Introduction
Gastric cancer is one of the major causes of
cancer-associated mortality worldwide, especially in Asia-Pacific
regions (1). The incidence rate is
high in Asian populations, including Korea, China and Japan.
Treatment usually involves complete resection by surgery. However,
only 20–30% of patients with advanced gastric cancer have their
survival rate improved by surgery alone, and these suffer a high
risk of recurrence and metachronous metastases after surgery
(2). It has therefore been
determined that adjuvant chemotherapy may have a subordinate role
in clinical trials for advanced or metastatic stomach cancers
(3). Therapeutic application of
several chemotherapeutic agents, including 5-fluorouracil (5-FU),
doxorubicin, mitomycin, carmustine and cisplatin led to a low
response rate, ranging from 15–30% in patients with advanced
gastric cancer, when used as single treatments. Combination
chemotherapy, such as the widely used regimen of 5-FU, doxorubicin,
and mitomycin, increased the response rate to 30–40% for a period
of 5 to 6 months, and patient overall survival time increased by 7
to 8 months (4).
From these different chemotherapeutic agents, the
present study specifically focused on the molecular mechanism of
5-FU treatment in gastric cancer (5). 5-FU is an analogue of uracil with a
fluorine atom at the C-5 position in place of a hydrogen, and is a
major chemotherapeutic agent used in therapies against
malignancies, including stomach, colon and breast carcinomas. It
has been previously determined that cytotoxicity may be induced
through a mechanism involving the incorporation of
fluoro-nucleotides into RNA and DNA and by inhibiting the
nucleotide synthetic enzyme thymidylate synthase (6). The cytotoxic effect of 5-FU may
activate programmed cell death pathways and induce apoptosis in
cancer cell lines, such as colorectal cancer cells (7). Autophagy induction by 5-FU has been
frequently observed in various cancer cells, including colon,
pancreatic and hepatocellular carcinoma cells (8–10).
Autophagy is an evolutionarily conserved process involving the
degradation of long-lived proteins, organelles and bulk cytoplasm
which occurs during cell development, stress or starvation
(11). However, its role in cancer
is controversial as previous studies (12,13)
have reported its role in tumor growth, whereas others report its
tumor suppressive function. It has also been previously reported
that autophagy delays apoptosis in response to chemotherapy and its
inhibition increases drug-induced apoptosis in human cancer cell
lines. However, anti-tumor agents in some conditions, such as at
high doses, appear to augment autophagic cell death (14).
The sirtuins (SIRT), a family of class III histone
deacetylase enzymes, are highly conserved mammalian homologues of
the yeast Sir2 gene. They have been identified to have an important
role in cancer biology, by acting as tumor promoters and tumor
suppressors in different cancer types (15). The sirtuin super family of proteins
has seven members (SIRT1-7) and SIRT1 has been identified to
modulate autophagy in gastric cancer (16). Previous studies have also
implicated other family members, including SIRT2, SIRT3, SIRT5 and
SIRT6 in autophagy regulation (17–20).
The present study aimed to determine the molecular
mechanism of 5-FU treatment in gastric cancer and to evaluate if a
combinatory therapeutic approach, including the use of autophagy
inhibitors may enhance the therapeutic efficacy of 5-FU against
gastric cancer.
Materials and methods
Chemical reagents and antibodies
5-FU was purchased from Xudong Haipu Pharmaceutics
(Shanghai, China), 3-methyladenine (3MA), sirtinol, rapamycin and
dimethyl sulfoxide (DMSO) were purchased from Sigma-Aldrich; Merck
Millipore (Darmstadt, Germany). The antibodies purchased were:
Microtubule-associated protein 1 light chain 3 (LC3; L7543;
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), which recognizes
human LC3-I and LC3-II by immunoblotting (18 kDa and 16 kDa
respectively), p70S6 kinase (9202) and Phospho-p70 S6 kinase
(Thr389; 9205; Cell Signaling Technology, Inc., Danvers, MA, USA),
Beclin 1 (sc-10086) and β-actin (sc-81178; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA). The GFP-LC3 plasmid was
provided by Dr Mengqiang Li (Peking University School of Medicine,
Peking, China).
Culturing of gastric cell lines
BGC-823 and AGS cells were purchased from American
Type Culture Collection (Manassas, VA, USA) and cultured in
RPMI-1640 medium, supplemented with 10% (v/v) fetal bovine serum,
100 kIU/l penicillin and 100 mg/l streptomycin (Invitrogen; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) at 37°C in a humidified
atmosphere incubator with 5% CO2. Cells were
sub-cultured every two days with a split ratio of 1:2.
To investigate the role of sirtuins in autophagy,
sirtinols was co-treated with 5-FU. The BGC-823 cells were either
treated individually or together with 5-FU, 3-MA, or sirtinol in
the following six groups: i) untreated (Ctr); ii) 5 mM 3MA (3MA);
iii) 40 µg/ml 5-FU (5-FU); iv) 40 µg/ml 5-FU+5 mM 3MA (5-FU+3MA);
v) 40 µg/ml of 5-FU+10 µM sirtinol (5-FU+sirtinol); and vi) 40
µg/ml of 5-FU+10 µM sirtinol+5 mM 3MA (5-FU+sirtinol+3MA).
