Introduction
The ETS variant 2 (ETV2) protein, also designated as
ETS-related 71 (ER71), is a DNA-binding transcription factor
(1,2); ETV2 belongs to the E26
transformation-specific (ETS) family of proteins, which are
characterized by a ~85 amino acid-long ETS DNA-binding domain and
comprises 28 members in humans (3). The knockout of ETV2 in mice has been
reported to cause embryonal lethality around day E9.5, indicating
that ETV2 is essential during early developmental stages; in
particular, ETV2 is required for the development of blood and
vascular structures (4,5). Mechanistically, ETV2 facilitates the
upregulation of fetal liver kinase (FLK1), a trans-membrane
tyrosine kinase and receptor for vascular endothelial growth factor
that is crucial for the aforementioned developmental processes.
Notably, a lack of FLK1 expression phenocopied ETV2 deficiency in
mice (6), emphasizing the
biological relevance of the ETV2-FLK1 axis in embryonal blood and
vessel formation.
In adult mice, ETV2 expression was reported to be
undetectable in endothelial cells and the conditional knockout of
ETV2 in the endothelial compartment did not affect adult
vasculature or viability. Upon injury, however, ETV2 expression was
rapidly upregulated in endothelial cells, and vascular regeneration
was dependent on ETV2 in several models, including the hindlimb
ischemic injury paradigm. In addition, the delivery of
lentivirus-expressing ETV2 into ischemic hindlimbs demonstrated
positive therapeutic effects by increasing capillary formation and
reducing tissue fibrosis (7).
Furthermore, the conditional deletion of ETV2 in adult
hematopoietic cells did not affect the viability of mice, but led
to a decrease in the number of hematopoietic stem cells. This may
not be relevant under normal conditions; however, the stem cell
repopulation potential appeared to be adversely affected following
damage (8). Thus ETV2 is
absolutely critical in hematopoietic and endothelial cells during
embryogenesis, and is not essential in these respective adult
cells, but may facilitate their response to injury.
Northern blot analyses have revealed elevated ETV2
expression levels in adult mouse testes, whereas other tissues did
not exhibit significant levels of ETV2 mRNA expression (1,9). In
addition, the ETV2 gene has been identified as a target of the
sex-determining region Y (SRY) protein that is encoded on the Y
chromosome. It has been reported that ETV2 expression becomes
upregulated within male mouse gonads following the transient
expression of SRY during embryogenesis (10), indicating that ETV2 may be involved
in the differentiation of the initially bipotential gonads into
testes. Furthermore, ETV2 can bind to and activate the promoter of
SOX9 (10,11), a transcription factor downstream of
SRY. SOX9 is important for male differentiation in the embryo, and
its expression persists in adult testes. Therefore, ETV2 may serve
a role in the development of male gonads and their function in
adults.
At present, very little is known about how ETV2 is
regulated at the molecular level. For instance, only a few
interaction partners of ETV2 have been identified. These include
ovo like zinc finger 2, a zinc-finger transcription factor that
cooperates with ETV2 in the regulation of the FLK1 gene promoter
(12), and Jumonji
domain-containing (JMJD)1A, a cofactor that may repress the
ETV2-mediated stimulation of matrix metalloproteinase (MMP) 1
transcription (13). Notably,
JMJD1A belongs to the JMJD protein family; most members of this
family have been demonstrated to be capable of demethylating
histone lysine residues (14,15).
The present study explored whether other members of the JMJD
protein family may modulate ETV2 function, and how these functions
may be associated with cancer.
Materials and methods
Bioinformatics
Data regarding the mRNA expression levels of ETV2 in
human tissues were obtained from the Genotype-Tissue Expression
project through the Human Protein Atlas (16), accessible from www.proteinatlas.org. ETV2 gene expression and
amplification in various cancers was identified with cbioportal
(www.cbioportal.org).
Coimmunoprecipitation experiments
293T cells (CRL-3216; American Type Culture
Collection, Manassas, VA, USA) were grown in poly-L-lysine-coated
6-cm plates to ~25% confluency in Dulbecco's modified Eagle's
medium (10–013-CV; Mediatech; Corning Inc, Corning, NY, USA)
supplemented with 10% fetal bovine serum (S11150; Atlanta
Biologicals, Flowery Branch, GA, USA), as previously described
(17). Subsequently, cells were
transfected by the calcium phosphate coprecipitation method
(18,19). The following amounts of DNA were
used for transfection: 1 µg 6Myc-ETV2 expression construct, as
previously described (9,13), or pCS3+-6Myc empty
vector; 2 µg Flag-tagged expression plasmids encoding various JMJD
proteins, or empty vector pEV3S; 6 µg pBluescript KS+.
At 10 h, the precipitate was removed by 2 washes in 2 ml PBS and
cells were incubated for 36 h in 4 ml growth medium (20). Cells were then lysed in 675 µl of
50 mM Tris-HCl (pH 7.4), 50 mM NaF, 150 mM NaCl, 0.5% Igepal
CA-630, 0.1 mM Na3VO4, 2 µg/ml aprotinin, 10
µg/ml leupeptin, 1 µg/ml pepstatin A, 1 mM phenylmethylsulfonyl
fluoride, 0.1 mM dithiothreitol and immunoprecipitations performed
with anti-Flag M2 (cat. no. F1804; Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) or anti-Myc 9E10 (cat. no. M4439;
Sigma-Aldrich; Merck KGaA) mouse monoclonal antibodies, as
previously described (21).
Immunoprecipitates were subjected to SDS-PAGE; proteins were
transferred onto a polyvinylidene fluoride membrane, as previously
described (22). Proteins on such
membranes were detected by incubation with the indicated primary
antibodies followed by incubation with goat anti-mouse (cat. no.
1706516; Bio-Rad Laboratories, Inc., Hercules, CA, USA) or goat
anti-rabbit (cat. no. 1706515; Bio-Rad Laboratories, Inc.)
polyclonal secondary antibodies coupled to horseradish-peroxidase
and visualized utilizing enhanced chemiluminescence, as previously
described (23).
Reporter gene experiments
Human LNCaP prostate cancer cells (CRL-1740;
American Type Culture Collection) were grown in
poly-L-lysine-coated 6-wells to ~30% confluency (24) and transfected with 1 µg pGL2-MMP1
(25) or pGL2-MMP7 (26) luciferase reporter plasmid, 1 µg
pBluescript KS+, pcDNA-ETV2 (9) or empty vector pcDNA3, and 100 ng
Flag-tagged JMJD2A or JMJD2D expression vector (27) or empty vector pEV3S, with 8 µg
polyethylenimine. At 8–10 h, cells were washed once with 2 ml PBS
and then incubated for another 40 h prior to lysis with 350 µl of
25 mM Tris, 2 mM EDTA (pH 7.8), 1% Triton X-100, 10% glycerol and 2
mM dithiothreitol, as previously described (28). Luciferase activities in 100 µl
lysate were determined using a luminometer ~25 min after lysis, as
previously described (29,30).
Statistical analysis
For luciferase reporter gene assays, the means of
independent replicates with standard deviations are presented.
