Introduction
Intrahepatic cholangiocarcinoma (IHCC) originates
from the biliary epithelial cells within the liver (1) and is the second most common type of
primary liver tumor, the first being hepatocellular carcinoma.
Although IHCC is a relatively rare cancer, accounting for 10–15% of
primary liver cancers (2), the
global incidence of IHCC has increased over the past few decades
(1). Currently, surgical resection
is the only curative method of IHCC treatment. However, there are
no specific markers for IHCC identified that may aid in early
diagnosis (3), and patients with
IHCC are usually asymptomatic until an advanced stage, with a
surgical resection rate of ~30% (4). Additionally, recurrence is reported
in 46–65% of patients following resection (5), and patients with unresectable IHCC
typically succumb within 12–24 months following diagnosis owing to
the limited adjuvant therapeutic strategies available (6). Therefore, it is crucial to elucidate
the exact molecular mechanisms of IHCC to identify more valuable
diagnostic markers and therapeutic targets.
Accumulating evidence has revealed that long
non-coding RNAs (lncRNAs) are involved in a broad range of cellular
processes, including the regulation of gene expression, genomic
imprinting, chromatin modification, transcription and
post-translational modification (7). lncRNAs have also been demonstrated to
be involved in cancer initiation and development (8,9). A
recent study revealed that lncRNAs are strongly associated with the
clinicopathological outcomes and prognosis of various cancers
(10). Notably, lncRNAs exhibit
cancer and lineage-specific expression patterns, indicating that
they may be drivers of cancer biology and may have potential as
clinical biomarkers (10,11). The lncRNA colon cancer-associated
transcript 2 (CCAT2) has been demonstrated to promote colorectal
cancer growth, metastasis and chromosomal instability through the
upregulation of MYC and WNT expression (12). CCAT2 is an lncRNA that encompasses
the rs6983267 single-nucleotide polymorphism (SNP) located on
chromosome 8q24.21. Increased cancer risk from this SNP variant has
been linked to several types of cancer, including prostate, ovarian
and inflammatory breast cancer (13,14).
Mutations in 8q24 have also been frequently detected in biliary
cancers and are included in the pancreaticobiliary probe set for
early cancer detection (15). A
meta-analysis of the ability of CCAT2 to predict metastasis and
poor prognosis confirmed that high CCAT2 expression was associated
with advanced tumor stage and may predict poor survival in cancers
and therefore may serve as a novel prognostic marker and
therapeutic target (16). However,
to the best of our knowledge, the functional role of CCAT2 in IHCC
has not been investigated.
The present study aimed to investigate the
expression levels of lncRNA CCAT2 in IHCC cells and clinical
specimens. The statistical association between CCAT2 levels and
IHCC clinicopathological characteristics was calculated to analyze
the clinical significance of CCAT2 in IHCC. The contribution of
CCAT2 in IHCC prognostic prediction was investigated and the
proliferative and invasive ability of IHCC cell with either CCAT2
knockdown or upregulated expression was evaluated by in
vitro assay.
Materials and methods
Patients and tissue samples
IHCC tissues and paired adjacent non-tumorous
tissues (n=106) were collected from the patients with IHCC who
underwent curative surgical resection between January 2008 and
October 2015 at The Fourth Hospital of Hebei Medical University
(Shijiazhuang, China). Tumor and adjacent non-tumorous tissues were
isolated. Non-tumorous tissues were obtained from ≥1 cm away from
the tumor border and were confirmed to contain no existing tumor
cells. Following resection, the collected paired tissue samples
were frozen with liquid nitrogen and stored at −80°C until further
use. No patients received anticancer therapies prior to surgery.
Patients diagnosed with more than two malignances were excluded
from the study. Written informed consent was obtained from each
patient. This study was approved by The Ethics Committee of The
Fourth Hospital of Hebei Medical University (Shijiazhuang,
China).
Cell culture and transfection
The IHCC cell lines HUH28, HuCCT1, RBE and HCCC9810
and the immortalized human cholangiocyte-derived cell line H69 were
all purchased from The Institute of Biochemistry and Cell Biology
of the Chinese Academy of Sciences (Shanghai, China). All cell
lines were cultured in RPMI-1640 medium (HuCCT1, HCCC9810 and RBE)
or Dulbecco's modified Eagle's medium (DMEM) (HUH28), supplemented
with 10% fetal bovine serum (all from Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) and 1% penicillin/streptomycin in an atmosphere
of 5% CO2 at 37°C.
lncRNA CCAT2 overexpression vector (pcDNA3.1-CCAT2),
control vector (pcDNA3.1-vector) and non-targeting small
interfering (si)RNA negative control (siNC) were all purchased from
Genewiz, Inc. (South Plainfield, NJ, USA). CCAT2 siRNA (iCCAT2.1
and siCCAT2.2) were designed using the Custom RNAi Design Tool on
the Integrated DNA Technologies (IDT) website (http://sg.idtdna.com/site) and synthesized by Genewiz,
Inc. Transfection was performed with Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA),
according to the manufacturer's instructions. Subsequent
experiments were performed 24 h after transfection.