MTT assay
Cells were seeded into 96-well flat bottom
microtiter plates, at a density of 104-105
cells/well in 100 µl culture medium. The seeded cells were
incubated for 24–48 h with 5-FU at concentrations of 5, 10, 25, 50,
100, and 200 µg/ml. Next, 100 µl MTT reagent (5 mg/ml,
Sigma-Aldrich; Merck KGaA) was added to each well, and the cells
were further incubated for 4 h at 37°C in a CO2
incubator. Finally, the crystals were dissolved in 100 µl DMSO, by
pipetting up and down, and the absorbance was quantified at 570 nm
in a microtiter plate reader (BD Biosciences, Franklin Lakes, NJ,
USA).
Confocal microscopy
In order to quantify the green fluorescent protein
(GFP)-microtubule-associated protein 1A/1B-light chain 3 (LC3)
puncta formation in cells treated with 5-FU and untreated cells
were used as controls, the cells at ~60% confluence were
transfected with GFP-LC3 cDNA using Lipofectamine 2000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). The cells were
exposed to treatment for 48, 24 h after the transfection.
Subsequently, LC3 puncta formation was assessed under confocal
microscopy (Olympus Corporation, Tokyo, Japan) and the distribution
was quantitatively evaluated using ImageJ version 1.48 (National
Institutes of Health, Bethesda, MD, USA). The percentage of GFP-LC3
puncta distribution was calculated from 5 non-overlapping fields
selected at random (magnification, ×600) and the statistical
significance was evaluated from three repeated independent
experiments.
Transmission electron microscopy
The cells were harvested by trypsin digestion
(Sigma-Aldrich; Merck KGaA) and washed twice with PBS before fixing
with ice-cold glutaraldehyde (3% in 0.1 M cacodylate buffer, pH
7.4; Sigma-Aldrich; Merck KGaA) for 30 min. After washing again
with PBS, the cells were post-fixed in OsO4 at room
temperature for 1 h and embedded in Epon resin overnight at room
temperature (Sigma-Aldrich; Merck KGaA). The 0.1-mm sections were
stained at room temperature for 10 min with uranyl acetate/lead
citrate (Fluka Chemie AG, Buchs, Switzerland) solution and then
observed under a JEM1230 electron microscope (JEOL, Ltd., Tokyo,
Japan).
Flow cytometry
After trypsinization, the cells were washed with PBS
twice, and then suspended (5×105 cells/500 µl) in the
binding buffer, prior to incubation with Annexin V-fluorescein
isothiocyanate (FITC) and propidium iodide (PI) solutions,
according to the manufacturer's protocol of Annexin V-FITC
apoptosis detection kit (Biovision Inc., Milpitas, CA, USA). The
cells were analyzed using a flow cytometer FACSAria (BD
Biosciences, Franklin Lakes, NJ, USA).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA (1 µg) was extracted by the TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) and then
reverse-transcribed into cDNA using a reverse transcription kit
(Invitrogen; Thermo Fisher Scientific, Inc.). Subsequently, the
cDNA was amplified using the following parameters: Pre-denaturation
at 95°C for 10 min, 40 cycles of 95°C for 15 sec and 60°C for 1
min, using an Applied Biosystems 7500 Real-Time PCR System (Thermo
Fisher Scientific, Inc.) with SYBR Green PCR Master Mix (Applied
Biosystems; Thermo Fisher Scientific, Inc.). Primers used for the
qPCR are presented in Table I. The
data was normalized against housekeeping gene GAPDH, and analysis
was performed using the 2−∆∆Cq method (21), where -∆Cq=∆Cq (treated sample)-∆Cq
(untreated), and ∆Cq=Cq (target gene)-Cq (housekeeping gene).
Changes in gene expression, reported as fold-change of relative
mRNA expression are expressed as the target/reference ratio of the
sample divided by the target/reference ratio of the control and the
experiments were repeated three times.
 | Table I.List of target primer pairs used for
gene specific amplification in quantitative polymerase chain
reaction. |
Table I.
List of target primer pairs used for
gene specific amplification in quantitative polymerase chain
reaction.
| Gene | Forward
(5′-3′) | Reverse
(5′-3′) |
|---|
| SIRT1 |
ACTTGTACGACGAAGACGAC |
GATCTGTGCCAATCATAAGA |
| SIRT2 |
GCAGGAGGCTCAGGACTCA |
TGACTCTGCGACAGCGTTC |
| SIRT3 |
CCTCCAGCAGTACGATCTCC |
GGTTCCATGAGCTTCAACCA |
| SIRT4 |
GGGTTATTTGTGCCAGCAAG |
AAGTTTCTCGCCCAGTACCG |
| SIRT5 |
GTTCAAGTATGGCAGATTTTCG |
CTCCAATAACCTCCAGCTCC |
| SIRT6 |
CCAAGTTCGACACCACCTTT |
CGGACGTACTGCGTCTTACA |
| SIRT7 |
ATCAGCACGGCAGCGTCTAT |
AGGTGGAGCCCGTCACAG |
| GAPDH |
ACGGATTTGGTCGTATTGGG |
TGATTTTGGAGGGATCTCGC |
RNA interference
The cells were seeded at 40% confluence in each well
(1.5×104/well) of 96-well plates overnight. The next
day, the cells were transfected with Beclin-1 siRNA or control
siRNA duplex (Santa Cruz Biotechnology, Inc.) using Lipofectamine
2000 reagent. The knockdown of the specific gene was confirmed
using western blot analysis 48 h after transfection.
