this study suggest that miR-34a
inhibits HCC glycolysis, cell proliferation, and invasion in
vitro by targeting LDHA.
References
|
1
|
Zhong GC, Liu Y, Chen N, Hao FB, Wang K,
Cheng JH, Gong JP and Ding X: Reproductive factors, menopausal
hormone therapies and primary liver cancer risk: A systematic
review and dose-response meta-analysis of observational studies.
Hum Reprod Update. 23:126–138. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nio K, Yamashita T and Kaneko S: The
evolving concept of liver cancer stem cells. Mol Cancer. 16:42017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rao CV, Asch AS and Yamada HY: Frequently
mutated genes/pathways and genomic instability as prevention
targets in liver cancer. Carcinogenesis. 38:2–11. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rabinel P, Dousse D, Muscari F and Suc B:
Management of liver cancer. The Surgeon's point of view. Rep Pract
Oncol Radiother. 22:176–180. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sia D, Villanueva A, Friedman SL and
Llovet JM: Liver cancer cell of origin, molecular class, and
effects on patient prognosis. Gastroenterology. 152:745–761. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Stroehl YW, Letzen BS, van Breugel JM,
Geschwind JF and Chapiro J: Intra-arterial therapies for liver
cancer: Assessing tumor response. Expert Rev Anticancer Ther.
17:119–127. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ma J, Lin J, Qian J, Qian W, Yin J, Yang
B, Tang Q, Chen X, Wen X, Guo H and Deng Z: miR-378 promotes the
migration of liver cancer cells by down-regulating Fus expression.
Cell Physiol Biochem. 34:2266–2274. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pang F, Zha R, Zhao Y, Wang Q, Chen D,
Zhang Z, Chen T, Yao M, Gu J and He X: miR-525-3p enhances the
migration and invasion of liver cancer cells by downregulating
ZNF395. PLoS One. 9:e908672014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yin J, Bai Z, Song J, Yang Y, Wang J, Han
W, Zhang J, Meng H, Ma X, Yang Y, et al: Differential expression of
serum miR-126, miR-141 and miR-21 as novel biomarkers for early
detection of liver metastasis in colorectal cancer. Chin J Cancer
Res. 26:95–103. 2014.PubMed/NCBI
|
|
10
|
Guo W, Qiu Z, Wang Z, Wang Q, Tan N, Chen
T, Chen Z, Huang S, Gu J, Li J, et al: miR-199a-5p is negatively
associated with malignancies and regulates glycolysis and lactate
production by targeting hexokinase 2 in liver cancer. Hepatology.
62:1132–1144. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu HB, Hua Y and Jin ZX: Effects of
MicroRNA-132 transfection on the proliferation and apoptosis of
human liver cancer cells in vitro and in vivo. Zhongguo Yi Xue Ke
Xue Yuan Xue Bao. 37:30–36. 2015.PubMed/NCBI
|
|
12
|
Lu Z, Zhang W, Gao S, Jiang Q, Xiao Z, Ye
L and Zhang X: miR-506 suppresses liver cancer angiogenesis through
targeting sphingosine kinase 1 (SPHK1) mRNA. Biochem Biophys Res
Commun. 468:8–13. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sun B, Li J, Shao D, Pan Y, Chen Y, Li S,
Yao X, Li H, Liu W, Zhang M, et al: Adipose tissue-secreted miR-27a
promotes liver cancer by targeting FOXO1 in obese individuals. Onco
Targets Ther. 8:735–744. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dong P, Xiong Y, Watari H, Hanley SJ,
Konno Y, Ihira K, Yamada T, Kudo M, Yue J and Sakuragi N: miR-137
and miR-34a directly target Snail and inhibit EMT, invasion and
sphere-forming ability of ovarian cancer cells. J Exp Clin Cancer
Res. 35:1322016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen W, Liu Y, Liang X, Huang Y and Li Q:
Chondroitin sulfate-functionalized polyamidoamine as a
tumor-targeted carrier for miR-34a delivery. Acta Biomater.
57:238–250. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen Y, Sun P, Guo X and Gao A: miR-34a, a
promising novel biomarker for benzene toxicity, is involved in cell
apoptosis triggered by 1,4-benzoquinone through targeting Bcl-2.
Environ Pollut. 221:256–265. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lin Y, Shen J, Li D, Ming J, Liu X, Zhang
N, Lai J, Shi M, Ji Q and Xing Y: miR-34a contributes to
diabetes-related cochlear hair cell apoptosis via SIRT1/HIF-1α
signaling. Gen Comp Endocrinol. 246:63–70. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ji Q, Hao X, Zhang M, Tang W, Yang M, Li
L, Xiang D, Desano JT, Bommer GT, Fan D, et al: MicroRNA miR-34
inhibits human pancreatic cancer tumor-initiating cells. PLoS One.