Reverse transcription
quantitative-polymerase chain reaction (RT-qPCR)
TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) was used to extract total RNA from clinical
specimens or cultured cells, according to the manufacturer's
instructions. The concentration and purity of the RNA were
determined with a NanoDrop 1000 spectrophotometer (Thermo Fisher
Scientific, Inc.). cDNA was reverse transcribed using 2 µg
extracted RNA with the PrimeScript One-Step RT-PCR kit (Takara
Biotechnology Co., Ltd., Dalian, China). qPCR was performed using a
SYBR PrimeScript RT-PCR kit (Takara Biotechnology Co., Ltd.) with
the Mx3005P qPCR System (Agilent Technologies, Inc., Santa Clara,
CA, USA), according to the manufacturer's instructions. The PCR
cycling conditions were as follows: initial denaturation at 95°C
for 5 min, 40 cycles of denaturation at 95°C for 30 sec, annealing
at 50°C for 30 sec and extension at 72°C for 30 sec. The expression
of lncRNA CCAT2 from tissue samples or cultured cells was
normalized to GAPDH and compared using the 2−ΔΔCq method
(17). The primer sequences were:
CCAT2 forward, 5′-CCCTGGTCAAATTGCTTAACCT-3′ and reverse,
5′-TTATTCGTCCCTTTTTATGGAT-3′; GADPH forward,
5′-ACCCACTCCTCCACCTTTGAC-3′ and reverse,
5′-TGTTGCTGTAGCCAAATTCGTT-3′.
MTT assay
Cells (2,000/well) were seeded into 96-well plates
for incubation at 37°C, and MTT (10 µl, 10 mg/ml) was added to the
medium at the indicated time (0, 24, 48, 72 and 96 h following the
initial incubation). The plates were incubated for another 4 h at
37°C, and dimethylsulfoxide (100 µl) was added to dissolve the
purple formazan crystals. The absorbance was subsequently measured
by a microplate spectrophotometer (Dynex Technologies, Inc.,
Sullyfield, VA, USA) at a wavelength of 490 nm. Each assay was
conducted in triplicate and repeated at least three times.
Colony formation assay
Cells (1,500 cells/dish) were seeded in 6 cm dishes
and incubated in an atmosphere of 5% CO2 at 37°C for 7
days until the colonies were large enough to be clearly discerned.
Colonies were defined as groups of >50 cells and were counted by
light microscopy (×10). Each assay was conducted in triplicate and
repeated at least three times.
Transwell and Matrigel assay
Cell migratory ability was evaluated using a
Transwell system (pore size, 8.0 µm; Sigma-Aldrich; Merck KGaA)
Cells (3×104) were suspended in serum-free medium
(RPMI-1640 medium for HuCCT1, HCCC9810 and RBE; DMEM medium for
HUH28) and plated in the upper chamber of the Transwell system, and
the lower chamber was filled with medium containing 10% FBS.
Following incubation for 24 h at 37°C, the cells in the upper
chamber were removed and the cells in the lower chamber were fixed
by pure cold methanol (−4°C) for 30 min and subjected to Giemsa
staining (1:10 dilution) for another 30 min at room temperature.
Cells were subsequently counted with a light microscope (×10). The
Matrigel assay procedure to assess cell invasive ability was
identical to the Transwell assay, except the Transwell membrane was
precoated with Matrigel (BD Biosciences, Franklin Lakes, NJ, USA).
All experiments were performed in triplicate.
Statistical analysis
Statistical analysis was performed using the SPSS
17.0 software package (SPSS, Inc., Chicago, IL, USA). All data is
expressed as the mean ± standard deviation. For statistical
analysis, categorical variables were compared using the
χ2 test and the Student's t-test was performed to
compare the differences between continuous data. One-way analysis
of variance and Bonferroni correction test were used to compare
multiple groups. The overall survival (OS) and progression free
survival (PFS) rate were analyzed with the Kaplan-Meier method and
differences between groups were assessed with the log-rank test.
The significance of the survival data was evaluated using
univariate and multivariate Cox regression methods. The
receiver-operating characteristic (ROC) curve was adopted to define
the cut-off score for discriminating the specimens with high or low
CCAT2 expression. The point on the curve with the shortest distance
to the coordinate (0, 1) was selected as the threshold value to
classify cases as high expression or low expression. P<0.05 was
considered to indicate a statistically significant difference.