Western blot analysis
5-FU-treated and control cells were lysed by
incubation in a lysis buffer (10 mM HEPES pH 7.4, 0.15 M NaCl, 1 mM
EDTA, 1 mM EGTA, 1% Triton X-100, 0.5% NP-40, with freshly added
proteinase inhibitor cocktail) on ice for 30 min. The pelleted cell
lysates were centrifuged at 20,817 × g for 15 min at 4°C and the
supernatant was collected. The protein concentration was quantified
using BCA assay reagent (Pierce; Thermo Fisher Scientific, Inc.)
and subsequently proteins were separated on 10% SDS-PAGE gel. This
was followed by their transfer to nitrocellulose membranes. The
membranes were blocked with 5% non-fat dry milk, the membranes were
incubated with the primary antibodies diluted at 1:1,000 overnight
at 4°C. The following day, the membranes were incubated with
horseradish peroxidase-conjugated goat anti-rat (7077; Cell
Signaling Technology, Inc.) at 1:5,000 dilution for 2 h at room
temperature. The protein bands were visualized on X-ray film using
an enhanced chemiluminescence system (Pierce; Thermo Fisher
Scientific, Inc.). To determine the linear range of the
chemiluminescent signals, X-ray films were quantitatively analyzed
via densitometry analysis by ImageJ software version 1.48 (National
Institutes of Health, Bethesda, MD, USA).
Statistical analysis
All statistical analyses were performed using SPSS
version 17.0 (SPSS, Inc., Chicago, IL, USA). Data were expressed as
mean ± standard error of the mean. Significant differences between
groups were analyzed using Student-Newman-Keuls analysis.
Non-normal distribution data were presented as mean without an
error bar and analyzed using the Mann-Whitney test to determine the
significance of the difference between two groups. P<0.05 was
considered to indicate statistically significant difference.
Results
5-FU inhibits gastric cancer cell
proliferation and induced apoptosis
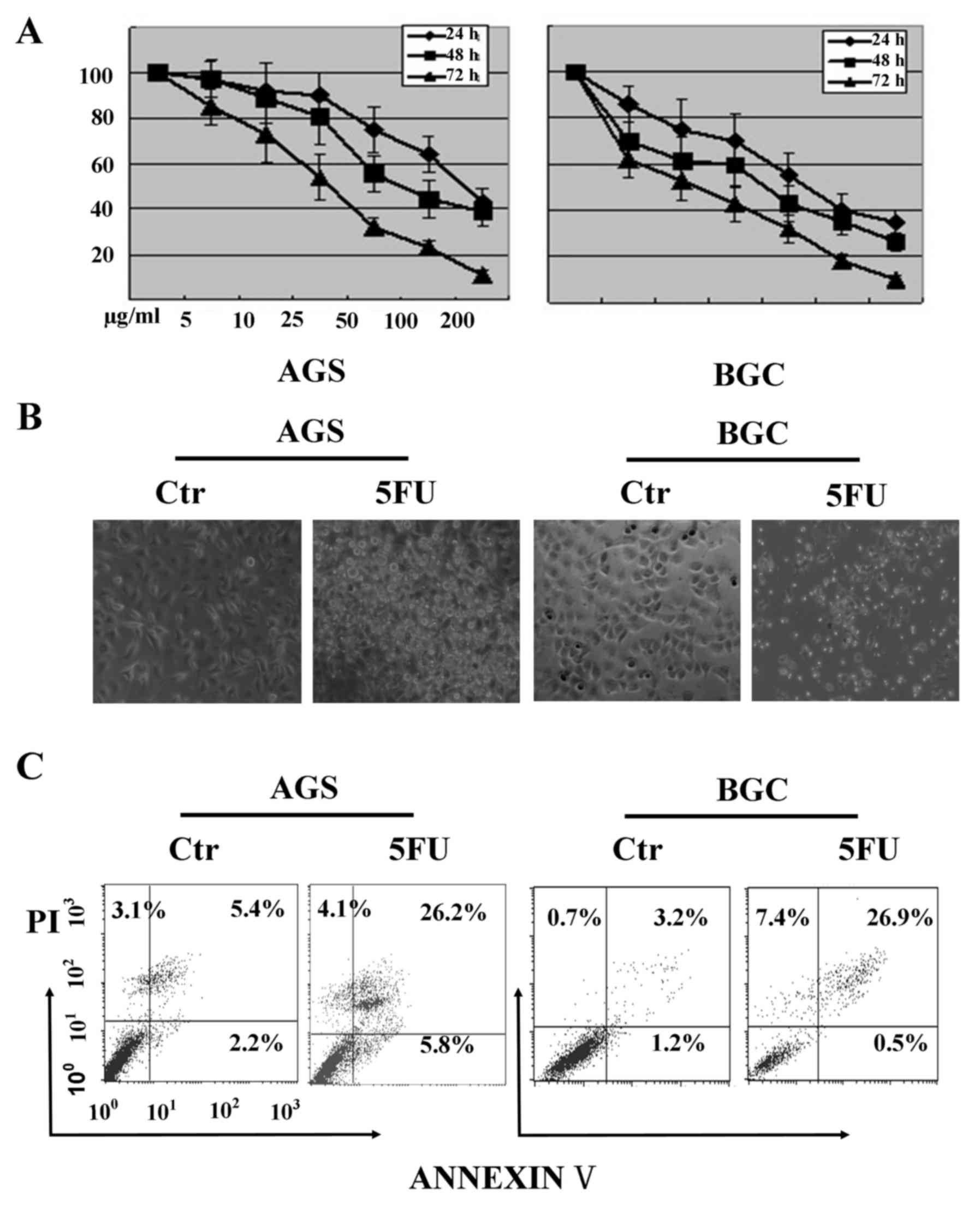
The MTT assay indicated that 5-FU treatment
inhibited BGC-823 and AGS cell viability in a dose- and
time-dependent manner. The 5-FU concentrations from 5 to 200 µg/ml
generated varied cellular cytotoxicity profiles after treatment
duration of 24, 48 and 72 h. An IC50 of 40 µg/ml after
48 h 5-FU treatment in BGC-823 cells and 60 µg/ml in AGS cells was
observed (Fig. 1A). Therefore,
these two 5-FU concentrations (40 µg/ml in BGC-823 and 60 µg/ml in
AGS cells) were used for subsequent experiments.
