Introduction
In mammals, the transition of the fetus from an
aqueous to an air breathing environment at birth is dependent upon
the formation and function of the lungs. Neonatal respiratory
distress, primarily resulting from the immaturity of fetal lungs is
a common cause of morbidity and mortality in preterm infants
(1). Synthesis and metabolism of
pulmonary surfactant (PS) in type II alveolar epithelial cells
(AECIIs) is closely associated with the state of maturation of the
fetal lung (2,3). PS is a complex lipid and protein
mixture, produced by AECIIs from late fetal development onwards,
and prevents alveolar collapse by regulating surface tension at the
pulmonary air-liquid interface (4). It is composed of 90% lipids and 10%
surfactant-associated proteins, including surfactant protein
(SP)-A, SP-B, SP-C and SP-D. SP-A and SP-D, hydrophilic collectin
proteins, participate in pulmonary host defense and modify immune
responses, whereas SP-B and SP-C, small hydrophobic proteins,
together with dipalmitoylphosphatidylcholine (DPPC), confer surface
tension lowering properties to the material (5–7).
MicroRNAs (miRNAs/miRs) are a class of small non-coding RNAs that
suppress gene translation, most commonly through promoting mRNA
degradation or disrupting mRNA translation. A total of >100
miRNAs have been identified to be crucial during lung development,
however detailed functions of the majority of them are unknown
(8). The authors previously
demonstrated that miR-26a was one of seven miRNAs that demonstrated
significant alterations in expression, as determined by miRNA
profiling, at three-time points in the developing rat lung
(9). It was also identified that
PS synthesis is regulated by miR-26a in fetal rat AECIIs (2). However, there is a lack of data
regarding exactly how miR-26a carries out its specific function
in vitro and in transgenic animal studies.
As the lung development and PS regulatory processes
are quite well conserved, the mouse is an ideal animal to study the
stages of lung development (10–12).
Lung development in mouse begins at pseudoglandular stage
(E9.5-E16.5). This is followed by the canalicular (E16.5-E17.5) and
saccular [E18.5-postnatal day (P) 5] stages, during which these
terminal branches narrow and form clusters of epithelial sacs that
later develop into alveoli in preparation for respiration at birth.
Finally, full maturation of the alveolus occurs during the
alveolarization stage (P0-P14) (8). The clustered regularly interspaced
short palindromic repeat/CRISPR-associated protein 9 (CRISPR/Cas9)
system is a useful gene knockout technique for investigating gene
function in vivo; since its development, this technique has
been utilized to generate knockout cell lines (13,14)
and animal models (15,16). An miR-26a-1/miR-26a-2 double
knockout mouse model has been successfully obtained by using the
CRISPR/Cas9 system (17). The
present study used the miR-26a double knockout mouse model to
investigate the role of miR-26a in different stages of lung
development and PS synthesis in in vitro and transgenic
animal studies.
Materials and methods
Animals
C57BL/6J (25 female, weight 11±2 g, 3–4 weeks old
and 3 male, weight 27±2 g, 13 weeks old) and FVB mice (12 female,
weight 20±2 g, 7–8 weeks old) were purchased from Nanjing
Biomedical Research Institute of Nanjing University, China. All the
mice were housed apart and were maintained on a normal 12-h
light/dark cycle at a temperature of 20–26°C) and 40–70% humidity.
Water and food was supplied ad libitum. The housing
conditions met the specific pathogen-free (SPF) standards. Lungs
were obtained at gestational days (E) 16.5 and 18.5, and P day 2
from double knockout mice and wild-type mice. The pregnant mice
were killed by cervical dislocation after anesthesia with 2%
chloral hydrate (0.2 ml/10 g). Fetal mice were removed and fixed to
a clean operating table. Lungs were dissected free of heart and
trachea, and placed into EP tubes. All procedures were approved by
the Nanjing Medical University Animal Ethical and Welfare Committee
(Nanjing, China).
Microinjection
A Cas9 expression plasmid (GenScript Biotech
Corporation, Piscataway Township, NJ, USA) containing the SP6
promoter was used as a template for in vitro transcription
(IVT) using an mMESSAGE mMACHINE SP6 kit (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) following linearization and
purification. As previously described (15), fusion crRNA and tracrRNA expression
vectors were constructed with a customizable synthetic (sg)RNA
template using miR-26a-1F/R and miR-26a-2F/R primers (Table I). The T7-sgRNA polymerase chain
reaction (PCR) product was purified and used as a template for IVT,
performed using a T7 kit (Takara Bio, Inc., Otsu, Japan). Female
C57BL/6J mice were superovulated using pregnant mare serum
gonadotropin (GenWay Biotech Inc., San Diego, CA, USA) and were
injected with human chorionic gonadotropin (GenWay Biotech Inc.)
following 48 h. The superovulated female mice were mated with
C57BL/6J stud males, and fertilized embryos were collected from
their oviducts. miR-26a-1 and miR-26a-2 sgRNAs (12.5 ng/µl each)
were mixed with Cas9 mRNA (25 ng/µl), and the mixture was
microinjected into the cytoplasm of the C57BL/6J mouse embryos at
the pronuclei stage. The injected zygotes were cultured in KSOM
(EMD Millipore, Billerica, MA, USA) with amino acids at 37°C and 5%
CO2 in air until the blastocyst stage (3.5 days).
Subsequently, ~15–25 blastocysts were transferred into the uterus
of each pseudopregnant FVB female.
 | Table I.Customizable synthetic RNA primer
sequences. |
Table I.