4:e68162009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Genovese G, Ergun A, Shukla SA, Campos B,
Hanna J, Ghosh P, Quayle SN, Rai K, Colla S, Ying H, et al:
microRNA regulatory network inference identifies miR-34a as a novel
regulator of TGF-β signaling in glioblastoma. Cancer Discov.
2:736–749. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tryndyak VP, Ross SA, Beland FA and
Pogribny IP: Down-regulation of the microRNAs miR-34a, miR-127, and
miR-200b in rat liver during hepatocarcinogenesis induced by a
methyl-deficient diet. Mol Carcinog. 48:479–487. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen Q, Li L, Tu Y, Zheng LL, Liu W, Zuo
XY, He YM, Zhang SY, Zhu W, Cao JP, et al: miR-34a regulates
apoptosis in liver cells by targeting the KLF4 gene. Cell Mol Biol
Lett. 19:52–64. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang XP, Zhou J, Han M, Chen CB, Zheng YT,
He XS and Yuan XP: MicroRNA-34a regulates liver regeneration and
the development of liver cancer in rats by targeting Notch
signaling pathway. Oncotarget. 8:13264–13276. 2017.PubMed/NCBI
|
|
23
|
Kaller M, Liffers ST, Oeljeklaus S,
Kuhlmann K, Röh S, Hoffmann R, Warscheid B and Hermeking H:
Genome-wide characterization of miR-34a induced changes in protein
and mRNA expression by a combined pulsed SILAC and microarray
analysis. Mol Cell Proteomics. 10:M111.0104622011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Du JY, Wang LF, Wang Q and Yu LD: miR-26b
inhibits proliferation, migration, invasion and apoptosis induction
via the downregulation of
6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase-3 driven
glycolysis in osteosarcoma cells. Oncol Rep. 33:1890–1898. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang J, Wang H, Liu A, Fang C, Hao J and
Wang Z: Lactate dehydrogenase A negatively regulated by miRNAs
promotes aerobic glycolysis and is increased in colorectal cancer.
Oncotarget. 6:19456–19468. 2015.PubMed/NCBI
|
|
26
|
Song K, Kwon H, Han C, Zhang J, Dash S,
Lim K and Wu T: Active glycolytic metabolism in CD133(+)
hepatocellular cancer stem cells: Regulation by MIR-122.
Oncotarget. 6:40822–40835. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ullmann P, Qureshi-Baig K, Rodriguez F,
Ginolhac A, Nonnenmacher Y, Ternes D, Weiler J, Gabler K, Bahlawane
C, Hiller K, et al: Hypoxia-responsive miR-210 promotes
self-renewal capacity of colon tumor-initiating cells by repressing
ISCU and by inducing lactate production. Oncotarget. 7:65454–65470.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xiao X, Huang X, Ye F, Chen B, Song C, Wen
J, Zhang Z, Zheng G, Tang H and Xie X: The miR-34a-LDHA axis
regulates glucose metabolism and tumor growth in breast cancer. Sci
Rep. 6:217352016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang Q, Jiang W, Zhuang C, Geng Z, Hou C,
Huang D, Hu L and Wang X: microRNA-22 downregulation of galectin-9
influences lymphocyte apoptosis and tumor cell proliferation in
liver cancer. Oncol Rep. 34:1771–1778. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Retraction notice to microarray analysis
of microRNA expression in liver cancer tissues and normal control
[GENE 523/2 (2014) 158–60]. Gene. 578:1372016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu C, Wang C, Wang J and Huang H:
miR-1297 promotes cell proliferation by inhibiting RB1 in liver
cancer. Oncol Lett. 12:5177–5182. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fang Z, He L, Jia H, Huang Q, Chen D and
Zhang Z: The miR-383-LDHA axis regulates cell proliferation,
invasion and glycolysis in hepatocellular cancer. Iran J Basic Med
Sci. 20:187–192. 2017.PubMed/NCBI
|
|
33
|
Kota J, Chivukula RR, O'Donnell KA,
Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P,
Torbenson M, Clark KR, et al: Therapeutic microRNA delivery
suppresses tumorigenesis in a murine liver cancer model. Cell.