The microscopic observation of cells after 5-FU
treatment revealed cells to be round, lifting off the culture
surface, and having multiple membrane blebs (Fig. 1B), indicating the probable onset of
apoptosis and necrosis. To verify these observations, the cells
were analyzed using flow cytometry to detect cell death, using an
Annexin V-FITC/PI assay. As presented in Fig. 1C, the number of Annexin V positive
cells increased significantly in AGS and BGC-823 cells,
respectively, after 5-FU treatment when compared with the untreated
group. This data indicated that 5-FU treatment promoted apoptosis
in gastric cancer cells.
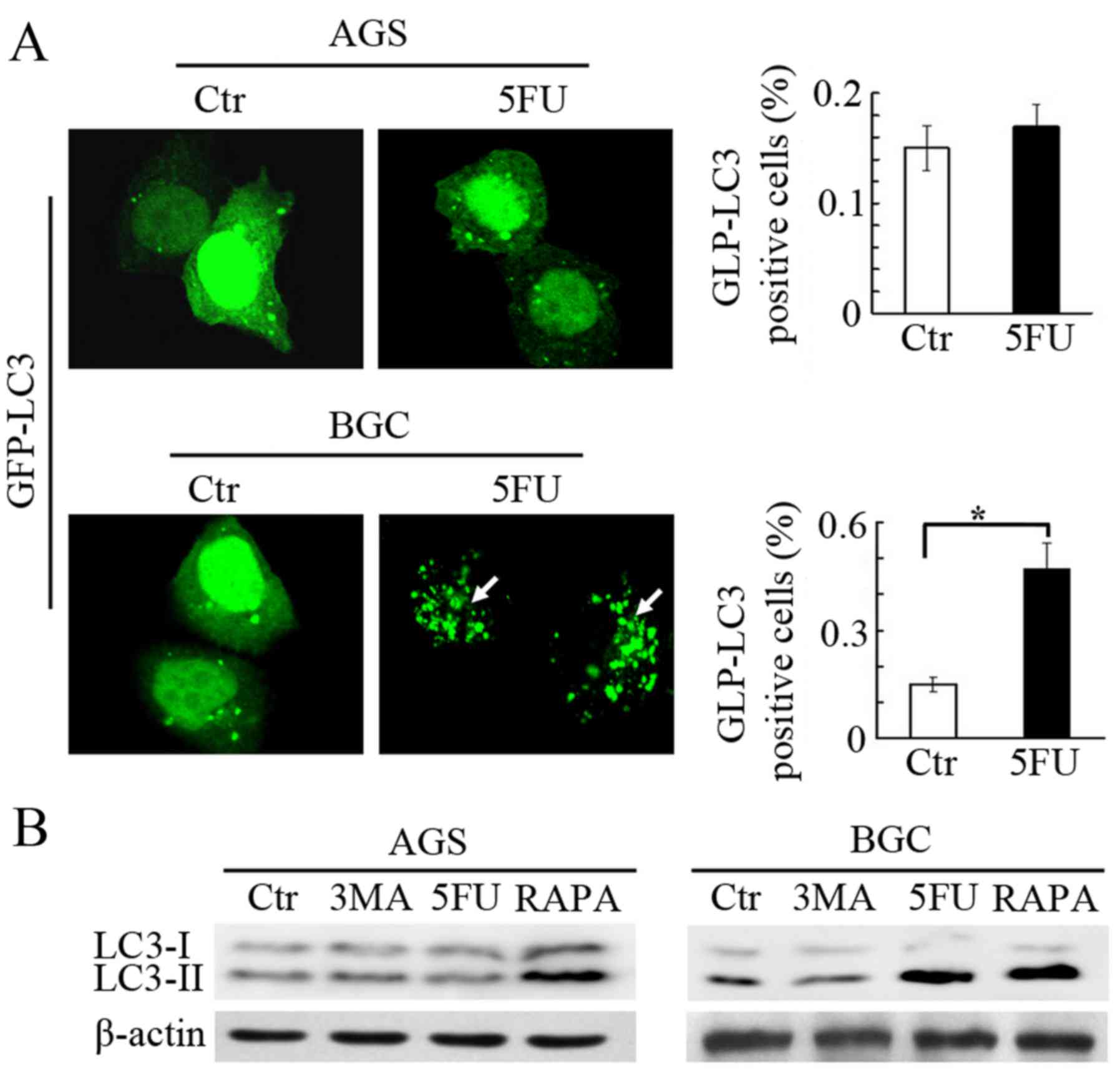
5-FU induces autophagy only in BGC-823
cells
A previous study determined that 5-FU induces
autophagosome formation in some cancer cells (22); therefore, the present study tested
this in both gastric cancer cell lines. LC3, a mammalian homologue
of the yeast Atg8 protein located in the autophagosome membrane,
acts as a specific marker of autophagy. Therefore, in order to
determine the formation of autophagosomes, both BGC-823 and AGS
cells were transfected with a GFP-LC3 plasmid. After 48 h of 5-FU
treatment, GFP-LC3 puncta were accumulated in BGC-823 cells
compared with untreated cells (Fig.