Customizable synthetic RNA primer
sequences.
| Variables | Sequence (5′-3′) |
|---|
| mmu-mir-26a-1 |
|
|
Forward |
GGGCTCTTTCCTTAGACTTGG |
|
Reverse |
GACCTGCTTTGCTCATAACACTC |
| mmu-mir-26a-2 |
|
|
Forward |
GTTGGTGCTGATGTGGGCTAG |
|
Reverse |
CTGGGAGACAGAGTGGATTGC |
Genotyping and breeding
Following ~19 days, DNA was extracted from the pups'
tails (founder mice), and the mice were genotyped by PCR and
sequencing. Then, male (7-week-old) and female (4-week-old) founder
mice were mated with wild-type mice. The next generation of mice
(F1 mice) was obtained and genotyped until mice positive for either
or both mutations in miR-26a-1 and/or miR-26a-2 were identified,
indicating the successful generation of gene knockout mouse
strains. Positive mice (>3 females and >3 males) were mated
with wild-type mice (SPF standard), and the PCR products from the
tail DNA samples were identified following restriction digestion
with T7 endonuclease I (T7EI) and resolution in a 1% agarose gel.
The 20 µl PCR mixture contained 10 µl master mix (Vazyme Biotech,
Nanjing, China), 8.2 µl ddH2O, 1 µl DNA, and 0.4 µl each
of the miR-26a-1 forward and reverse primers or 0.4 µl each of the
miR-26a-2 forward and reverse primers (Table I). The PCR amplification program
was as follows: 94°C for 5 min; 35 cycles at 94°C for 30 sec, 55°C
for 30 sec, and 72°C for 60 sec; and 72°C for 7 min. The amplicons
were separated by 1.0% agarose gel electrophoresis. The bands were
visualized by ultraviolet projection and a Gel Doc XR+ Imaging
System with Image Lab Software version 5.2.1 (Bio-Rad Laboratories,
Inc., Hercules, CA, USA).
Morphological analysis
Lung tissue was fixed with 4% buffered
paraformaldehyde at 4°C overnight, dehydrated and embedded in
paraffin. Sections at 3–4 µm thickness were prepared for
hematoxylin and eosin (H&E) staining and immunohistochemistry.
Sections were dewaxed with xylene and hydrated in a series of
ethanol/water solutions. Then, the sections were stained with
hematoxylin for 5 min, differentiated with 1% ethanol hydrochloride
for 3 sec and transferred to eosin solution for 2 min at room
temperature. Then the sections were dehydrated and mounted.
Analysis was conducted under a microscope (BX51; Olympus
Corporation, Tokyo, Japan). The sections pretreated for
immunohistochemistry were dewaxed and rehydrated with xylene and
ethanol/water. The sections were immersed in sodium citrate antigen
retrieval solution (maintained at a sub-boiling temperature for 8
min, standing for 8 min and then followed by another sub-boiling
temperature for 7 min). To block endogenous peroxidase, sections
were immersed in 3% H2O2, kept in a dark
place and incubated at room temperature for 15 min. Then, sections
were blocked with 3% BSA at room temperature for 30 min. The
sections were incubated at 4°C overnight with primary antibodies,
rinsed with PBS, incubated with HRP-conjugated goat-anti-rabbit
secondary antibody (K5007; Dako; Agilent Technologies, Inc., Santa
Clara, CA, USA), and developed using diaminobenzidine.
Subsequently, Hematoxylin staining solution was used for
counterstaining the nucleus. Finally, the sections were dehydrated
and mounted. Analysis was conducted under a microscope (BX51;
Olympus Corporation). Primary antibodies used included rabbit
anti-SP-A antibody (ab115791; 1:100; Abcam, Cambridge, MA, USA),
rabbit anti-pro and mature SP-B antibody (ab40876; 1:200; Abcam),
rabbit anti-pro-SP-C antibody (AB3786; 1:1,000; EMD Millipore).
Electron microscopy was performed on lung tissues
obtained from E16.5, E18.5 and P2 mutant mice and littermate
controls. Lungs were fixed in glutaraldehyde at 4°C for 2–4 h,
post-fixed with 1% OsO4 in PBS for 2 h at room
temperature, dehydrated with ethanol/water and acetone, infiltrated
and embedded by acetone and EMBed 812 (90529-77-4; SPI Supplies,
West Chester, PA, USA), heated in a 60°C oven for 48 h. Sections of
60–80 nm thickness were prepared using an ultramicrotome, stained
with uranyl acetate in pure ethanol for 15 min and lead citrate for
15 min, and dried overnight at room temperature. Finally, electron
microscopy (Hitachi, Ltd., Tokyo, Japan) was performed.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from lung tissues according
to the manufacturer's protocol using TRIzol® reagent
(Thermo Fisher Scientific, Inc.). The RNA quantity control and
concentration were detected by NanoDrop 2000 Spectrophotometer
(Thermo Fisher Scientific, Inc.). A High-Capacity cDNA Reverse
Transcription kit (Applied Biosystems; Thermo Fisher Scientific,
Inc.) was used for RT. qPCR was performed using SYBR Green (Roche,
Shanghai, China), and specific primers for SP-A (Sftpa1), SP-B
(Sftpb), and SP-C (Sftpc) were synthesized according to published
cDNA sequences (Table II). The
PCR was performed with an ABI 7500 thermal cycler (Applied
Biosystems; Thermo Fisher Scientific, Inc.) and the reaction
conditions were 95°C for 10 min, 50°C for 2 min, then 40 cycles at
60°C for 1 min, and 95°C for 15 sec. Dissociation curves were
generated for genes under the following conditions: 95°C for 15
sec, 60°C for 60 sec, and 95°C for 15 sec. Relative quantification
of gene expression in multiple samples was achieved by
normalization against an endogenous control, GAPDH. Then, the
relative expression levels were compared between the knockout mice
and wild-type mice using the 2−ΔΔCq method (18).
 | Table II.Reverse transcription-quantitative
polymerase chain reaction primer sequences. |
Table II.