Statistical significance was evaluated with a one-way analysis of
variance followed by Tukey's multiple comparison test, and
corresponding P-values were calculated with GraphPad Prism 6.0 h
software (GraphPad Software, Inc., La Jolla, CA, USA). For Pearson
correlations, statistical significance was estimated using a
two-tailed Student's t-test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Expression levels of ETV2 in normal
tissue and cancer
Previously, it was reported that ETV2 expression
levels in adult mouse tissues, apart from the testes, were
undetectable by northern blotting (1,9);
however, analysis was not previously conducted in human tissue. In
the present study, downloaded RNA sequencing datasets were employed
to analyze ETV2 expression via bioinformatics analyses, as RNA
sequencing is more sensitive than northern blotting. As expected,
it was revealed that the highest levels of human ETV2 mRNA
expression were observed in the testes; however, significant levels
of ETV2 mRNA were also detected in a variety of other human tissues
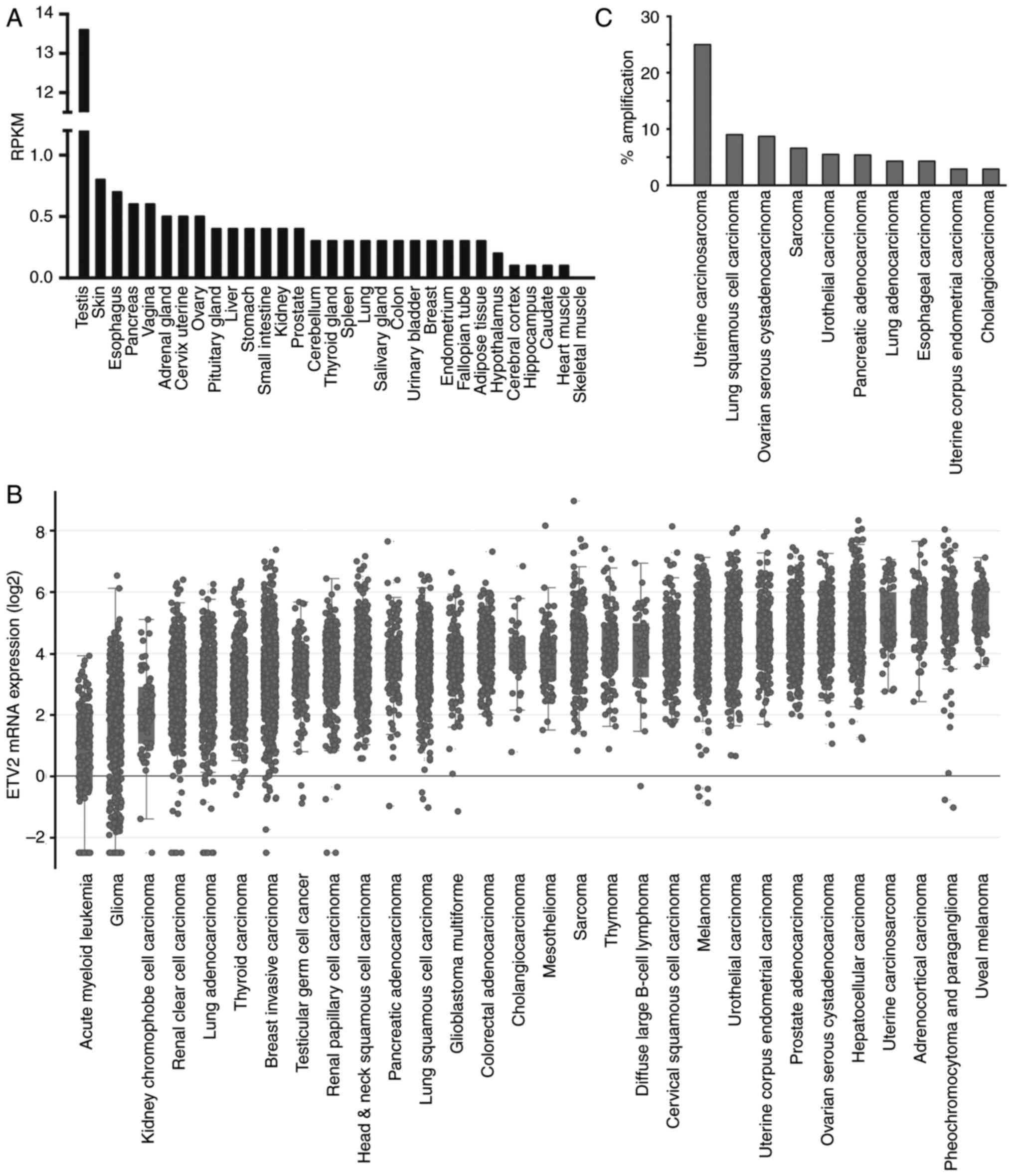
(Fig. 1A), suggesting that ETV2
may function in multiple organs in adult humans.
Similarly, the ETV2 expression levels in datasets
from numerous types of human cancer from the provisional Cancer
Genome Atlas (TCGA) were characterized. As presented in Fig. 1B, numerous cancer types, including
uveal melanoma, pheochromocytoma and paraganglioma, or
adrenocortical carcinoma displayed higher median ETV2 mRNA levels
compared with other types of cancer, including acute myeloid
leukemia and glioma. Notably, the high expression level of ETV2 in
many types of cancer was associated with its high expression in
normal tissue of that type, e.g., adrenocortical carcinoma with the
adrenal gland, or hepatocellular carcinoma with the liver (compare
Fig. 1B to A). In addition, the
analysis of the provisional TCGA revealed that the ETV2 gene is
amplified in various types of cancer, with the highest
amplification rate (25%) in uterine carcinosarcoma (Fig. 1C). Collectively, these data suggest
that ETV2 potentially contributes to tumorigenesis.
Our laboratory is focused on prostate cancer
research; it was identified that ETV2 was robustly expressed in
normal prostate tissue, as well as prostate adenocarcinomas
(Fig. 1A and B). In a study by
Beltran et al (31) where
datasets other than the provisional TCGA were used, neuroendocrine
prostate cancer exhibited an ETV2 gene amplification rate of 17.8%,
indicating that ETV2 may be involved in prostate cancer. Previous
studies have demonstrated that several JMJD2 histone demethylases,
also known as lysine demethylase 4 (KDM4) proteins, are
overexpressed in prostate cancer (32,33).
Furthermore, JMJD2A was reported to exert oncogenic functions in
the prostate (33–35). Therefore, the association between
ETV2, JMJD2A/KDM4A and the homolog JMJD2D/KDM4D was investigated in
the present study. JMJD2B/KDM4B and JMJD2C/KDM4C were excluded from
the analysis, as these proteins are highly homologous to
JMJD2A/KDM4A and the scope of the study was limited by resource
restraints.
Binding of ETV2 to JMJD2A and
JMJD2D
To assess whether ETV2 interacts with JMJD2
proteins, Myc-tagged ETV2 and Flag-tagged JMJD2A or JMJD2D were
coexpressed within 293T cells. Following immunoprecipitation with
anti-Flag antibodies, any coprecipitated ETV2 was detected by
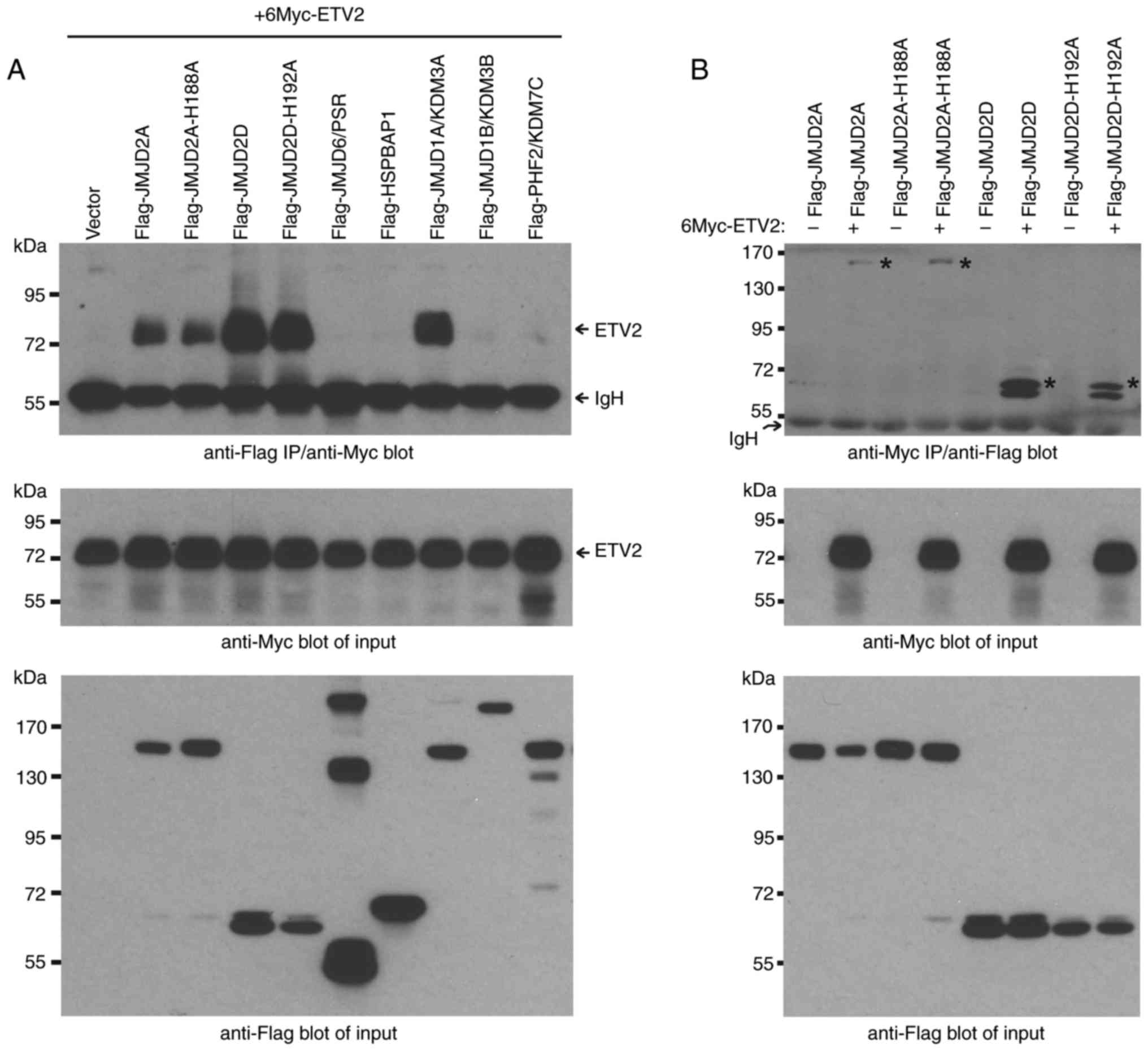
anti-Myc western blotting. It was revealed that ETV2
coimmunoprecipitated with JMJD2A and JMJD2D (Fig. 2A, top panel). Notably, an increased
amount of ETV2 was coimmunoprecipitated with JMJD2D compared with
JMJD2A, despite similar expression levels of the JMJD2 proteins and
indistinguishable ETV2 expression levels (Fig. 2A, bottom panels). This indicated
that ETV2 may interact more strongly with JMJD2D than JMJD2A.