2A). Subsequently, the levels of LC3-II (16 kDa), a mature and
spliced form of LC3 that represents autophagic activity (23), were also assessed by western
blotting. It is of note that increased expression of LC3-II was
observed only in BGC-823 cells, treated with 5-FU for 48 h. This
was consistent with GFP-LC3 immunofluorescence data. Rapamycin
treatment with 10 nM concentration at room temperature for 48 h (a
notable agonist of autophagy) was used as control, upregulated
LC3-II levels in both cell lines were observed as expected, and 3MA
(a well-known inhibitor of autophagy) treatment downregulated
LC3-II levels in BGC-823 cells, and no change in AGS cells was
observed (Fig. 2B). The present
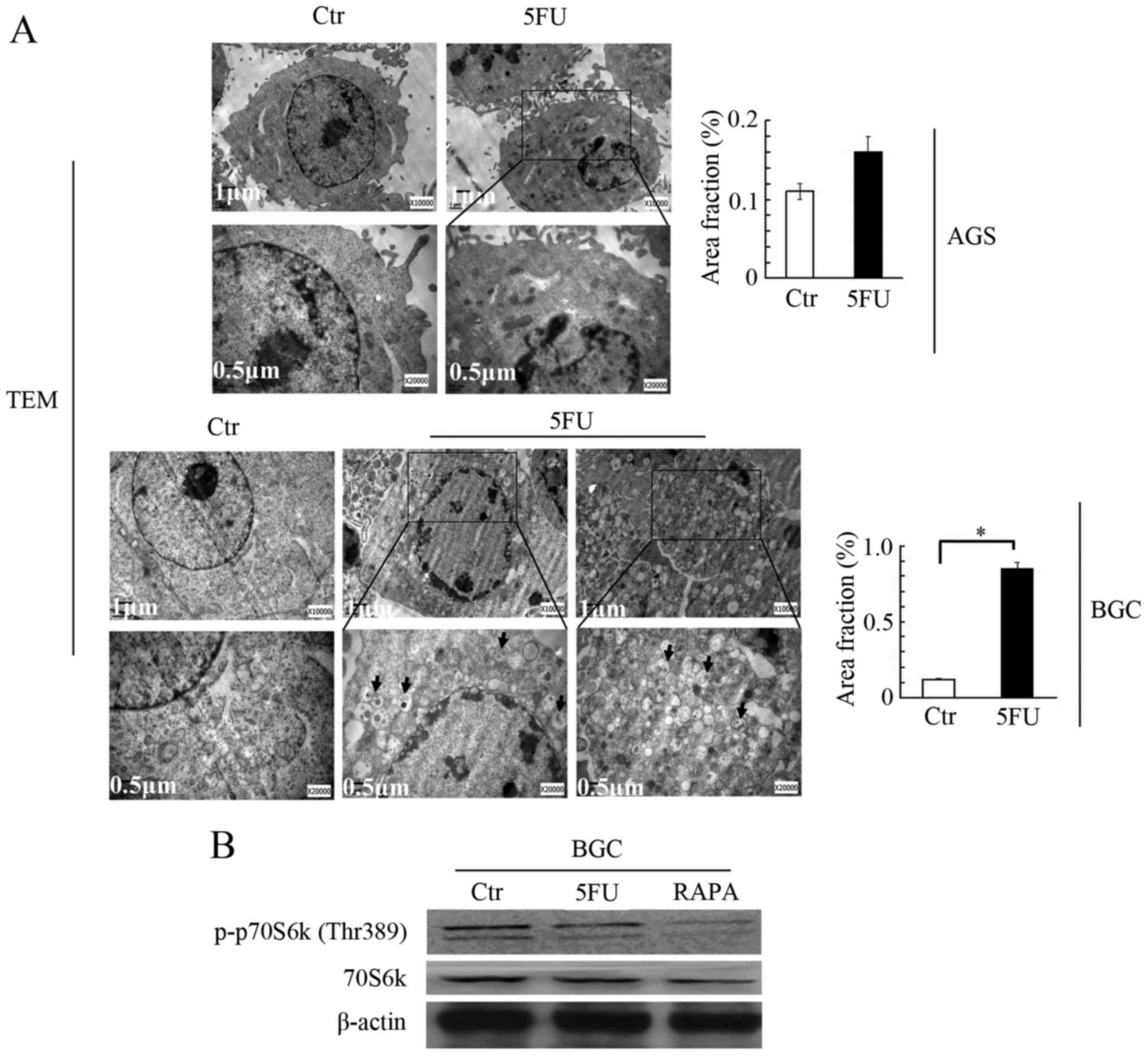
study also verified the effect of 5-FU on autophagosome formation
induction using transmission electron microscopy (TEM). The present
study observed multiple autophagosomes (a double membrane
structure) and autolysosomes (a single membrane structure) in the
cytoplasm of BGC-823 cells treated with 5-FU when compared to
untreated cells (Fig. 3A).