Reverse transcription-quantitative
polymerase chain reaction primer sequences.
| Variables | Sequence (5′-3′) |
|---|
| m-SP-A |
|
|
Forward |
ACCTCCTTCTGCTTGGAACC |
|
Reverse |
CCAGGAGCAGGCACTTTCTA |
| m-SP-B |
|
|
Forward |
CTGCTTCCTACCCTCTGCTG |
|
Reverse |
ATCCTCACACTCTTGGCACA |
| m-SP-C |
|
|
Forward |
TTGTCGTGGTGATTGTAGGG |
|
Reverse |
AGGTAGCGATGGTGTCTGCT |
| m-GAPDH |
|
|
Forward |
GGTGAAGGTCGGTGTGAACG |
|
Reverse |
CTCGCTCCTGGAAGATGGTG |
Protein extraction and western blot
analysis
Briefly, total proteins from tissues were extracted
using radioimmunoprecipitation assay buffer (Beyotime Institute of
Biotechnology, Nantong, China) containing protease inhibitors and
the concentrations were measured by bicinchoninic acid (BCA)
solution (Beyotime Institute of Biotechnology, Nantong, China). The
western blotting was performed under the standard program. Proteins
(60 µg/lane) were loaded on 12% SDS-PAGE gels, and the
separated proteins were transferred onto PVDF membrane (EMD
Millipore). The membrane was then blocked for non-specific binding
for 2 h by incubating with 5% fat-free dry milk in 100 mM
Tris-buffered saline plus 0.1% Tween-18 (TBS-T). Primary antibodies
were diluted in blocking buffer and incubated with membrane
overnight at 4°C. Primary antibodies used included rabbit anti-SP-A
antibody (ab115791; 1:4,000; Abcam), rabbit anti-pro and mature
SP-B antibody (ab40876; 1:5,000; Abcam), rabbit anti-pro-SP-C
antibody (AB3786; 1:500; EMD Millipore), and rabbit anti-GAPDH
antibody (KGAA002; 1:1,000; Nanjing KeyGen Biotech Co., Ltd.,
Nanjing, China). Following washing in TBST 3 times, the membrane
was incubated with HRP-conjugated goat-anti-rabbit secondary
antibody (bs-0295G-HRP; 1:2,000; BIOSS, Beijing, China) for 1.5 h
at room temperature. The antibody-detected protein bands were
visualized by an enhanced chemiluminescence detection system
(ChemiDoc XRS + Imaging System; Bio-Rad Laboratories, Inc.)
Statistical analysis
All quantitative data were expressed as the mean ±
standard error of the mean. All experiments were repeated 3 times,
and data represent consistent results. The results were analyzed
using the Student's t-test with SPSS software, version 17.0 (SPSS,
Inc., Chicago, IL, USA) and GraphPad Prism 5.0 (GraphPad Software,
Inc., La Jolla, CA, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
Successful establishment of
miR-26a-1/miR-26a-2 double knockout mouse model
CRISPR/Cas9 plasmids targeting miR-26a-1 and
miR-26a-2 were constructed. Mice harboring mutations in the two
genes were produced by coinjection of Cas9 with miR-26a-1 and
miR-26a-2 sgRNAs into zygotes. The efficiency of obtaining mice
carrying mutations in the two targeted genes reached 28%, and ~56%
of the mice harbored a biallelic mutation in one of the targeted
genes (17). Phenotypic analysis
revealed that the majority of the double knockout mice did not
demonstrate any differences in phenotype, body weight, pregnancy
rate, birth rate, or growth rate compared with the wild-type mice
(17). A number of the knockout
mice were blind and had white abdominal hair, however the reason
was unknown and requires further investigation.
Knockout of miR-26a promotes
maturation in lung formation
H&E staining was performed to observe
differences in lung formation at E16.5, E18.5, and P2 between the
miR-26a double knockout mice and wild-type mice. At the beginning
of the canalicular stage, numerous epithelial tubules were
observed, and the high columnar epithelial cells altered to a low
cuboidal shape in lungs from the two groups, however dilated lumens
and a more prominent aerated area was observed in the knockout mice
lungs (E16.5; Fig. 1A). At the
latter stage (E18.5; Fig. 1B), a
large number of AECIIs were produced and lungs from miR-26a
knockout mice demonstrated an increase in the maturation of
alveolar structure compared with wild-type mice lungs. Lung tissues
from the two groups demonstrated further alveolar maturation
characterized by further-expanded alveolar air spaces and thinning
air sac wall at P2, however more AECIIs and fewer type I alveolar
epithelial cells (AECIs) were observed in knockout mice lungs
(Fig. 1C and D). These
observations indicated increased maturity of lung tissues from the
double knockout mice in lung development.