Results
lncRNA CCAT2 is overexpressed in
IHCC
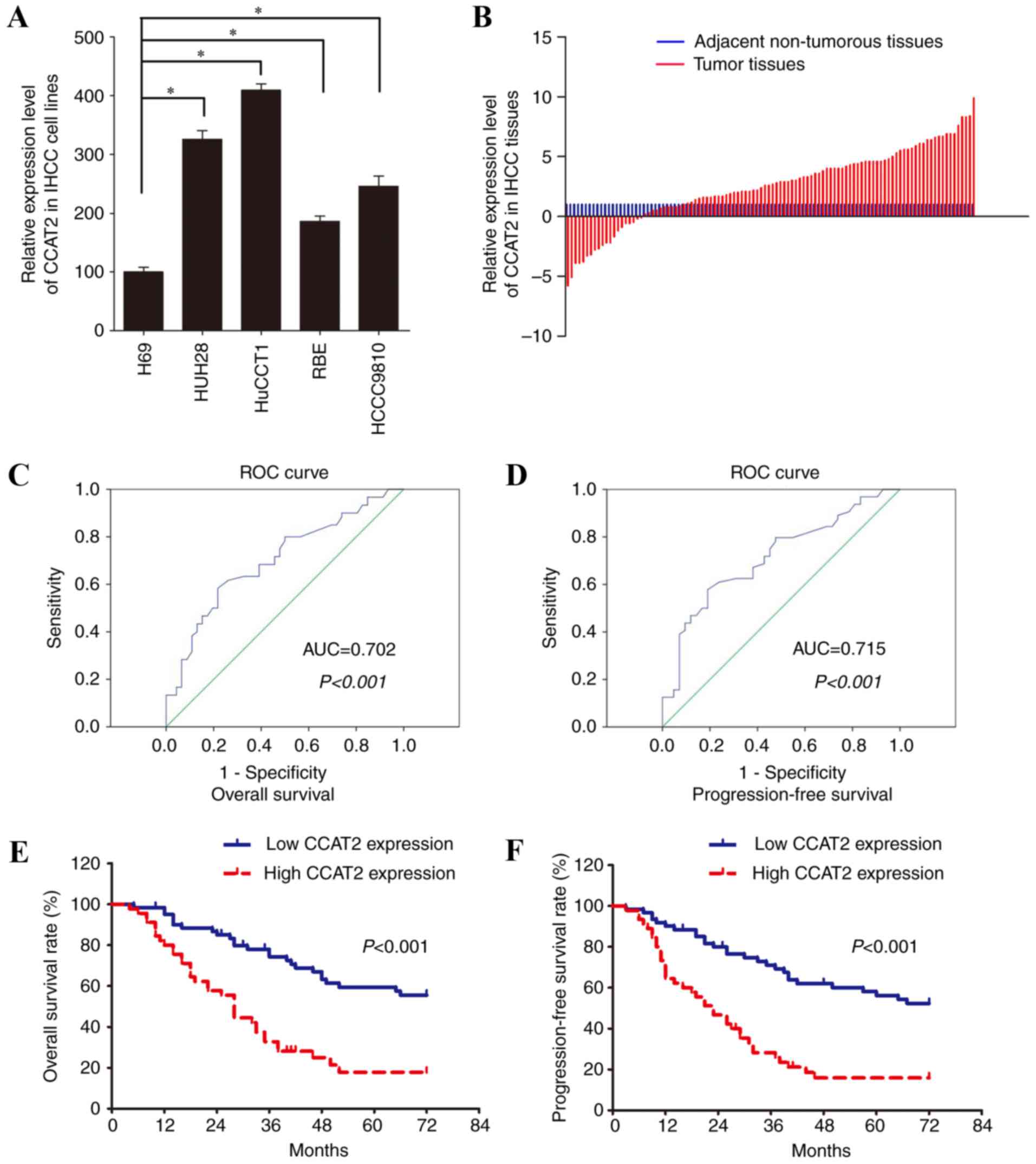
The analysis of lncRNA CCAT2 expression in IHCC
cells lines by RT-qPCR assay revealed that CCAT2 expression was
upregulated in IHCC cell lines compared with normal biliary
epithelial cells (Fig. 1A). CCAT2
expression was higher in 70.8% (75/106) of IHCC samples compared
with the paired adjacent non-tumorous tissues (Fig. 1B). Further statistical analysis
confirmed that IHCC tissues had significantly higher relative CCAT2
expression compared with paired adjacent non-tumorous tissues (1
vs. 2.52±3.17; P<0.001; Fig.
1B), which indicated that lncRNA CCAT2 may be oncogenic in
IHCC.
Expression level of lncRNA CCAT2 is
correlated with IHCC clinical progression
To better define the value of CCAT2 expression level
in predicting the prognosis of patients with IHCC, the ROC curve
was adopted to identify the optimal cut-off score for high/low
CCAT2 expression (Fig. 1C and D).
The area under the curves were 0.702 and 0.715 for OS and PFS,
respectively, which suggested that CCAT2 may be a useful biomarker
for the prognostic prediction of IHCC (P<0.001). The cutoff
score was 4.4 for both OS and PFS.
To investigate the clinical significance of CCAT2 in
IHCC, the association between CCAT2 expression and
clinicopathological characteristics was statistically analyzed.
High CCAT2 expression was revealed to be associated with
microvascular invasion (P<0.001), differentiation grade
(P=0.040), and tumor (T; P=0.012), lymph node (N; P<0.001),
metastasis (M; P=0.045) and combined TNM (P<0.001) stages of
IHCC (Table I). However, other
clinicopathological parameters including age, sex, hepatitis B
viral protein, hepatitis B surface antigen, hepatitis C virus,
cancer antigen 19-9, tumor number, cirrhosis and encapsulation were
not indicated to be statistically significant (P>0.05; Table I). These results suggested that
CCAT2 overexpression may be associated with the development of
IHCC.
 | Table I.Correlation between long non-coding
RNA CCAT2 expression and the clinicopathological characteristics of
intrahepatic cholangiocarcinoma. |
Table I.