137:1005–1017. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cui H, Ge J, Xie N, Banerjee S, Zhou Y,
Antony VB, Thannickal VJ and Liu G: miR-34a inhibits lung fibrosis
by inducing lung fibroblast senescence. Am J Respir Cell Mol Biol.
56:168–178. 2017.PubMed/NCBI
|
|
35
|
Fu BC, Lang JL, Zhang DY, Sun L, Chen W,
Liu W, Liu KY, Ma CY, Jiang SL, Li RK and Tian H: Suppression of
miR-34a expression in the myocardium protects against
ischemia-reperfusion injury Through SIRT1 protective pathway. Stem
Cells Dev. 26:1270–1282. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jiang L and Hermeking H: miR-34a and
miR-34b/c suppress intestinal tumorigenesis. Cancer Res.
77:2746–2758. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu YP, Hu H, Xu F and Wen JJ: Relation of
miR-34a expression in diffuse large B cell lymphoma with clinical
prognosis. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 25:455–459. 2017.(In
Chinese). PubMed/NCBI
|
|
38
|
Maroni P, Puglisi R, Mattia G, Care A,
Matteucci E, Bendinelli P and Desiderio MA: In bone metastasis
miR-34a-5p absence inversely correlates with Met expression, while
Met oncogene is unaffected by miR-34a-5p in non-metastatic and
metastatic breast carcinomas. Carcinogenesis. 38:492–503. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Song C, Lu P, Sun G, Yang L and Wang Z and
Wang Z: miR-34a sensitizes lung cancer cells to cisplatin via
p53/miR-34a/MYCN axis. Biochem Biophys Res Commun. 482:22–27. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chen AH, Qin YE, Tang WF, Tao J, Song HM
and Zuo M: miR-34a and miR-206 act as novel prognostic and therapy
biomarkers in cervical cancer. Cancer Cell Int. 17:632017.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sukata T, Sumida K, Kushida M, Ogata K,
Miyata K, Yabushita S and Uwagawa S: Circulating microRNAs,
possible indicators of progress of rat hepatocarcinogenesis from
early stages. Toxicol Lett. 200:46–52. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Pineau P, Volinia S, McJunkin K, Marchio
A, Battiston C, Terris B, Mazzaferro V, Lowe SW, Croce CM and
Dejean A: miR-221 overexpression contributes to liver
tumorigenesis. Proc Natl Acad Sci USA. 107:pp. 264–269. 2010;
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Pogribny IP, Tryndyak VP, Boyko A,
Rodriguez-Juarez R, Beland FA and Kovalchuk O: Induction of
microRNAome deregulation in rat liver by long-term tamoxifen
exposure. Mutat Res. 619:30–37. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Dang Y, Luo D, Rong M and Chen G:
Underexpression of miR-34a in hepatocellular carcinoma and its
contribution towards enhancement of proliferating inhibitory
effects of agents targeting c-MET. PLoS One. 8:e610542013.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chen B, Li H, Zeng X, Yang P, Liu X, Zhao
X and Liang S: Roles of microRNA on cancer cell metabolism. J
Transl Med. 10:2282012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhao E, Maj T, Kryczek I, Li W, Wu K, Zhao
L, Wei S, Crespo J, Wan S, Vatan L, et al: Cancer mediates effector
T cell dysfunction by targeting microRNAs and EZH2 via glycolysis
restriction. Nat Immunol. 17:95–103. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Xu W, Zhang Z, Zou K, Cheng Y, Yang M,
Chen H, Wang H, Zhao J, Chen P, He L, et al: miR-1 suppresses tumor
cell proliferation in colorectal cancer by inhibition of
Smad3-mediated tumor glycolysis. Cell Death Dis. 8:e27612017.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Han RL, Wang FP, Zhang PA, Zhou XY and Li
Y: miR-383 inhibits ovarian cancer cell proliferation, invasion and
aerobic glycolysis by targeting LDHA. Neoplasma. 64:244–252. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhang R, Su J, Xue SL, Yang H, Ju LL, Ji
Y, Wu KH, Zhang YW, Zhang YX, Hu JF and Yu MM: HPV E6/p53 mediated
down-regulation of miR-34a inhibits Warburg effect through
targeting LDHA in cervical cancer. Am J Cancer Res. 6:312–320.
2016.PubMed/NCBI
|
|
50
|
Wang J, Yan S, Zhang W, Zhang H and Dai J:
Integrated proteomic and miRNA transcriptional analysis reveals the
hepatotoxicity mechanism of PFNA exposure in mice. J Proteome Res.
14:330–341. 2015. View Article : Google Scholar : PubMed/NCBI
|