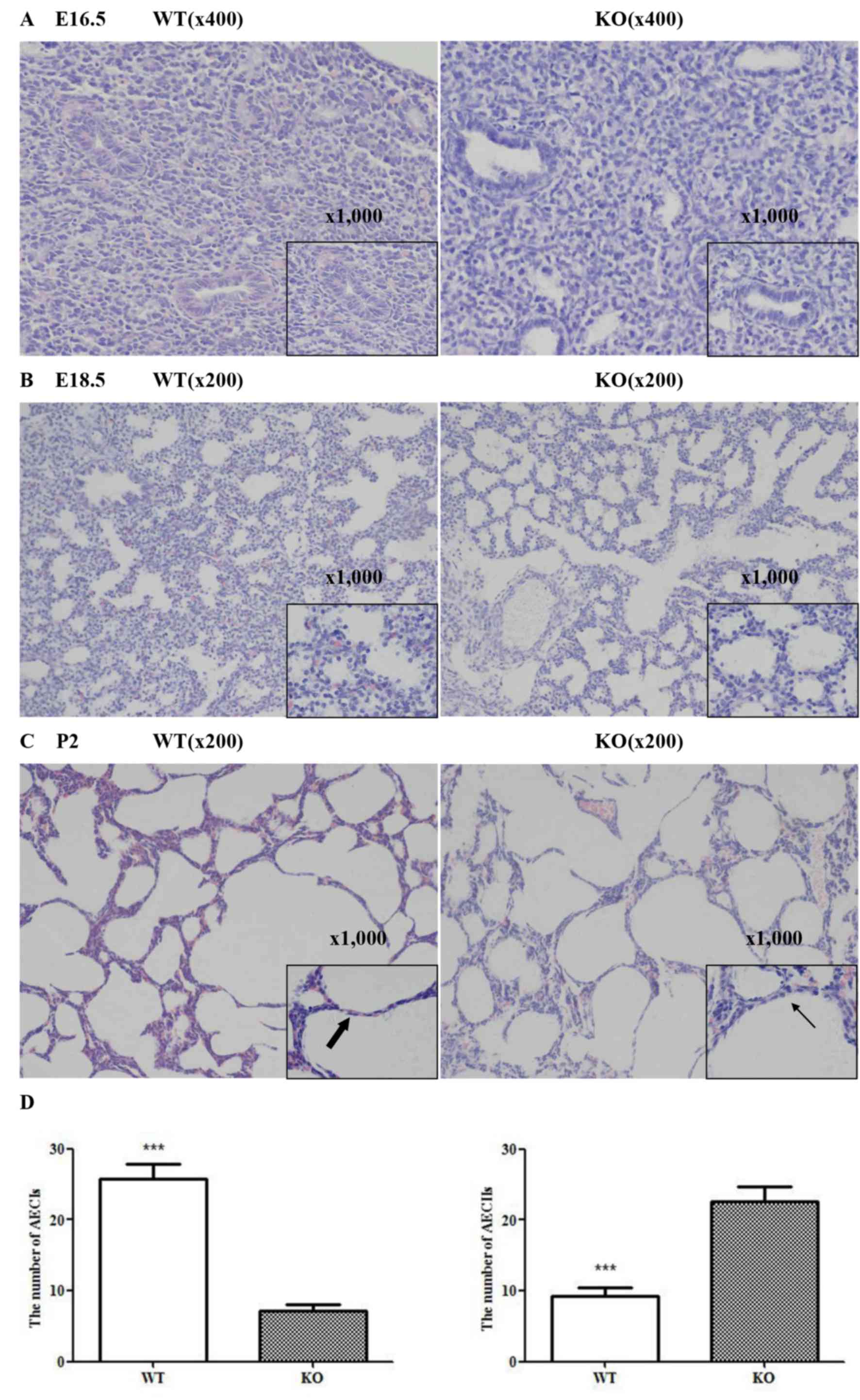 | Figure 1.Effects of miR-26a on maturation of
lung formation, observed by Hematoxylin and eosin staining (n=3).
(A) At an early stage (E16.5), dilated lumens and a prominent
aerated area were observed in lungs of KO mice compared with WT
mice (magnification, ×400 and ×1,000). (B) Numerous AECIIs were
produced (magnification, ×1,000) and the alveolar structure of KO
mice lungs was more mature compared with WT mice lungs at E18.5
(magnification, × 200). (C) Although lung tissues from the two
groups demonstrated further alveolar maturation (magnification,
×200), more AECIIs (fine arrows) and fewer AECIs (thick arrows)
were detected in KO mice compared with WT mice (magnification,
×1,000) and (D) the difference was statistically significant. Data
are presented as the mean ± standard deviation (n=6, ***P<0.001
vs. KO mice). miR, microRNA; AEC, alveolar epithelial cell; KO,
knockout mice; WT, wild-type mice; P, postnatal day. |
miR-26a serves an important role in
the production process of PS
Compared with wild-type mice lungs, the epithelial
cells of knockout mice lungs had more glucogen at the beginning of
the canalicular stage (E16.5, Fig.
2A). PS production and secretion was increasingly detected in
knockout mice lungs at E18.5d and P2, as demonstrated by more
mature lamellar bodies inside AECIIs (Fig. 2B and C).