Correlation between long non-coding
RNA CCAT2 expression and the clinicopathological characteristics of
intrahepatic cholangiocarcinoma.
|
|
| CCAT2 |
|
|---|
|
|
|
|
|
|---|
| Parameter | No. of patients | High/Low | P-value |
|---|
| Age (years) |
|
|
0.204 |
|
<60 | 50 | 18/32 |
|
| ≥60 | 56 | 27/29 |
|
| Sex |
|
|
0.620 |
| Male | 78 | 32/46 |
|
|
Female | 28 | 13/15 |
|
| HBeAg |
|
|
0.855 |
|
Negative | 72 | 31/41 |
|
|
Positive | 34 | 14/20 |
|
| HBsAg |
|
|
0.919 |
|
Negative | 43 | 18/25 |
|
|
Positive | 63 | 27/36 |
|
| HCV |
|
|
0.290 |
|
Negative | 87 | 39/48 |
|
|
Positive | 19 | 6/13 |
|
| CA19-9 (U/ml) |
|
|
0.327 |
|
<37 | 46 | 22/24 |
|
| ≥37 | 60 | 23/37 |
|
| No. of tumors |
|
|
0.851 |
|
Single | 88 | 37/51 |
|
|
Multiple | 18 | 08/10 |
|
| Microvascular
invasion |
|
| <0.001 |
| No | 60 | 16/44 |
|
| Yes | 46 | 29/17 |
|
| Cirrhosis |
|
|
0.179 |
| No | 35 | 11/24 |
|
|
Yes | 71 | 32/39 |
|
| Encapsulation |
|
|
0.821 |
| No | 67 | 29/38 |
|
|
Complete | 39 | 16/23 |
|
| Differentiation
grade |
|
|
0.040 |
| Low +
intermediate | 57 | 19/38 |
|
|
High | 49 | 26/23 |
|
| Tumor size
(cm) |
|
|
0.326 |
|
<5 | 53 | 20/33 |
|
| ≥5 | 53 | 25/28 |
|
| T stage |
|
|
0.012 |
|
T1+T2 | 77 | 27/50 |
|
|
T3+T4 | 29 | 18/11 |
|
| N stage |
|
| <0.001 |
| N0 | 69 | 19/50 |
|
| N1 | 37 | 26/11 |
|
| M stage |
|
|
0.045 |
| M0 | 99 | 39/60 |
|
| M1 | 7 | 6/1 |
|
| TNM stage |
|
| <0.001 |
| I +
II | 55 | 13/42 |
|
| III +
IV | 51 | 32/19 |
|
CCAT2 is a prognostic marker and an
independent risk factor for IHCC prognosis
The significance of CCAT2 in the prediction of
prognosis was evaluated by Kaplan-Meier analysis. Patients with
high CCAT2 expression were demonstrated to have a lower OS rate
(P<0.001) and a shorter PFS period (P<0.001; Fig. 1E and F)
Furthermore, univariate analysis demonstrated that
high CCAT2 expression was a risk factor for OS [hazard ratio (HR) =
3.184; 95% confidence interval (CI) = 1.882–5.385; P<0.001] and
for PFS (HR = 2.926; 95% CI = 1.771–4.834; P<0.001) of patients
with IHCC (Table II).
Multivariate analysis also identified high CCAT2 expression as an
independent risk factor of OS (HR=1.894; 95% CI=1.060–3.384;
P=0.031) and PFS (HR=2.134; 95% CI=1.232–3.699; P=0.007) of
patients with IHCC (Table III).
Thus, lncRNA CCAT2 may be useful as a prognostic marker for OS and
PFS of patients with IHCC.
 | Table II.Univariate analysis of
clinicopathological features for overall survival and
progression-free survival of patients with intrahepatic
cholangiocarcinoma. |
Table II.
Univariate analysis of
clinicopathological features for overall survival and
progression-free survival of patients with intrahepatic
cholangiocarcinoma.
|
| Overall
survival | Progression-free
survival |
|---|
|
|
|
|
|---|
| Parameter | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age (years) |
|
|
|
|
|
|
| <60
vs. ≥60 | 1.255 | 0.750–2.100 | 0.387 | 1.261 | 0.766–2074 | 0.362 |
| Sex |
|
|
|
|
|
|
| Female
vs. male | 0.596 | 0.322–1.104 | 0.100 | 0.609 | 0.336–1.103 | 0.102 |
| HBeAg |
|
|
|
|
|
|
|
Positive vs. negative | 0.824 | 0.470–1.446 | 0.500 | 0.873 | 0.510–1.493 | 0.619 |
| HBsAg |
|
|
|
|
|
|
|
Positive vs. negative | 1.252 | 0.746–2.101 | 0.395 | 1.218 | 0.739–2.008 | 0.439 |
| HCV |
|
|
|
|
|
|
|
Positive vs. negative | 1.707 | 0.938–3.108 | 0.080 | 1.788 | 1.002–3.190 | 0.049 |
| CA19-9 (U/ml) |
|
|
|
|
|
|
| <37
vs. ≥37 | 0.919 | 0.551–1.533 | 0.747 | 0.857 | 0.523–1.402 | 0.538 |
| Tumor number |
|
|
|
|
|
|
| Single
vs. multiple | 1.535 | 0.830–2.839 | 0.172 | 1.495 | 0.813–2.751 | 0.196 |
| Microvascular
invasion |
|
|
|
|
|
|
| Yes vs.
no | 0.485 | 0.288–0.816 | 0.006 | 0.512 | 0.307–0.853 | 0.010 |
| Cirrhosis |
|
|
|
|
|
|
| Yes vs.
no | 1.428 | 0.848–2.405 | 0.181 | 1.360 | 0.823–2.249 | 0.231 |
| Encapsulation |
|
|
|
|
|
|
|
Complete vs. none | 1.023 | 0.605–1.730 | 0.931 | 1.042 | 0.628–1.730 | 0.872 |
|
Differentiation |
|
|
|
|
|
|
| Low +
intermediate vs. high | 1.945 | 1.155–3.276 | 0.012 | 2.133 | 1.283–3.547 | 0.003 |
| Tumor size
(cm) |
|
|
|
|
|
|
| <5
vs. ≥5 | 1.351 | 0.812–2.250 | 0.247 | 1.195 | 0.731–1.953 | 0.478 |
| T stage |
|
|
|
|
|
|
| T1+T2
vs. T3+T4 | 2.337 | 1.378–3.964 | 0.002 | 2.353 | 1.406–3.936 | 0.001 |
| N stage |
|
|
|
|
|
|
| N0 vs.