Additionally, the present study identified a significant difference
in the ratio of autophagosome area to cytoplasm between 5-FU
treated cells and control cells (Fig.
3A, lower panel). However, autophagosomes and autolysosomes
were rarely observed under TEM in AGS cells following 5-FU
treatment. Overall, this data indicated that 5-FU promoted
autophagosome formation in BGC-823 cells only and had no effect on
AGS cells.
5-FU reduces the phosphorylation of
p70S6 kinase in BGC-823 cells
Considering that 5-FU promoted the autophagic
process only in BGC-823 cells, and not in AGS cells, the present
study aimed to determine the molecular mechanism behind its
process. Among the various signaling pathways regulating autophagy,
the mTOR pathway inhibition is the one that has been widely
reported to promote autophagy (24). Therefore, this pathway was
monitored by analyzing the phosphorylation of downstream kinase
70S6 kinase at the Threonine 389 site. As presented in Fig. 3B, 5-FU treatment reduced the
phosphorylation at Threonine 389 in BGC-823 cells compared with
untreated cells. Rapamycin treatment was used as a positive
control. These results indicated that 5-FU-induced autophagy may be
dependent on mTOR inhibition.
3-MA augments 5-FU-induced cell death
in BGC-823 cells
Autophagy tends to execute a dual function
(promotion of cell death or cell survival) in a context-dependent
manner (25). The present study
aimed to assess the influence of the autophagy inhibitor 3-MA (5
mM) on 5-FU effects in BGC-823 cells (Fig. 4). GFP-LC3-transfected BGC-823 cells
were treated with 5-FU in the presence or absence of 3-MA, to test
autophagosome formation. As presented in Fig. 4A, the number of GFP-LC3 puncta was
significantly lower in cells treated with a combination of 5-FU and
3-MA compared with those subjected to 5-FU treatment alone.
Microscopic assessment of cells treated with 5-FU and 3-MA
presented variable cell viability, where the majority of the cells
appeared round, lifted off from the culture surface, with multiple
membrane blebs (Fig. 4C, upper
panel), indicating the possible onset of apoptosis and necrosis. In
order to verify these observations, cell death was investigated
using flow cytometry. As presented in Fig. 4C (lower panel), the number of
Annexin V and PI positive cells in the 5-FU+3MA group was 19.9%
higher compared with the 5-FU group alone. This indicated that
autophagy inhibition by 3-MA treatment increased 5-FU-induced cell
death.
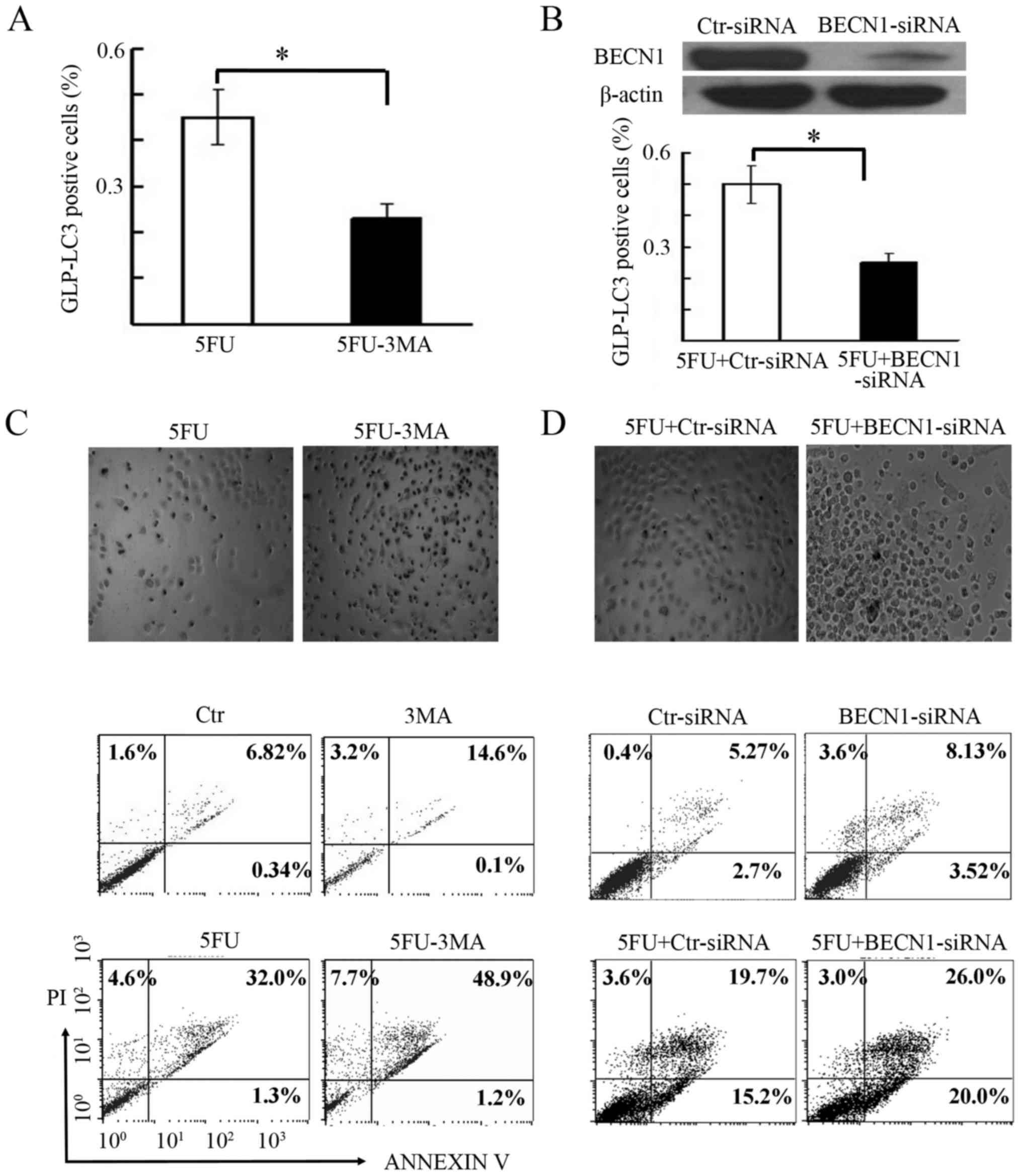 | Figure 4.Autophagy inhibition enhanced
5-FU-induced cell death. (A) Assessment of GFP-LC3 positive puncta
(%) in BGC-823 cells treated with 5-FU and 5-FU+3MA. *P<0.05.