As a positive control, JMJD1A (also known as KDM3A)
was employed; it has previously been demonstrated to interact with
ETV2 (13). This interaction was
confirmed in the present study (Fig.
2A). However, ETV2 did not coimmunoprecipitate with several
other JMJD proteins (including JMJD6/PSR, HSPBAP1, JMJD1B/KDM3B and
PHF2/KDM7C), which indicated that ETV2 does not universally
interact with JMJD proteins. Additionally, the catalytically
inactive mutants JMJD2A-H188A and JMJD2D-H192A, as previously
described (36,37), were employed. These inactive
mutants had an equivalent likelihood of coimmunoprecipitation as
their wild-type counterparts, indicating that the catalytic
activity of JMJD2A and JMJD2D may not be required for ETV2 complex
formation (Fig. 2A).
To corroborate these results, reverse order
coimmunoprecipitation experiments were conducted by the pull-down
of Myc-tagged ETV2 and subsequent probing for coprecipitated
Flag-tagged JMJD2 proteins (Fig.
2B). The interaction of ETV2 with JMJD2A and JMJD2D was
confirmed; once again, ETV2 complexes with JMJD2D were more readily
formed compared with JMJD2A, and the catalytic activity of the
JMJD2 proteins was dispensable for their interaction with ETV2.
JMJD2D presented as a doublet in the western blot images of
Fig. 2; the higher molecular
weight form appeared to be enriched in the ETV2 immunoprecipitates
(compare Fig. 2B, top to bottom
blot), suggesting that the higher molecular weight form of JMJD2D
bound more readily to ETV2 than the lower molecular weight form. It
has not been determined why a doublet was observed in the present
study, or why its degree of appearance was variable from experiment
to experiment. However, we hypothesize that this was due to the
posttranslational modification of JMJD2D, the degree of which could
be affected by the variable density of the 293T cells, their
passage number and the lot of serum that was utilized to cultivate
the cells. Altogether, the data presented in Fig. 2 indicate that ETV2 can form
complexes with JMJD2A and JMJD2D in vivo.
Identification of interaction
domains
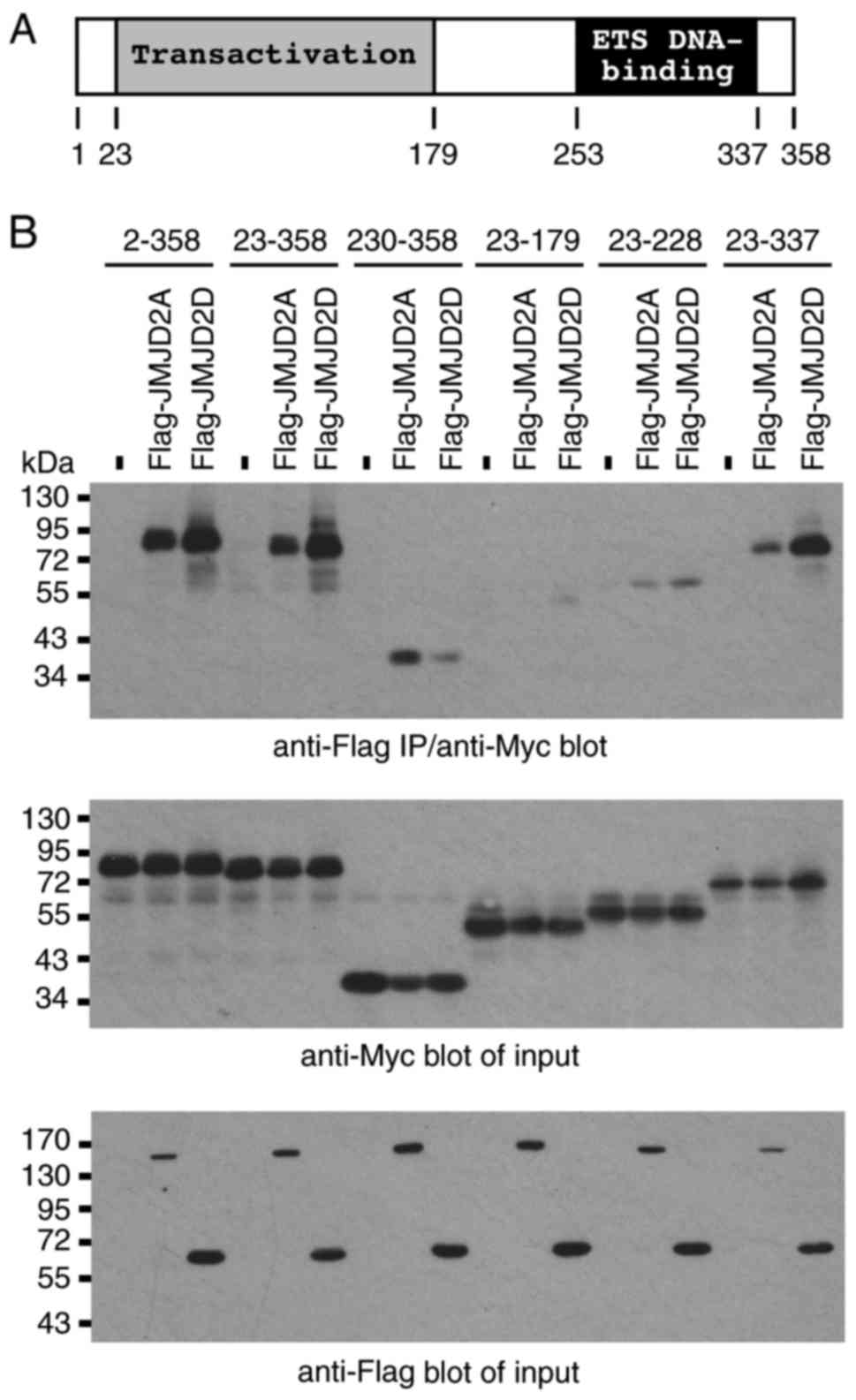
To identify which region(s) of ETV2 may be required
for the interaction with JMJD2 proteins, various ETV2 fragments
(Fig. 3A) were coexpressed with
JMJD2A or JMJD2D, and their interactions were assessed by
coimmunoprecipitation. Similar to full-length ETV2 (amino acids
2–358), amino acids 23–358 interacted with JMJD2A and JMJD2D
(Fig. 3B). However, further
truncating ETV2 from the N-terminus down to amino acids 230–358 led
to a marked reduction in the interaction with JMJD2D, whereas the
interaction with JMJD2A was only slightly affected. Deletion of the
21 C-terminal amino acids from EVT2 did not affect the interaction
with JMJD2A and JMJD2D (Fig. 3B;
residues 23–358 and 23–337). However, a deletion of further
C-terminal amino acids, including the ETS DNA-binding domain,
severely compromised the ability of ETV2 to interact with JMJD2A
and JMJD2D; only a low level of interaction was observable with the
ETV2 amino acids 23–228, whereas amino acids 23–179 were
essentially unable to form complexes with JMJD2A and JMJD2D
(Fig. 3B). Collectively, this data
indicated that the ETV2 amino acids 230–337 primarily mediated the
interaction with JMJD2A; however, further amino acids in the 23–229
region were required for a strong interaction, in particular with
JMJD2D.