N1 | 1.526 | 1.190–1.957 | 0.001 | 1.533 | 1.201–1.956 | 0.001 |
| M stage |
|
|
|
|
|
|
| M0 vs.
M1 | 5.364 | 2.250–12.788 | <0.001 | 4.643 | 1.958–11.011 | <0.001 |
| TNM stage |
|
|
|
|
|
|
| I + II
vs. III + IV | 4.647 | 2.613–8.262 | <0.001 | 4.617 | 2.656–8.025 | <0.001 |
| CCAT2 |
|
|
|
|
|
|
| Low vs.
high | 3.184 | 1.882–5.385 | <0.001 | 2.926 | 1.771–4.834 | <0.001 |
 | Table III.Multivariate analysis of
clinicopathological features for overall survival and
progression-free survival of patients with intrahepatic
cholangiocarcinoma. |
Table III.
Multivariate analysis of
clinicopathological features for overall survival and
progression-free survival of patients with intrahepatic
cholangiocarcinoma.
|
| Overall
survival | Progression-free
survival |
|---|
|
|
|
|
|---|
| Parameter | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| HCV | – | – | – | 2.091 | 1.154–3.788 |
0.015 |
|
Positive vs. negative |
|
|
|
|
|
|
| Microvascular
invasion | 0.485 | 0.279–0.843 |
0.010 | 0.502 | 0.292–0.863 |
0.013 |
| Yes vs.
no |
|
|
|
|
|
|
|
Differentiation | 0.938 | 0.503–1.751 |
0.842 | 1.107 | 0.606–2.021 |
0.742 |
| Low +
intermediate vs. high |
|
|
|
|
|
|
| T stage | 0.607 | 0.299–1.231 |
0.166 | 0.527 | 0.261–1.063 |
0.074 |
| T1+T2
vs. T3+T4 |
|
|
|
|
|
|
| N stage | 0.890 | 0.517–1.529 |
0.672 | 0.767 | 0.465–1.264 |
0.298 |
| N0 vs.
N1 |
|
|
|
|
|
|
| M stage | 3.078 | 1.193–7.943 |
0.020 | 2.642 | 1.035–6.746 |
0.042 |
| M0 vs.
M1 |
|
|
|
|
|
|
| TNM stage | 4.972 |
2.108–12.250 | <0.001 | 5.797 |
2.431–13.824 | <0.001 |
| I + II
vs. III + IV |
|
|
|
|
|
|
| CCAT2 | 1.894 | 1.060–3.384 |
0.031 | 2.134 | 1.232–3.699 |
0.007 |
| Low vs.
high |
|
|
|
|
|
|
CCAT2 promotes the proliferation and
metastasis of IHCC cells
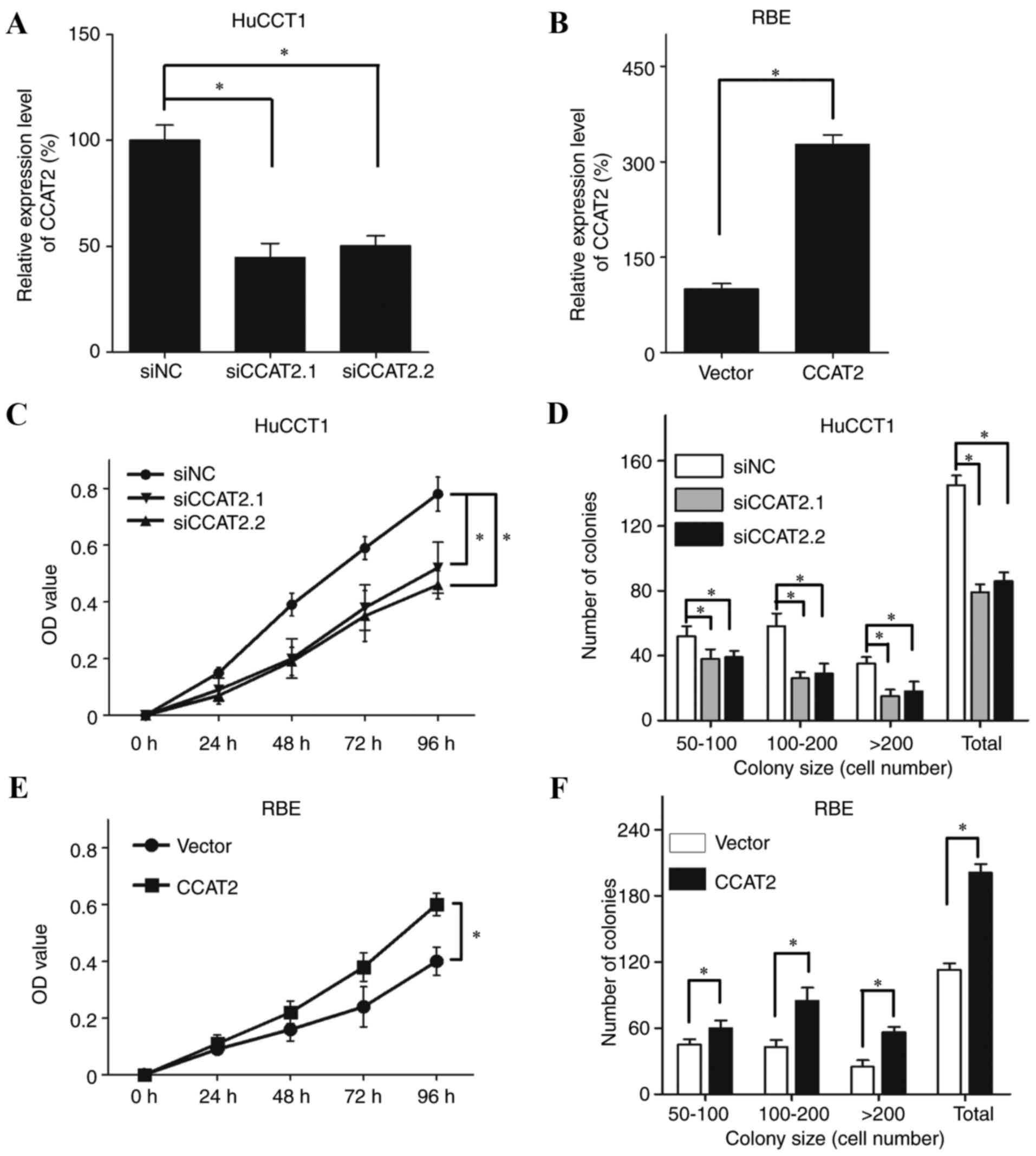
To extend our understanding on the role of CCAT2 in
IHCC, in vitro assays were performed in IHCC cells with
either CCAT2 silencing and overexpression (Fig. 2A and B). As CCAT2 expression was
relatively high in HuCCT1 cells and low in RBE cells (Fig. 1A) these cell lines were used to
confirm the effects of siRNA-induced CCAT2 silencing and CCAT2
overexpression, respectively (Fig. 2A
and B, respectively). The MTT assay revealed that the