The GFP-LC3 positive cells (%) are presented as the mean ± standard
error of the mean from three independent experiments. (B) Beclin-1
ablation by its specific siRNA in BGC-823 cells, in comparison to
control siRNA (Ctr-siRNA). β-actin expression was used as the
protein loading control. The lower panel shows the effect of
Beclin-1 ablation on GFP-LC3 positive cells, after 5-FU treatment.
*P<0.05. (C) Microscopy (×100) and flow cytometry-based
detection of apoptosis after autophagy inhibition by 3MA. The upper
panel shows a representative image of apoptotic cells obtained
through microscopy, whereas the lower panel shows the percentage of
Annexin V and/or PI positive cells, after treatment with 5-FU alone
and with 3MA co-treatment (5-FU+3MA). (D) Microscopy (×100) and
flow cytometry based detection of apoptosis after autophagy
inhibition by Beclin-1 ablation. Upper panel shows a representative
image of apoptotic cells obtained through microscopy, whereas the
lower panel shows the percentage of Annexin V and/or PI positive
cells, after treatment with 5-FU and Beclin-1 siRNA
(5-FU+BECN1-siRNA) or control siRNA (5-FU+CtrsiRNA). Ctr, control;
5-FU, 5-fluorouracil; BECN1-siRNA, Beclin-1 small interfering RNA;
3MA, 3-methyladenine; PI, propidium iodide, LC3,
microtubule-associated protein 1A/1B-light chain 3; GFP, green
fluorescent protein. |
Inhibition of autophagy by Beclin-1
siRNA enhances 5-FU-induced cell death
To independently confirm the induction of cell death
by 5-FU following autophagy inhibition, the present study blocked
Beclin-1 expression, an important component of the autophagy
pathway (26–27). The BGC-823 cells transfected with
Beclin-1 siRNA or scrambled siRNA for 24 h were later treated with
5-FU for another 48 h. As presented in Fig. 4B (upper panel), the Beclin-1
expression was ablated by 90% with its specific siRNA duplex
compared to scrambled siRNA. The effect of Beclin-1 siRNA on
autophagy inhibition was confirmed by monitoring GFP-LC3 puncta
(Fig. 4B, lower panel). Further
examination of siRNA-transfected cells by microscopy and flow
cytometry confirmed these findings. The cells exhibited rounded
morphology, were lifted off the culture surface and had multiple
membrane blebs, after treatment with 5-FU in combination with
Beclin-1 ablation (Fig. 4D, upper
panel). Flow cytometry analysis also revealed a higher percentage
(49%) of Annexin V and PI positive cells after 5-FU treatment and
Beclin-1 ablation than control ablation (38.5%; Fig. 4D, lower panel). This confirmed the
observation that 5-FU-induced gastric cancer cell death was
enhanced by autophagy inhibition.