Conversely, the regions within JMJD2 proteins that
mediate the interaction with ETV2 were investigated by sectioning
JMJD2A into three parts, and ETV2's interaction with these sections
was assessed. The N-terminal JMJD2A amino acids 2–350, which
encompass the conserved JmjN and JmjC domains (Fig. 4A), did not interact with ETV2
(Fig. 4B), whereas amino acids
301–703 and 704–1,064 did (Fig.
4B). The latter region encompasses two protein-protein
interaction motifs, the double PHD and Tudor domains (Fig. 4A). These data indicate that two
regions within JMJD2A contributed to its interaction with ETV2. The
input levels for JMJD2A amino acids 704–1,064 were the lowest, but
the amount of immunoprecipitated ETV2 was higher than with
full-length JMJD2A (amino acids 2–1,064; Fig. 4B); therefore, the amino acids at
the N-terminal from 704–1,064 may negatively affect the ability to
bind to ETV2. Similar to JMJD2A, the 2–354 N-terminal amino acids
of JMJD2D, which contain the catalytic domains (Fig. 4C), did not bind to ETV2, whereas
amino acids 241–523 did (Fig. 4D).
In addition, the interaction of amino acids 241–523 with ETV2
appeared to be stronger compared with full-length JMJD2D,
suggesting that the N-terminus of JMJD2D may exert a negative
regulatory effect on the binding to ETV2. Collectively, these data
reveal that the JmjN and JmjC domains of JMJD2A and JMJD2D may not
be required for complex formation with ETV2. This may explain why
ETV2 does not universally interact with all members of the JMJD
protein family, which is characterized by the conserved JmjC
domain.
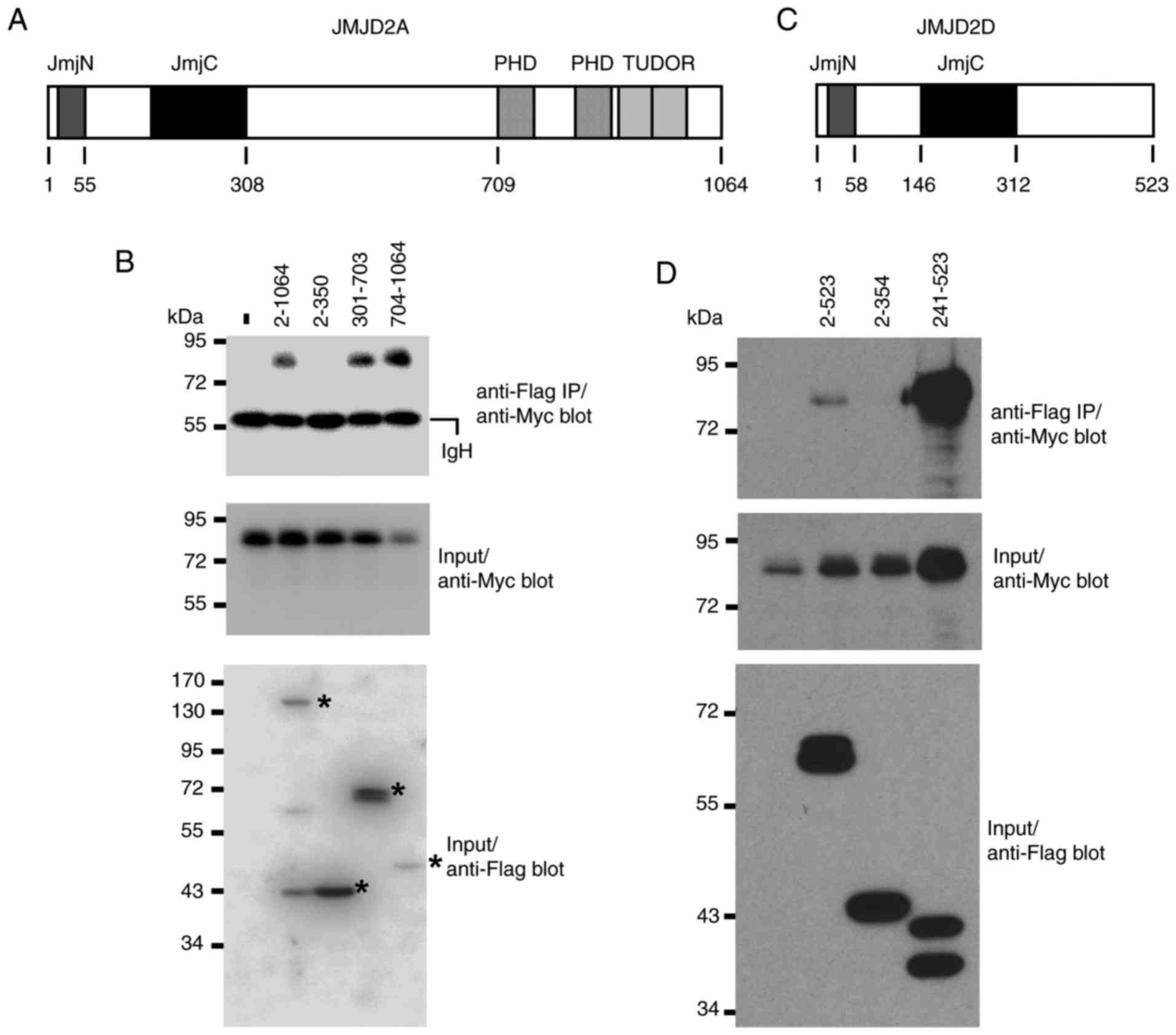 | Figure 4.Determination of JMJD2 amino acids
mediating complex formation with ETV2. (A) Domain structure of
human JMJD2A; JmjC catalytic center is modulated by the JmjN
domain. The double PHD and TUDOR domains that may bind to
methylated histone lysine residues are also presented. (B)
Flag-tagged JMJD2A, or indicated truncations thereof, were
cotransfected with 6Myc-ETV2 into 293T cells, anti-Flag IPs
performed and coprecipitated ETV2 was detected by anti-Myc western
blotting (top panel); the input levels of 6Myc-ETV2 and Flag-tagged
JMJD2A proteins are presented in the bottom two panels. (C) As (A),
but for Flag-tagged JMJD2D; (D) as (B), but for Flag-tagged JMJD2D.