proliferative ability of HuCCT1 cells was significantly decreased
following transfection with siCCAT2.1 and siCCAT2.2 compared with
cells transfected with the siNC (Fig.
2C; P<0.05). The colony formation assay also revealed that
CCAT2 silencing in HuCCT1 cells significantly decreased colony
numbers compared with the siNC-transfected cells (Fig. 2D; P<0.05). In addition, CCAT2
overexpression significantly increased the proliferative ability
(Fig. 2E; P<0.05) and the
number of colonies (Fig. 2F;
P<0.05) of RBE cells compared with cells transfected with the
empty vector.
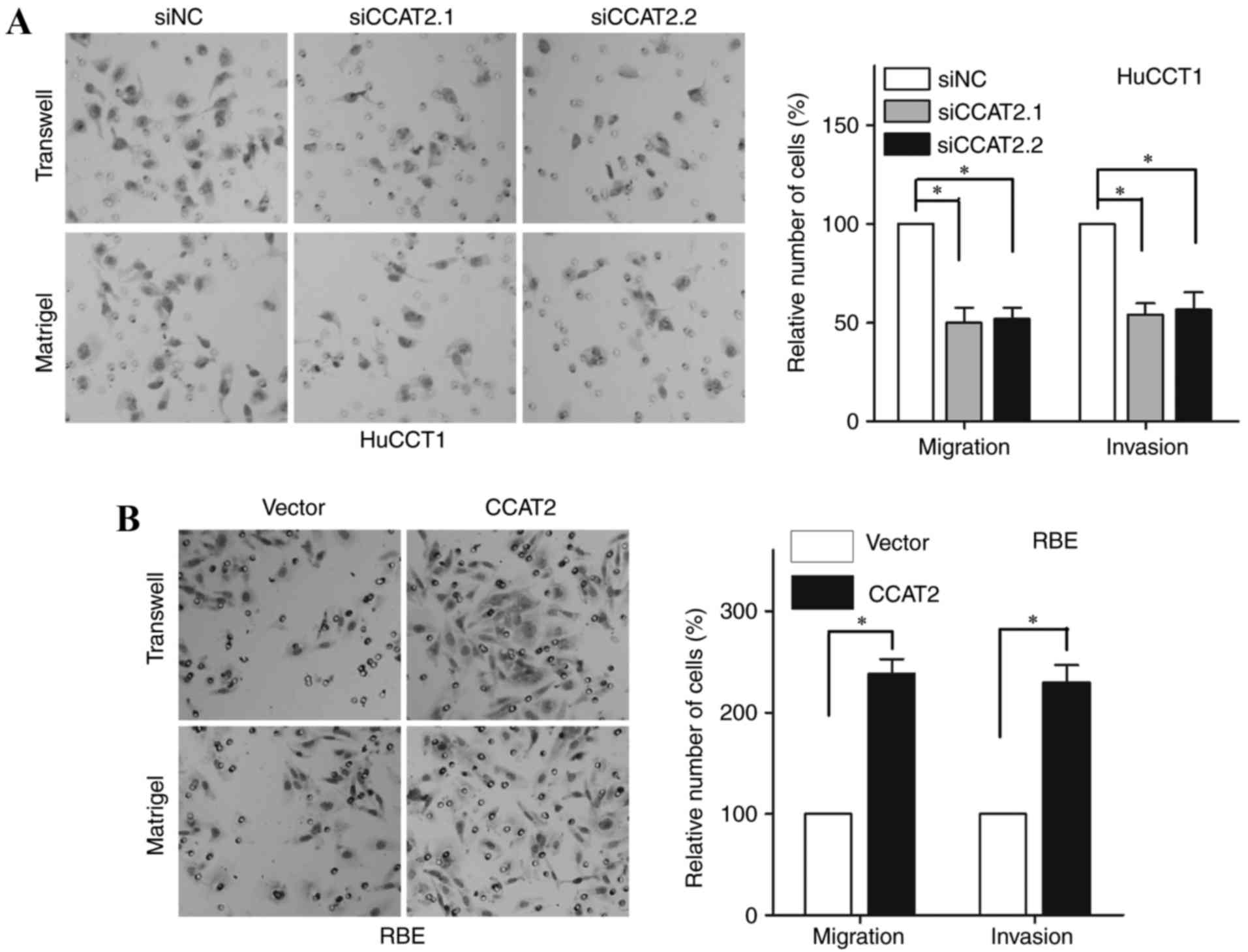
The migratory and invasive abilities of HuCCT1 cells
were significantly inhibited in cells transfected with either CCAT2
siRNA compared with siNC-transfected cells (Fig. 3A; P<0.05) By contrast, the
migratory and invasive abilities were significantly increased in
RBE cells overexpressing CCAT2 compared with vector control cells
(Fig. 3B; P<0.05). These data
suggested that CCAT2 may promote proliferation and metastasis of
IHCC cells.
Discussion
Previous studies have reported the involvement of
lncRNA CCAT2 in the development and metastasis of a number of
cancers. For example, Ling et al (12) reported that CCAT2 expression was
markedly increased in microsatellite-stable colorectal cancer and
that CCAT2 promoted tumor growth, metastasis and chromosomal
instability through the regulation of MYC and WNT expression. Qiu
et al (18) demonstrated
that CCAT2 was upregulated in non-small cell lung cancer and
increased the proliferation and metastasis of tumor cells. The
functional role of CCAT2 in promoting tumor growth and metastasis
has also been demonstrated in esophageal squamous cell carcinoma,
glioma, breast cancer, hepatocellular carcinoma and ovarian cancer
(19–22). Furthermore, a previous
meta-analysis revealed that high CCAT2 expression was significantly
correlated with OS (HR=2.30; 95% CI=1.62–3.25; P<0.001) and PFS
(HR=2.76; 95% CI=1.74–4.37; P<0.001) in various cancers
(16).
The present study confirmed that CCAT2 was
overexpressed in IHCC cell lines and tissues. Statistical analysis
of CCAT2 expression and IHCC clinicopathological features revealed