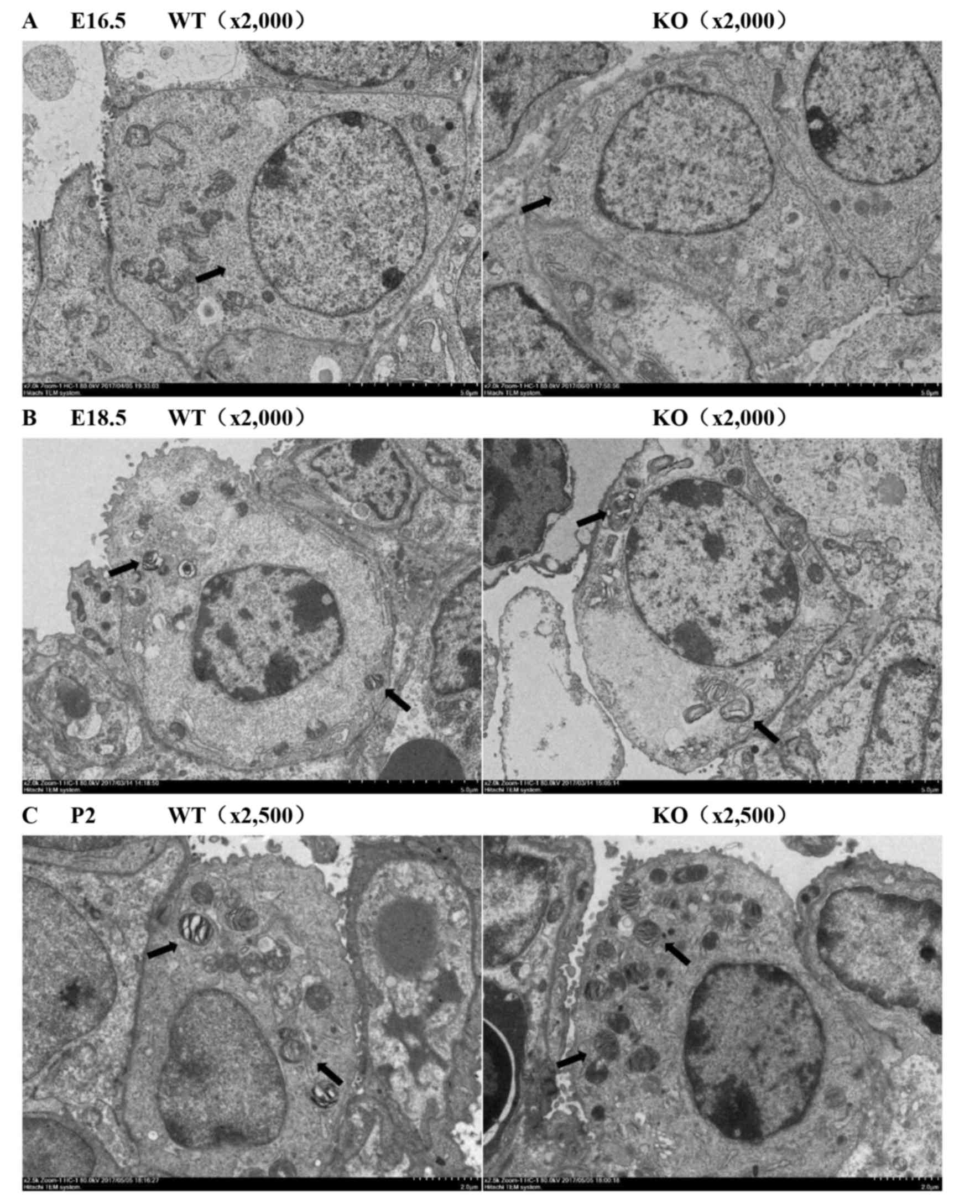 | Figure 2.Importance of miR-26a in the
production process of PS was detected by transmission electron
microscopy (n=3). (A) Increased intracellular glucogen (arrows) was
observed in KO mice lungs compared with WT mice lungs at E16.5
(magnification, ×2,000). (B and C) Increased mature lamellar bodies
(arrows) inside AECIIs were detected in the KO mice compared with
the WT mice at E18.5 (magnification, ×2,000) and P2 (magnification,
×2,500). PS, pulmonary surfactant; miR, microRNA; KO, knockout
mice; WT, wild-type mice; AEC, alveolar epithelial cell; P,
postnatal day. |
Knockout of miR-26a increases the
expression of SP-A, SP-B and SP-C
To explore the effect of miR-26a on PS synthesis,
the expression of SP-A, SP-B and SP-C were examined by
immunohistochemistry, RT-qPCR and western blot analysis. The SP-A,
SP-B and SP-C protein levels, detected by immunohistochemistry at
E18.5 and P2, were significantly increased in knockout mice
compared with the wild-type mice (Fig.
3A-C). Results consistent with immunohistochemistry were
obtained by western blotting (E18.5, P2; Fig. 3D). Furthermore, the RT-qPCR
analyses of the mRNA levels of SP-A, SP-B and SP-C also
demonstrated a significant increase in miR-26a knockout E18.5 and
P2 lungs (Fig. 3E). These results
supported the hypothesis that knockout of miR-26a induced an
increase in PS synthesis.