JMJD, Jumonji domain-containing; ETV2, ETS variant 2; JmjC,
catalytic Jumonji C domain; JmjN, Jumonji N-terminal domain; PHD,
plant homeodomain; IP, immunoprecipitation. |
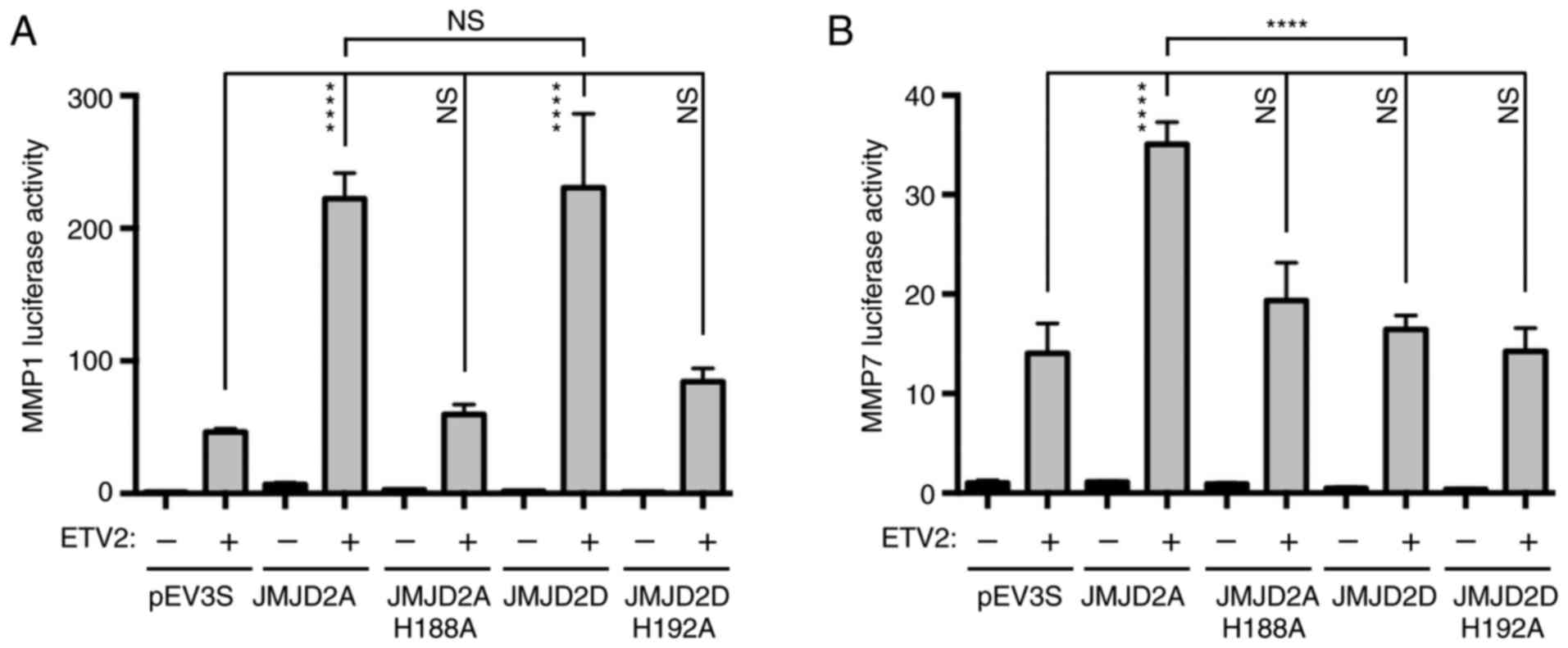
Activation of MMP promoters by
ETV2
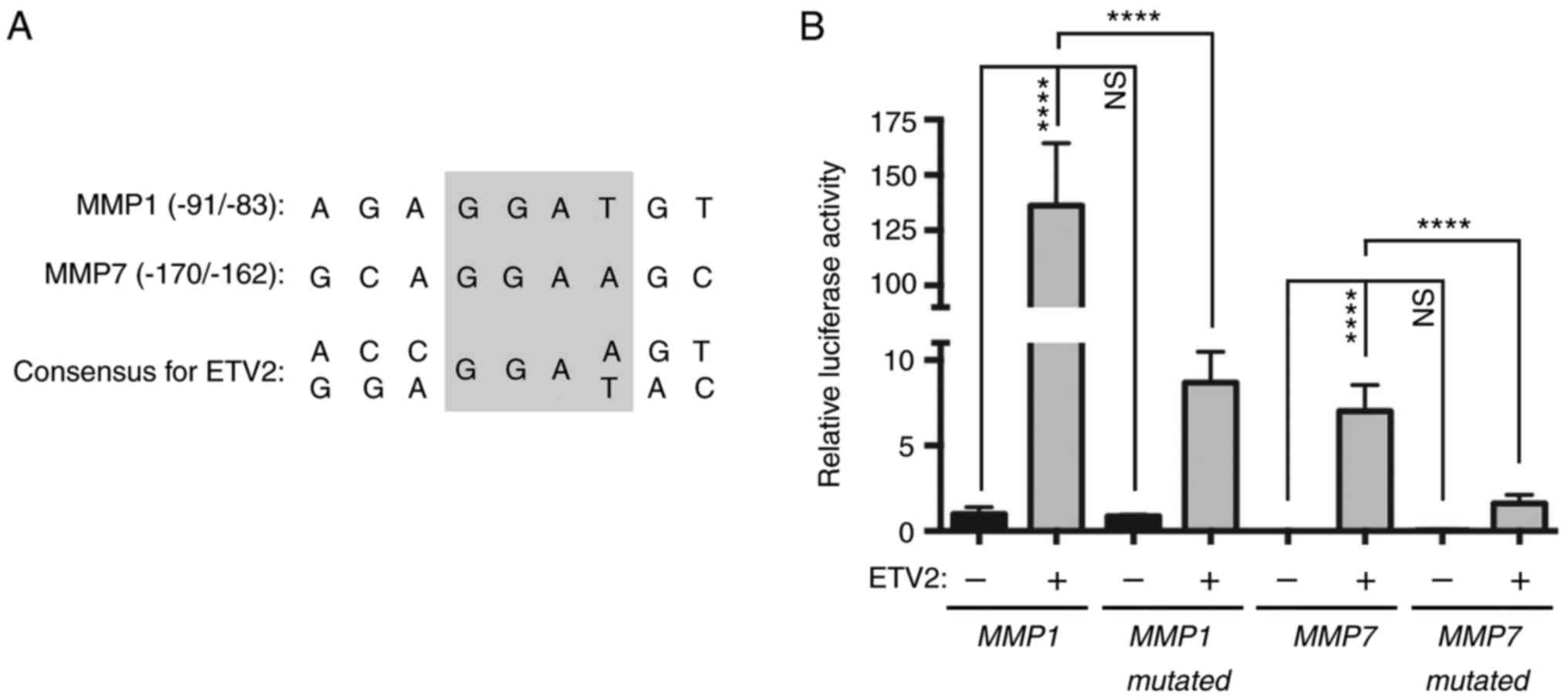
ETV2 was previously reported to bind to the human
MMP1 gene promoter and stimulate its activity (2). MMP7 is another MMP reported to be
regulated by multiple ETS proteins, but not ETV2 (38). In addition, analysis of the MMP7
promoter for the presence of consensus binding sites for human
ETV2, [(A/G)(C/G)(C/A)GGA(A/T)(G/A)(T/C)] (39), revealed a match that was similar to
the reported binding site for ETV2 in the MMP1 promoter (Fig. 5A). Therefore, the regulation of
MMP1 and MMP7 transcriptional activity by ETV2 in human LNCaP
prostate cancer cells was investigated. ETV2 markedly stimulated
the expression of an MMP1 luciferase reporter construct (Fig. 5B). Additionally, mutation of the
known ETV2 binding site of the MMP1 reporter markedly reduced the
ETV2-dependent transcription, but not basal transcription. The
residual induction of this mutated MMP1 promoter by ETV2 may be due
to further potential ETS binding sites in the MMP1 luciferase
reporter construct (2), which may
interact with ETV2. Regardless, these findings demonstrated that
the −91/-83 ETS binding site in the MMP1 gene promoter
predominantly mediated its response to ETV2 in LNCaP prostate
cancer cells.
Analysis of the MMP7 gene promoter in LNCaP cells
revealed reduced activity compared with the MMP1 gene promoter
(Fig. 5B). However, a marked
induction in luciferase activity was nonetheless observed upon the
overexpression of ETV2. Additionally, mutations in the −170/-162
ETS site of the MMP7 promoter significantly decreased
transactivation by ETV2 (Fig. 5B).
Therefore, it was concluded that MMP1 and MMP7 are bona fide target
genes of ETV2 in LNCaP prostate cancer cells.
Cooperation between ETV2 and JMJD2
proteins
Subsequently, the effects of ETV2 and JMJD2
coexpression on the activity of the MMP1 gene promoter were
investigated. Although ETV2 markedly induced the MMP1 luciferase
activity within LNCaP cells, this did not occur when JMJD2A or
JMJD2D were expressed in the absence of ETV2 (Fig. 6A). The co-expression of ETV2 with
either JMJD2A or JMJD2D raised luciferase activity by ~5-fold
compared with ETV2 expression alone, indicating that JMJD2A or
JMJD2D synergized with ETV2 in activating transcription.
Additionally, no significant difference between JMJD2A and JMJD2D
was observed, despite the stronger interaction between JMJD2D and
ETV2 compared with JMJD2A. In contrast to wild-type JMJD2 proteins,
the catalytically inactive JMJD2A-H188A and JMJD2D-H192A mutants
did not engage with ETV2 (Fig.
6A), suggesting that catalytic activity may be required for
JMJD2 proteins to function as coactivators of ETV2.
The cooperation of ETV2 with JMJD2A or JMJD2D at the
MMP7 gene promoter was also investigated. As presented in Fig. 6B, JMJD2A, but not its catalytic
H188A mutant, elevated ETV2-dependent transcription by ~2.5-fold,
which was lower than the interaction at the MMP1 promoter. In
addition, JMJD2D did not cooperate at all with ETV2 on the MMP7
promoter (Fig. 6B), indicating
that JMJD2A and JMJD2D are not identical in their abilities to
coactivate ETV2.