that high CCAT2 expression was associated with microvascular
invasion, differentiation grades, T, N, M and TNM stages of IHCC
(P<0.05). Based on these data and results from in vitro
assays, it was hypothesized that CCAT2 may promote IHCC progression
and metastasis. Survival analysis demonstrated that patients with
high CCAT2 expression had a lower OS rate and shorter PFS period
(P<0.05). Furthermore, high CCAT2 expression was identified as
an independent risk factor of poor OS (HR=3.184; 95%
CI=1.882–5.385; P<0.001) and PFS (HR=2.926; 95% CI=1.771–4.834;
P<0.001) of patients with IHCC in the multivariate Cox
regression analysis.
Mechanistically, several studies have concentrated
on identifying the downstream mechanisms of CCAT2 function.
Previous studies reported that the mechanism of CCAT2 in promoting
cancer development is mainly due to the activation of WNT signaling
by enhancing the transcriptional activity of transcription factor
7-like 2 and the subsequent increase in MYC expression (22–24).
Another study reported that CCAT2 regulates epithelial ovarian
cancer progression by functioning as a competing endogenous RNA, or
sponge, and negatively targeting miR-424 (25). Further investigations are required
to fully understand the mechanisms of CCAT2 function and its
contribution to IHCC progression and metastasis.
In conclusion, results from the present study
demonstrated that lncRNA CCAT2 expression was upregulated in IHCC
and this high expression level may be correlated with IHCC
progression and metastasis. High CCAT2 expression predicted a poor
OS rate and shorter PFS period. CCAT2 overexpression was also an
independent risk factor for both OS and PFS. The functional role of
CCAT2 in facilitating proliferation and metastasis of IHCC cells
was further confirmed in in vitro assays and CCAT2 may be a
promising prognostic biomarker and therapeutic target in IHCC.
References
|
1
|
Razumilava N and Gores GJ:
Cholangiocarcinoma. Lancet. 383:2168–2179. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Aljiffry M, Abdulelah A, Walsh M,
Peltekian K, Alwayn I and Molinari M: Evidence-based approach to
cholangiocarcinoma: A systematic review of the current literature.