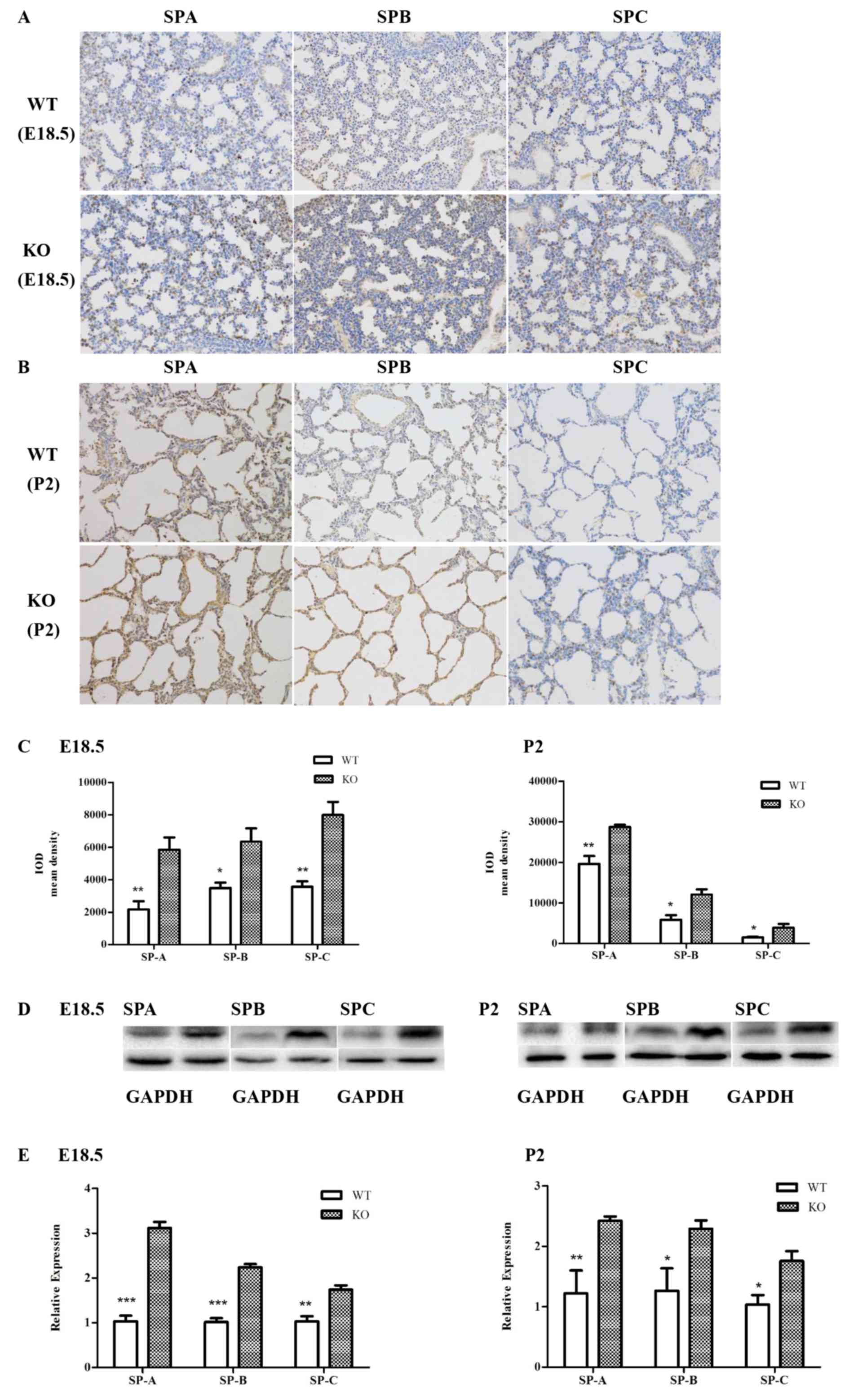 | Figure 3.Effects of miR-26a on the expression
of SP-A, SP-B, and SP-C. (A and B) SP-A, SP-B and SP-C were
identified by immunohistochemistry (representative images;
magnification, ×200). (C) Analysis of the mean IOD demonstrated
that the SP-A, SP-B and SP-C protein levels were significantly
increased in the KO mice at E18.5 and P2. Data are presented as the
mean ± standard deviation (n=4; *P<0.05; **P<0.01 vs. KO
mice). (D) Gene expression at protein level was detected by western
blotting and was significantly increased in the KO mice compared
with the WT mice at E18.5 and P2 (n=6). (E) Increased mRNA levels
of SP-A, SP-B, and SP-C were quantified by reverse
transcription-quantitative polymerase chain reaction in E18.5 and
P2 lungs of KO mice versus WT mice (n=6, *P<0.05; **P<0.01;
***P<0.001 vs. KO mice). miR, microRNA; SP, surfactant protein;
KO, knockout mice; WT, wild-type mice; IOD, integrated optical
density. |
Discussion
The fetal lung undergoes extensive physiological and
biochemical maturation prior to birth in preparation for its
postnatal function as an organ for gas exchange (19). PS, a unique developmentally
regulated, phospholipid-rich lipoprotein, is synthesized by AECIIs
of the pulmonary alveolus and is stored in organelles termed
lamellar bodies (2,7). PS lines the alveoli with a role to
decrease surface tension at the air-liquid interface and so
facilitate the expansion of the lungs (20). PS is markedly increased during late
lung development, and is required for proper lung development and
the adaptation to air breathing following birth (6,12,19).
Underdevelopment of lung structure and/or any reduction in the
ability of AECIIs to produce PS contribute significantly to the
morbidity and mortality of infants born prematurely (1,19,20).