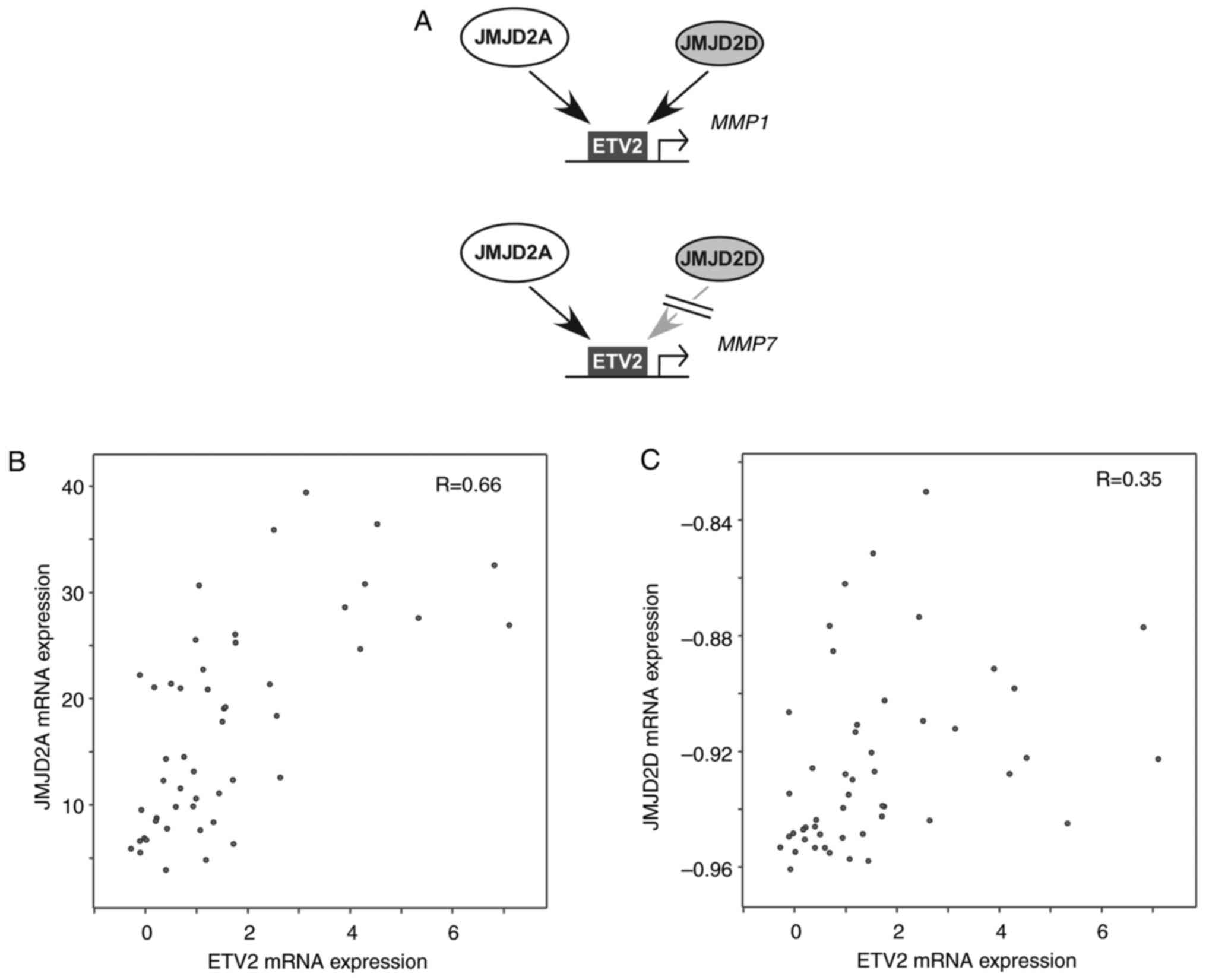
The present data suggest that ETV2 has the potential
to cooperate with JMJD2 proteins to regulate transcription, as in
the case of MMP1 and MMP7 genes (Fig.
7A), in LNCaP prostate cancer cells. Notably, analysis of
published mRNA expression data (31) revealed that ETV2 expression was
significantly correlated with expression of JMJD2A (Fig. 7B) and JMJD2D (Fig. 7C) in neuroendocrine prostate
tumors, implying that the transcriptional cooperation of these
factors could be relevant in this prostate cancer subtype.
Discussion
In the present study, it was identified that ETV2
forms complexes with JMJD2A or JMJD2D and that these two histone
demethylases can cooperate with ETV2 to induce gene transcription
within human LNCaP prostate cancer cells. JMJD2A synergized with
ETV2 on the MMP1 and MMP7 promoters, whereas JMJD2D only cooperated
with ETV2 on the MMP1 gene promoter (Fig. 7A). A recent study reported that
JMJD2A, and not JMJD2D, may coactivate ETV1-dependent YAP1 gene
transcription (33); however, both
JMJD2A and JMJD2D coactivated the androgen receptor on the
prostate-specific antigen and the mouse mammary tumor virus
promoter (40). JMJD2A and JMJD2D
may even have antagonistic activities, as exemplified by their
opposing functions in p53-dependent p21 promoter regulation
(41,42). Thus, it would be expected in
genome-wide studies for JMJD2A and JMJD2D to display different
activities at a significant proportion of ETV2 target gene
promoters. In addition, the ratio of JMJD2A to JMJD2D may determine
the extent to which a target gene will be regulated by ETV2.
The JMJD2 gene family of histone demethylases
encompasses six members in humans (43–45),
suggesting that family members other than JMJD2A and JMJD2D may
also cooperate with ETV2. A comparison between JMJD2A and JMJD2D
revealed that they target a different spectrum of substrates: For
instance, JMJD2A may target methylated lysines 9 and 36 on histone
H3, whereas JMJD2D can only catalyze the demethylation of histone
H3 lysine 9 (36,46). This may be one reason why JMJD2A
and JMJD2D behaved differently at the MMP7 gene promoter. Notably,
trimethylation of histone H3 on lysine 9 represses the initiation
of gene transcription (47),
providing one explanation why demethylation of this histone mark by
JMJD2A and JMJD2D may upregulate gene promoters upon recruitment by
ETV2. In addition, JMJD2A contains double PHD and Tudor domains,
but JMJD2D lacks any of these domains involved in binding to
histone modifications, which may explain the different behaviors of
JMJD2A and JMJD2D at the MMP7 promoter. For instance, the Tudor
domain of JMJD2A can bind trimethylated lysine 4 on histone H3
(48,49), which may protect this activating
histone mark from demethylation and thereby contribute to the
coactivator function of JMJD2A.
Another important finding presented in this report
is that ETV2 is robustly expressed, or its gene is amplified, in
various types of cancer. This suggests that ETV2 may serve a role
in cancer. As MMPs are important modulators of tumor formation,
invasion, angiogenesis and metastasis (50,51),
the activation of MMP1 and MMP7 transcription by ETV2 may
contribute to its tumor-promoting function. Notably, ETV2 gene
amplification was observed in lung squamous cell carcinomas and
adenocarcinomas, and ETV2 expression was evident in prostate,
breast and lung cancer, the three carcinomas most frequently
diagnosed in humans in the western hemisphere (52). As JMJD2A has been reported to be
overexpressed in prostate (32,33),
breast (53–56) and lung (57,58)
tumors, JMJD2A and ETV2 may cooperate in such tumors. Analysis of
neuroendocrine prostate tumors revealed that the ETV2 gene
amplification rate was 17.8% (31); in the present study, ETV2
expression was significantly correlated with JMJD2A, as well as
JMJD2D. These data supported the notion that the ETV2-JMJD2A/D
interaction may serve an important role in neuroendocrine prostate
tumors.