J Am Coll Surg. 208:134–147. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sia D, Tovar V, Moeini A and Llovet JM:
Intrahepatic cholangiocarcinoma: Pathogenesis and rationale for
molecular therapies. Oncogene. 32:4861–4870. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Endo I, Gonen M, Yopp AC, Dalal KM, Zhou
Q, Klimstra D, D'Angelica M, DeMatteo RP, Fong Y, Schwartz L, et
al: Intrahepatic cholangiocarcinoma: Rising frequency, improved
survival, and determinants of outcome after resection. Ann Surg.
248:84–96. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dodson RM, Weiss MJ, Cosgrove D, Herman
JM, Kamel I, Anders R, Geschwind JF and Pawlik TM: Intrahepatic
cholangiocarcinoma: Management options and emerging therapies. J Am
Coll Surg. 217:736–750.e4. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sirica AE, Dumur CI, Campbell DJ, Almenara
JA, Ogunwobi OO and Dewitt JL: Intrahepatic cholangiocarcinoma
progression: Prognostic factors and basic mechanisms. Clin
Gastroenterol Hepatol. 7 11 Suppl:S68–S78. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Isin M and Dalay N: lncRNAs and neoplasia.
Clin Chim Acta. 444:280–288. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sahu A, Singhal U and Chinnaiyan AM: Long
noncoding RNAs in cancer: From function to translation. Trends
Cancer. 1:93–109. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bartonicek N, Maag JL and Dinger ME: Long
noncoding RNAs in cancer: Mechanisms of action and technological
advancements. Mol Cancer. 15:432016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fatima R, Akhade VS, Pal D and Rao SM:
Long noncoding RNAs in development and cancer: Potential biomarkers
and therapeutic targets. Mol Cell Ther. 3:52015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Schmitt AM and Chang HY: Long noncoding
RNAs in cancer pathways. Cancer Cell. 29:452–463. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ling H, Spizzo R, Atlasi Y, Nicoloso M,
Shimizu M, Redis RS, Nishida N, Gafà R, Song J, Guo Z, et al:
CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic
progression and chromosomal instability in colon cancer. Genome
Res. 23:1446–1461. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ghoussaini M, Song H, Koessler T, Al Olama
AA, Kote-Jarai Z, Driver KE, Pooley KA, Ramus SJ, Kjaer SK, Hogdall
E, et al: Multiple loci with different cancer specificities within
the 8q24 gene desert. J Natl Cancer Inst. 100:962–966. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bertucci F, Lagarde A, Ferrari A, Finetti
P, Charafe-Jauffret E, Van Laere S, Adelaide J, Viens P, Thomas G,
Birnbaum D and Olschwang S: 8q24 cancer risk allele associated with
major metastatic risk in inflammatory breast cancer. PLoS One.
7:e379432012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Barr Fritcher EG, Voss JS, Brankley SM,
Campion MB, Jenkins SM, Keeney ME, Henry MR, Kerr SM, Chaiteerakij
R, Pestova EV, et al: An optimized set of fluorescence in situ
hybridization probes for detection of pancreatobiliary tract cancer
in cytology brush samples. Gastroenterology. 149:1813–1824.e1.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fan YH, Fang H, Ji CX, Xie H, Xiao B and
Zhu XG: Long noncoding RNA CCAT2 can predict metastasis and poor
prognosis: A meta-analysis. Clin Chim Acta. 466:120–126. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Qiu M, Xu Y, Yang X, Wang J, Hu J, Xu L
and Yin R: CCAT2 is a lung adenocarcinoma-specific long non-coding
RNA and promotes invasion of non-small cell lung cancer. Tumour
Biol. 35:5375–5380. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang S, Qing C, Huang Z and Zhu Y: The
long non-coding RNA CCAT2 is up-regulated in ovarian cancer and
associated with poor prognosis. Diagn Pathol. 11:492016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou N, Si Z, Li T, Chen G, Zhang Z and Qi
H: Long non-coding RNA CCAT2 functions as an oncogene in
hepatocellular carcinoma, regulating cellular proliferation,
migration and apoptosis. Oncol Lett. 12:132–138. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang J, Qiu M, Xu Y, Li M, Dong G, Mao Q,
Yin R and Xu L: Long noncoding RNA CCAT2 correlates with smoking in
esophageal squamous cell carcinoma. Tumour Biol. 36:5523–5528.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zeng J, Du T, Song Y, Gao Y, Li F, Wu R,
Chen Y, Li W, Zhou H, Yang Y and Pei Z: Knockdown of long noncoding
RNA CCAT2 inhibits cellular proliferation, invasion, and
epithelial-mesenchymal transition in glioma cells. Oncol Res.
25:913–921. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kasagi Y, Oki E, Ando K, Ito S, Iguchi T,
Sugiyama M, Nakashima Y, Ohgaki K, Saeki H, Mimori K and Maehara Y:
The expression of CCAT2, a novel long noncoding RNA transcript, and
rs6983267 single-nucleotide polymorphism genotypes in colorectal
cancers. Oncology. 92:48–54. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cai Y, He J and Zhang D: Long noncoding
RNA CCAT2 promotes breast tumor growth by regulating the Wnt
signaling pathway. Onco Targets Ther. 8:2657–2664. 2015.PubMed/NCBI
|
|
25
|
Hua F, Li CH, Chen XG and Liu XP: Long
noncoding RNA CCAT2 knockdown suppresses tumorous progression by
sponging miR-424 in epithelial ovarian vancer. Oncol Res. May
21–2017.(Epub ahead of print).
|