The present study aimed to explore the potential role of miR-26a in
lung formation and PS synthesis at different stages of lung
development using a miR-26a double knockout mouse model.
miRNAs have been increasingly recognized to serve
crucial roles in the regulation of lung maturation and PS
synthesis, including miR-17, miR-20a, miR-106b and miR-127
(21–23). In the authors' previous study,
miRNA profiling was performed at three-time points (E16, E19 and
E21) of the developing fetal rat lung and the results suggested
that miR-26a was upregulated during late lung development,
indicating that miR-26a may serve an important role in lung
development (9). Previous studies
have indicated that miR-26a serves a critical role in
tumorigenesis, either as a tumor suppressor or as an oncogenic
miRNA, depending on different tumor types, however there have been
few studies on the association between miR-26a and lung development
(24–27). The authors' previous study
identified that the overexpression of miR-26a in AECIIs inhibited
PS synthesis by regulating SMAD1-associated bone morphogenic
protein (BMP) signaling pathways, which serve essential roles in
regulating lung development and function (2,11,28).
A miR-26a-1/miR-26a-2 double knockout mouse model was successfully
created using the CRISPR/Cas9 system and the homozygous double
knockout mice represented an ideal experimental model to analyze
the function of miR-26a in vivo. Furthermore, it was
identified that PS synthesis and the number of AECIIs were
significantly increased in miR-26a knockout mice (17). However, data from in vitro
and transgenic animal studies remain lacking. To the best of the
authors' knowledge, the present study was the first to explore the
effects of miR-26a during different stages of lung development
in vitro using knockout mice.
The present study was concerned with three critical
time points (E16.5, E18.5 and P2). To characterize the lung
formation in miR-26a double knockout mice, the prenatal and
neonatal lung morphology at different developmental stages was
examined by H&E staining. At E16.5, lungs progressed to the
canalicular stage and epithelial cells were altered to a low
cuboidal shape, observed in mutant mice and wild-type mice lungs,
however dilated lumens and more aerated area were observed in
mutant mice indicating increased maturity of fetal lungs. At the
saccular stage, a large number of AECIIs were produced. An increase
in the maturation of alveolar structure at E18.5 was observed and
there were more AECIIs in double knockout mice lungs at P2,
compared with wild-type mice lungs. Lamellar bodies are the
intracellular storage form of PS and serve an important role in the
production of PS. By using transmission electron microscopy, it was
identified that the number of lamellar bodies increased at E18.5
and P2 in mutant mice. In order to further understand the role of
miR-26a in PS synthesis, immunohistochemistry, RT-qPCR and western
blot analysis were performed to detect the expression of SP-A, SP-B
and SP-C, which serve important roles in the metabolism and surface
characteristics of PS. mRNA and protein levels of SP-A, SP-B and
SP-C increased in mutant mice compared with wild-type mice at the
late stages of lung development. These findings indicated that
knockout of miR-26a increases the maturation in lung formation and
the expression of surfactant-associated proteins, which is crucial
for gas exchange following birth.
Although the present study considered the role of
miR-26a in lung development in vivo, there remain a number
of deficiencies. A previous study identified that miR-26a is
selectively expressed in bronchial and alveolar epithelial cells of
the murine lung (10). However,
the experimental model employed in the present study was not a
tissue-specific knockout mice model. A previous in vitro
study suggested that miR-26a in fetal rat AECIIs inhibits PS
synthesis by regulating SMAD1-associated BMP signal pathways
(2), however the mechanism by
which miR-26a regulates PS synthesis and lung development in
vivo remains to be elucidated and further research is
required.
In conclusion, miR-26a serves an essential role
during lung development. Notably, the findings of the present study
indicated that knockout of miR-26a may promote the maturation in
lung formation and increase the expression of surfactant-associated
proteins in PS synthesis, which implicated the potential
application of miR-26a in the therapy of neonatal respiratory
distress.
Acknowledgements
The authors would like to thank the Nanjing Key
Laboratory of Pediatrics for technical assistance.
Funding
The present study was supported by funding from the
National Natural Science Foundation of China (grant no.
81270725).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YFS, QK and XYZ designed the experiments. YFS, YY,
YHZ, JXS and CZ performed experiments, collected the data presented
in the present study and interpreted results. YFS, QK and XYZ wrote
the manuscript.
Ethics approval and consent to
participate
All procedures were approved by the Nanjing Medical
University Animal Ethical and Welfare Committee (Nanjing,
China).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Martis PC, Whitsett JA, Yan X, Perl AT,
Wan HJ and Ikegami M: C/EBPalpha is required for lung maturation at
birth. Development. 133:1155–1164. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhang XQ, Zhang P, Yang Y, Qiu J, Kan Q,
Liang HL, Zhou XY and Zhou XG: Regulation of pulmonary surfactant
synthesis in fetal rat type II alveolar epithelial cells by
microRNA-26a. Pediatr Pulmonol. 49:863–872. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Brasch F, Schimanski Sven, Muhlfeld C,
Barlage S, Langmann T, Aslanidis C, Boettcher A, Dada A, Schroten
H, Mildenberger E, et al: Alteration of the pulmonary surfactant
system in full-term infants with hereditary ABCA3 deficiency. Am J
Respir Crit Care Med. 174:571–580. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gesche J, Fehrenbach H, Koslowski R, Ohler
FM, Pynn CJ, Griese M, Poets CF and Bernhard W: rhKGF stimulates
lung surfactant production in neonatal rats in vivo. Pediatr
Pulmonol. 46:882–895. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Han S and Mallampalli RK: The Role of
Surfactant in lung disease and host defense against pulmonary
infections. Ann Am Thorac Soc. 12:765–774. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jin J, Li YC, Ren JG, Lam SM, Zhang YD,
Hou Y, Zhang XJ, Xu R, Shui GH and Ma RZ: Neonatal respiratory
failure with retarded perinatal lung maturation in mice caused by
reticulocalbin 3 disruption. Am J Respir Cell Mol Biol. 54:410–423.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
El-Gendy N, Kaviratna A, Berkland C and
Dhar P: Delivery and performance of surfactant replacement
therapies to treat pulmonary disorders. Ther Deliv. 4:951–980.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Herriges M and Morrisey EE: Lung
development: Orchestrating the generation and regeneration of a
complex organ. Development. 141:502–513. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang Y, Kai G, Pu XD, Qing K, Guo XR and
Zhou XY: Expression profile of microRNAs in fetal lung development
of sprague-dawley rats. Int J Mol Med. 29:393–402. 2012.PubMed/NCBI
|
|
10
|
Williams AE, Moschos SA, Perry MM, Barnes
PJ and Lindsay MA: Maternally imprinted microRNAs are
differentially expressed during mouse and human lung development.