In hematopoietic stem cells, ETV2 is capable of
promoting their regeneration by stimulating cell proliferation
(8,59). In addition, JMJD2A activity appears
to promote embryonic stem cell self-renewal (60). Further investigation is required to
understand whether ETV2 in cooperation with JMJD2A/D may be
involved in cancer stem cell maintenance and therefore, contribute
to tumorigenesis. In addition, ETV2 has been identified as a
reprogramming factor, either working with other factors (61–64)
or alone (65–67), in converting amniotic cells or
fibroblasts into endothelial cells. As cell reprogramming entails
epigenetic alterations, the interactions of ETV2 with the
epigenetic modifiers JMJD2A and JMJD2D, or the previously
identified interacting JMJD1A (13), may be required for the generation
of endothelial cells from other differentiated cells. Finally,
JMJD2D is highly expressed in adult testes and regulates the
methylation status of lysine 9 on histone H3 during spermatogenesis
(68). As ETV2 is also
preferentially expressed in testes (1,9), the
ETV2-JMJD2D complex may plausibly contribute to testicular
function; however, further investigation is required.
Acknowledgements
The present study was supported by grants from the
National Cancer Institute (to RJ; grant no. R01 CA154745) and
funding from the Graduate School of Jilin University and the
China-Japan Union Hospital of Jilin University (to XL). The content
is solely the responsibility of the authors and does not
necessarily represent the official views of the granting
agencies.
References
|
1
|
Brown TA and McKnight SL: Specificities of
protein-protein and protein-DNA interaction of GABP alpha and two
newly defined ets-related proteins. Genes Dev. 6:2502–2512. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
De Haro L and Janknecht R: Functional
analysis of the transcription factor ER71 and its activation of the
matrix metalloproteinase-1 promoter. Nucleic Acids Res.
30:2972–2979. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hollenhorst PC, McIntosh LP and Graves BJ:
Genomic and biochemical insights into the specificity of ETS
transcription factors. Annu Rev Biochem. 80:437–471. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lee D, Park C, Lee H, Lugus JJ, Kim SH,
Arentson E, Chung YS, Gomez G, Kyba M, Lin S, et al: ER71 acts
downstream of BMP, Notch, and Wnt signaling in blood and vessel
progenitor specification. Cell Stem Cell. 2:497–507. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ferdous A, Caprioli A, Iacovino M, Martin
CM, Morris J, Richardson JA, Latif S, Hammer RE, Harvey RP, Olson
EN, et al: Nkx2-5 transactivates the Ets-related protein 71 gene
and specifies an endothelial/endocardial fate in the developing
embryo. Proc Natl Acad Sci USA. 106:pp. 814–819. 2009; View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shalaby F, Rossant J, Yamaguchi TP,
Gertsenstein M, Wu XF, Breitman ML and Schuh AC: Failure of
blood-island formation and vasculogenesis in Flk-1-deficient mice.
Nature. 376:62–66. 1995. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Park C, Lee TJ, Bhang SH, Liu F, Nakamura
R, Oladipupo SS, Pitha-Rowe I, Capoccia B, Choi HS, Kim TM, et al:
Injury-mediated vascular regeneration requires endothelial
ER71/ETV2. Arterioscler Thromb Vasc Biol. 36:86–96. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lee D, Kim T and Lim DS: The Er71 is an
important regulator of hematopoietic stem cells in adult mice. Stem
Cells. 29:539–548. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
De Haro L and Janknecht R: Cloning of the
murine ER71 gene (Etsrp71) and initial characterization of its
promoter. Genomics. 85:493–502. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
DiTacchio L, Bowles J, Shin S, Lim DS,
Koopman P and Janknecht R: Transcription factors ER71/ETV2 and SOX9
participate in a positive feedback loop in fetal and adult mouse
testis. J Biol Chem. 287:23657–23666. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Otake T and Kuroiwa A: Molecular mechanism
of male differentiation is conserved in the SRY-absent mammal,
Tokudaia osimensis. Sci Rep. 6:328742016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kim JY, Lee RH, Kim TM, Kim DW, Jeon YJ,
Huh SH, Oh SY, Kyba M, Kataoka H, Choi K, et al: OVOL2 is a
critical regulator of ER71/ETV2 in generating FLK1+,
hematopoietic, and endothelial cells from embryonic stem cells.
Blood. 124:2948–2952. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Knebel J, De Haro L and Janknecht R:
Repression of transcription by TSGA/Jmjd1a, a novel interaction
partner of the ETS protein ER71. J Cell Biochem. 99:319–329. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yamane K, Toumazou C, Tsukada Y,
Erdjument-Bromage H, Tempst P, Wong J and Zhang Y: JHDM2A, a
JmjC-containing H3K9 demethylase, facilitates transcription
activation by androgen receptor. Cell. 125:483–495. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kooistra SM and Helin K: Molecular
mechanisms and potential functions of histone demethylases. Nat Rev
Mol Cell Biol. 13:297–311. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Uhlén M, Fagerberg L, Hallström BM,
Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C,
Sjöstedt E, Asplund A, et al: Proteomics. Tissue-based map of the
human proteome. Science. 347:12604192015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mooney SM, Grande JP, Salisbury JL and
Janknecht R: Sumoylation of p68 and p72 RNA helicases affects
protein stability and transactivation potential. Biochemistry.
49:1–10. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dowdy SC, Mariani A and Janknecht R:
HER2/Neu- and TAK1-mediated up-regulation of the transforming
growth factor beta inhibitor Smad7 via the ETS protein ER81. J Biol
Chem. 278:44377–44384. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Janknecht R: Regulation of the ER81
transcription factor and its coactivators by mitogen- and
stress-activated protein kinase 1 (MSK1). Oncogene. 22:746–755.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kim TD, Fuchs JR, Schwartz E, Abdelhamid
D, Etter J, Berry WL, Li C, Ihnat MA, Li PK and Janknecht R:
Pro-growth role of the JMJD2C histone demethylase in HCT-116 colon
cancer cells and identification of curcuminoids as JMJD2
inhibitors. Am J Transl Res. 6:236–247. 2014.PubMed/NCBI
|
|
21
|
Berry WL, Kim TD and Janknecht R:
Stimulation of β-catenin and colon cancer cell growth by the KDM4B
histone demethylase. Int J Oncol. 44:1341–1348. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Papoutsopoulou S and Janknecht R:
Phosphorylation of ETS transcription factor ER81 in a complex with
its coactivators CREB-binding protein and p300. Mol Cell Biol.
20:7300–7310. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shin S, Kim TD, Jin F, van Deursen JM,
Dehm SM, Tindall DJ, Grande JP, Munz JM, Vasmatzis G and Janknecht
R: Induction of prostatic intraepithelial neoplasia and modulation
of androgen receptor by ETS variant 1/ETS-related protein 81.
Cancer Res. 69:8102–8110. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Oh S, Shin S, Lightfoot SA and Janknecht
R: 14-3-3 proteins modulate the ETS transcription factor ETV1 in
prostate cancer. Cancer Res. 73:5110–5119. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bosc DG, Goueli BS and Janknecht R:
HER2/Neu-mediated activation of the ETS transcription factor ER81
and its target gene MMP-1. Oncogene. 20:6215–6224. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shin S, Oh S, An S and Janknecht R: ETS
variant 1 regulates matrix metalloproteinase-7 transcription in
LNCaP prostate cancer cells. Oncol Rep. 29:306–314. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shin S and Janknecht R: Diversity within
the JMJD2 histone demethylase family. Biochem Biophys Res Commun.
353:973–977. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mooney SM, Goel A, D'Assoro AB, Salisbury
JL and Janknecht R: Pleiotropic effects of p300-mediated
acetylation on p68 and p72 RNA helicase. J Biol Chem.
285:30443–30452. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wu J and Janknecht R: Regulation of the
ETS transcription factor ER81 by the 90-kDa ribosomal S6 kinase 1
and protein kinase A. J Biol Chem. 277:42669–42679. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Goel A and Janknecht R: Concerted
activation of ETS protein ER81 by p160 coactivators, the
acetyltransferase p300 and the receptor tyrosine kinase HER2/Neu. J
Biol Chem. 279:14909–14916. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Beltran H, Prandi D, Mosquera JM, Benelli
M, Puca L, Cyrta J, Marotz C, Giannopoulou E, Chakravarthi BV,
Varambally S, et al: Divergent clonal evolution of
castration-resistant neuroendocrine prostate cancer. Nat Med.
22:298–305. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cloos PA, Christensen J, Agger K, Maiolica
A, Rappsilber J, Antal T, Hansen KH and Helin K: The putative
oncogene GASC1 demethylates tri- and dimethylated lysine 9 on
histone H3. Nature. 442:307–311. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kim TD, Jin F, Shin S, Oh S, Lightfoot SA,
Grande JP, Johnson AJ, van Deursen JM, Wren JD and Janknecht R:
Histone demethylase JMJD2A drives prostate tumorigenesis through
transcription factor ETV1. J Clin Invest. 126:706–720. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kim TD, Shin S and Janknecht R: ETS
transcription factor ERG cooperates with histone demethylase KDM4A.
Oncol Rep. 35:3679–3688. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kim TD, Oh S, Lightfoot SA, Shin S, Wren
JD and Janknecht R: Upregulation of PSMD10 caused by the JMJD2A
histone demethylase. Int J Clin Exp Med. 9:10123–10134.
2016.PubMed/NCBI
|
|
36
|
Whetstine JR, Nottke A, Lan F, Huarte M,
Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M and Shi
Y: Reversal of histone lysine trimethylation by the JMJD2 family of
histone demethylases. Cell. 125:467–481. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Klose RJ, Yamane K, Bae Y, Zhang D,
Erdjument-Bromage H, Tempst P, Wong J and Zhang Y: The
transcriptional repressor JHDM3A demethylates trimethyl histone H3
lysine 9 and lysine 36. Nature. 442:312–316. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Crawford HC, Fingleton B, Gustavson MD,
Kurpios N, Wagenaar RA, Hassell JA and Matrisian LM: The PEA3
subfamily of Ets transcription factors synergizes with
beta-catenin-LEF-1 to activate matrilysin transcription in
intestinal tumors. Mol Cell Biol. 21:1370–1383. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wei GH, Badis G, Berger MF, Kivioja T,
Palin K, Enge M, Bonke M, Jolma A, Varjosalo M, Gehrke AR, et al:
Genome-wide analysis of ETS-family DNA-binding in vitro and in
vivo. EMBO J. 29:2147–2160. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Shin S and Janknecht R: Activation of
androgen receptor by histone demethylases JMJD2A and JMJD2D.
Biochem Biophys Res Commun. 359:742–746. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kim TD, Shin S, Berry WL, Oh S and
Janknecht R: The JMJD2A demethylase regulates apoptosis and
proliferation in colon cancer cells. J Cell Biochem. 113:1368–1376.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kim TD, Oh S, Shin S and Janknecht R:
Regulation of tumor suppressor p53 and HCT116 cell physiology by
histone demethylase JMJD2D/KDM4D. PLoS One. 7:e346182012.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Katoh M and Katoh M: Identification and
characterization of JMJD2 family genes in silico. Int J Oncol.
24:1623–1628. 2004.PubMed/NCBI
|
|
44
|
Berry WL and Janknecht R: KDM4/JMJD2
histone demethylases: Epigenetic regulators in cancer cells. Cancer
Res. 73:2936–2942. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Labbe RM, Holowatyj A and Yang ZQ: Histone
lysine demethylase (KDM) subfamily 4: Structures, functions and
therapeutic potential. Am J Transl Res. 6:1–15. 2013.PubMed/NCBI
|
|
46
|
Hillringhaus L, Yue WW, Rose NR, Ng SS,
Gileadi C, Loenarz C, Bello SH, Bray JE, Schofield CJ and Oppermann
U: Structural and evolutionary basis for the dual substrate
selectivity of human KDM4 histone demethylase family. J Biol Chem.
286:41616–41625. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Black JC, Van Rechem C and Whetstine JR:
Histone lysine methylation dynamics: Establishment, regulation, and
biological impact. Mol Cell. 48:491–507. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kim J, Daniel J, Espejo A, Lake A, Krishna
M, Xia L, Zhang Y and Bedford MT: Tudor, MBT and chromo domains
gauge the degree of lysine methylation. EMBO Rep. 7:397–403.
2006.PubMed/NCBI
|
|
49
|
Huang Y, Fang J, Bedford MT, Zhang Y and
Xu RM: Recognition of histone H3 lysine-4 methylation by the double
tudor domain of JMJD2A. Science. 312:748–751. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Hadler-Olsen E, Winberg JO and
Uhlin-Hansen L: Matrix metalloproteinases in cancer: Their value as
diagnostic and prognostic markers and therapeutic targets. Tumour
Biol. 34:2041–2051. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Shay G, Lynch CC and Fingleton B: Moving
targets: Emerging roles for MMPs in cancer progression and
metastasis. Matrix Biol 44–46. 1–206. 2015.
|
|
52
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Patani N, Jiang WG, Newbold RF and Mokbel
K: Histone-modifier gene expression profiles are associated with
pathological and clinical outcomes in human breast cancer.
Anticancer Res. 31:4115–4125. 2011.PubMed/NCBI
|
|
54
|
Slee RB, Steiner CM, Herbert BS, Vance GH,
Hickey RJ, Schwarz T, Christan S, Radovich M, Schneider BP,
Schindelhauer D and Grimes BR: Cancer-associated alteration of
pericentromeric heterochromatin may contribute to chromosome
instability. Oncogene. 31:3244–3253. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Berry WL, Shin S, Lightfoot SA and
Janknecht R: Oncogenic features of the JMJD2A histone demethylase
in breast cancer. Int J Oncol. 41:1701–1706. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Li LL, Xue AM, Li BX, Shen YW, Li YH, Luo
CL, Zhang MC, Jiang JQ, Xu ZD, Xie JH and Zhao ZQ: JMJD2A
contributes to breast cancer progression through transcriptional
repression of the tumor suppressor ARHI. Breast Cancer Res.
16:R562014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Mallette FA and Richard S: JMJD2A promotes
cellular transformation by blocking cellular senescence through
transcriptional repression of the tumor suppressor CHD5. Cell Rep.
2:1233–1243. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Xu W, Jiang K, Shen M, Qian Y and Peng Y:
SIRT2 suppresses non-small cell lung cancer growth by targeting
JMJD2A. Biol Chem. 396:929–936. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Xu CX, Lee TJ, Sakurai N, Krchma K, Liu F,
Li D, Wang T and Choi K: ETV2/ER71 regulates hematopoietic
regeneration by promoting hematopoietic stem cell proliferation. J
Exp Med. 214:1643–1653. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Pedersen MT, Kooistra SM, Radzisheuskaya
A, Laugesen A, Johansen JV, Hayward DG, Nilsson J, Agger K and
Helin K: Continual removal of H3K9 promoter methylation by Jmjd2
demethylases is vital for ESC self-renewal and early development.
EMBO J. 35:1550–1564. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Ginsberg M, James D, Ding BS, Nolan D,
Geng F, Butler JM, Schachterle W, Pulijaal VR, Mathew S, Chasen ST,
et al: Efficient direct reprogramming of mature amniotic cells into
endothelial cells by ETS factors and TGFβ suppression. Cell.
151:559–575. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Han JK, Chang SH, Cho HJ, Choi SB, Ahn HS,
Lee J, Jeong H, Youn SW, Lee HJ, Kwon YW, et al: Direct conversion
of adult skin fibroblasts to endothelial cells by defined factors.
Circulation. 130:1168–1178. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Ginsberg M, Schachterle W, Shido K and
Rafii S: Direct conversion of human amniotic cells into endothelial
cells without transitioning through a pluripotent state. Nat
Protoc. 10:1975–1985. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wong WT and Cooke JP: Therapeutic
transdifferentiation of human fibroblasts into endothelial cells
using forced expression of lineage-specific transcription factors.
J Tissue Eng. 7:2016. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Morita R, Suzuki M, Kasahara H, Shimizu N,
Shichita T, Sekiya T, Kimura A, Sasaki K, Yasukawa H and Yoshimura
A: ETS transcription factor ETV2 directly converts human
fibroblasts into functional endothelial cells. Proc Natl Acad Sci
USA. 112:pp. 160–165. 2015; View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Van Pham P, Vu NB, Nguyen HT, Huynh OT and
Truong MT: Significant improvement of direct reprogramming efficacy
of fibroblasts into progenitor endothelial cells by ETV2 and
hypoxia. Stem Cell Res Ther. 7:1042016. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Lee S, Park C, Han JW, Kim JY, Cho K, Kim
EJ, Kim S, Lee SJ, Oh SY, Tanaka Y, et al: Direct reprogramming of
human dermal fibroblasts into endothelial cells using ER71/ETV2.
Circ Res. 120:848–861. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Iwamori N, Zhao M, Meistrich ML and Matzuk
MM: The testis-enriched histone demethylase, KDM4D, regulates
methylation of histone H3 lysine 9 during spermatogenesis in the
mouse but is dispensable for fertility. Biol Reprod. 84:1225–1234.
2011. View Article : Google Scholar : PubMed/NCBI
|