Dev Dyn. 236:572–580. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sun JP, Chen H, Chen C, Whitsett JA,
Mishina Y, Bringas P Jr, Ma JC, Warburton D and Shi W: Prenatal
lung epithelial cell-specific abrogation of Alk3-bone morphogenetic
protein signaling causes neonatal respiratory distress by
disrupting distal airway formation. Am J Pathol. 172:571–582. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Orgeig S, Morrison JL and Daniels CB:
Prenatal development of the pulmonary surfactant system and the
influence of hypoxia. Respir Physiol Neurobiol. 178:129–145. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jinek M, Chylinski K, Fonfara I, Hauer M,
Doudna JA and Charpentier E: A programmable dual-RNA-guided DNA
endonuclease in adaptive bacterial immunity. Science. 337:816–821.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wiedenheft B, Sternberg SH and Doudna JA:
RNA-guided genetic silencing systems in bacteria and archaea.
Nature. 482:331–338. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li D, Qiu Z, Shao Y, Chen Y, Guan Y, Liu
M, Li Y, Gao N, Wang L, Lu X, et al: Heritable gene targeting in
the mouse and rat using a CRISPR-Cas system. Nat Biotechnol.
31:681–683. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang
L, Kang Y, Zhao X, Si W, Li W, et al: Generation of gene-modified
cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell
embryos. Cell. 156:836–843. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang YH, Wu LZ, Liang HL, Yang Y, Qiu J,
Kan Q, Zhu W, Ma CL and Zhou XY: Pulmonary surfactant synthesis in
miRNA-26a-1/miRNA-26a-2 double knockout mice generated using the
CRISPR/Cas9 system. Am J Transl Res. 9:355–365. 2017.PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2 (-Dalta Dalta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
King G, Maker GL, Berryman D, Trengove RD
and Cake MH: Role of neuregulin-1beta in dexamethasone-enhanced
surfactant synthesis in fetal type II cells. FEBS Lett.
588:975–980. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Smith LJ, McKay KO, Asperen PP, Selvadurai
H and Fitzgerald DA: Normal development of the lung and premature
birth. Paediatr Respir Rev. 11:135–142. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Carraro G, El-Hashash A, Guidolin D,
Tiozzo C, Turcatel G, Young BM, Langhe SP, Bellusci S, Shi W,
Parnigotto PP and Warburton D: miR-17 family of microRNAs controls
FGF10-mediated embryonic lung epithelial branching morphogenesis
through MAPK14 and STAT3 regulation of E-Cadherin distribution. Dev
Biol. 333:238–250. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nana-Sinkam SP, Karsies T, Riscili B,
Ezzie M and Piper M: Lung microRNA: From development to disease.
Expert Rev Respir Med. 3:373–385. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bhaskaran M, Wang Y, Zhang HH, Weng TT,
Baviskar P, Guo YJ, Gou DM and Liu L: MicroRNA-127 modulates fetal
lung development. Physiol Genomics. 37:268–278. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Deng JJ, He MX, Chen LZ, Chen C, Zheng JM
and Cai ZL: The loss of miR-26a-mediated post-transcriptional
regulation of cyclin E2 in pancreatic cancer cell proliferation and
decreased patient survival. PLoS One. 8:e764502013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tan YW, Ge GH, Pan TL, Wen DF, Chen L, Yu
XJ, Zhou XB and Gan JH: A serum microRNA panel as potential
biomarkers for hepatocellular carcinoma related with hepatitis B
virus. PLoS One. 9:e1079862014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Salvatori B, Iosue I, Mangiavacchi A,
Loddo G, Padula F, Chiaretti S, Peragine N, Bozzoni I, Fazi F and
Fatica A: The microRNA-26a target E2F7 sustains cell proliferation
and inhibits monocytic differentiation of acute myeloid leukemia
cells. Cell Death Dis. 3:e4132012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ichikawa T, Sato F, Terasawa K, Tsuchiya
S, Toi M, Tsujimoto G and Shimizu K: Trastuzumab produces
therapeutic actions by upregulating miR-26a and miR-30b in breast
cancer cells. PLoS One. 7:e314222012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen C, Chen H, Sun J, Bringas P Jr, Chen
Y, Warburton D and Shi W: Smad1 expression and function during
mouse embryonic lung branching morphogenesis. Am J Physiol Lung
Cell Mol Physiol. 288:L1033–1039. 2005. View Article : Google Scholar : PubMed/NCBI
|

