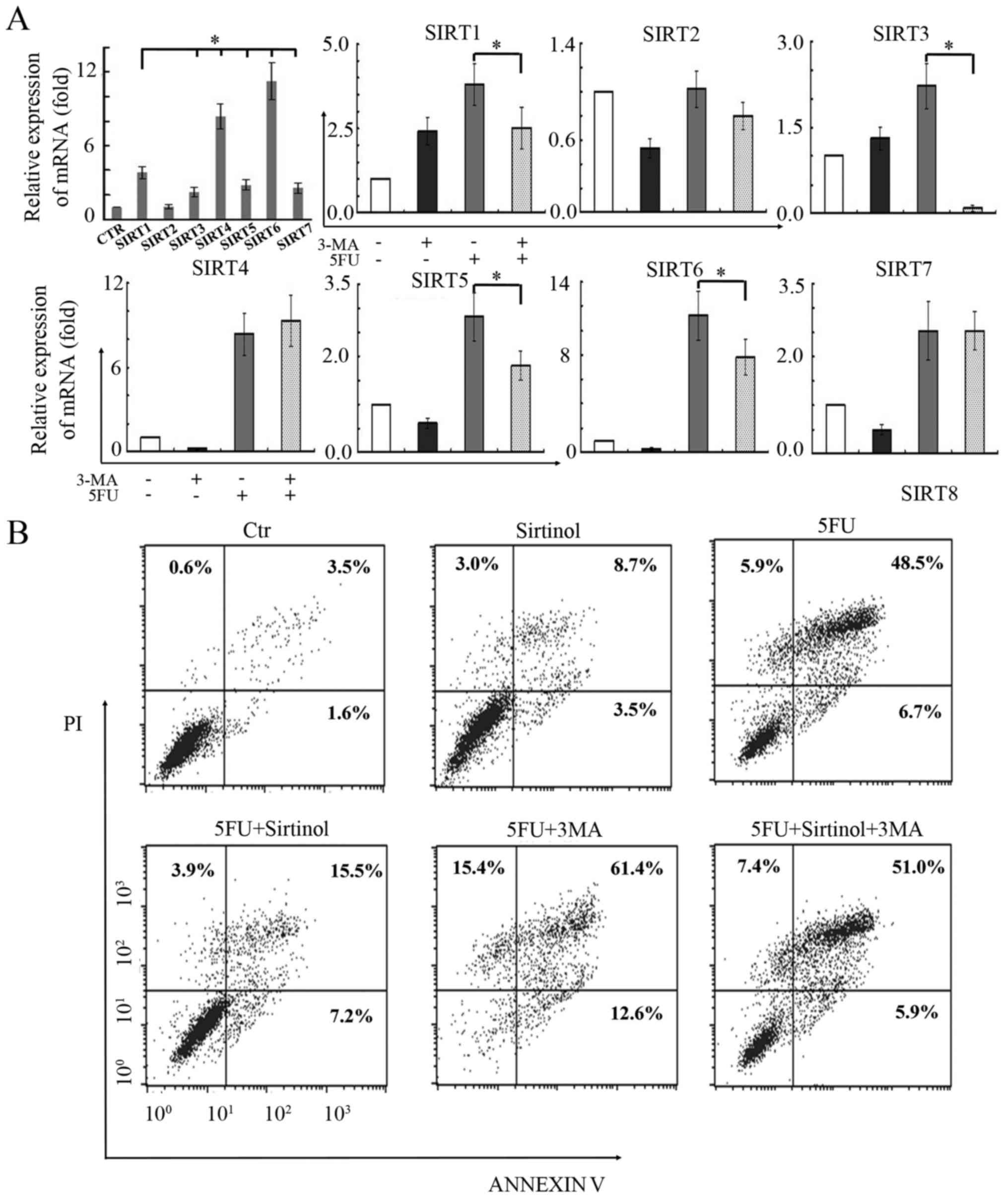
Sirtuins are involved in 5-FU-induced
autophagy-mediated cell death in BGC-823 cells
The involvement of the sirtuin family of proteins
was determined in 5-FU-induced autophagy induction in gastric
carcinoma cells. The RT-qPCR data revealed that the mRNA levels of
six sirtuins, including SIRT1 and SIRT3-7, were upregulated after
5-FU treatment in BGC-823 cells (Fig.
5A, top left). It is of note that the levels of SIRT4 and SIRT6
mRNA had the highest expression compared with the remaining
sirtuins. Subsequently, the present study assessed the sirtuin
levels in BGC-823 cells treated with 5-FU in the presence of 3-MA
(5 mM). RT-qPCR analysis indicated that increases in SIRT1, SIRT3,
SIRT5 and SIRT6 mRNA expression induced by 5-FU was attenuated by
3-MA (Fig. 5A). These findings
indicate that four sirtuins, SIRT1, SIRT3, SIRT5, and SIRT6 may be
involved in 5-FU-induced autophagy in BGC-823 cells. Additionally,
the present study tested the effect of sirutin inhibition, using
sirtinol (a specific inhibitor of SIRT), on 5-FU-induced cell
death. The BGC-823 cells were co-treated with 5-FU, 3-MA and
sirtinol. Following these treatments, the cells were analyzed by
flow cytometry for Annexin V-FITC/PI staining to detect cell death.
The findings of the present study demonstrated that sirtinol
treatment abrogated both 5-FU-induced and 5-FU+3MA-induced cell
death (Fig. 5B). The percentage of
cell death decreased from 61.1% in the 5-FU treatment group to
26.6% in the 5-FU+sirtinol treatment group, whereas it decreased
from 89.4% in the 5-FU+3MA treatment group to 64.3% in the
5-FU+sirtinol+3MA treatment group. Therefore, the protective role
of SIRT proteins in 5-FU-induced cell death is evident.
Discussion
Despite reports of 5-FU inducing apoptosis and
inhibiting cell proliferation in gastric cancer cells (28), its exact molecular mechanism
remains to be elucidated. Consistent with these previously
published studies, the present study also observed that 5-FU
promoted gastric cancer cell apoptosis in BGC-823 and AGS cell
lines and inhibited their proliferation. Microscopic observation of
cells following 5-FU treatment revealed rounded morphology, cells
starting to lift off the culture surface and exhibited multiple
membrane blebs. This is indicative of very early apoptosis and
necrotic behavior and then confirmed by the Annexin V-FITC/PI
assay. In terms of the molecular mechanisms of many
chemotherapeutic drugs, it has been previously suggested that they
induce autophagy in cancer cell lines and animal models (25). 5-FU induced autophagy, was not
observed under conventional microscopy, GFP-LC3 fluorescence
signals (indicative of puncta formation), LC3 immunoblotting, and
TEM were used instead (29).
Similarly, the present study determined that 5-FU induced
autophagosome formation in the BGC-823 cell line, which may be
dependent on mTOR inhibition. Conversely, autophagosome formation
through 5-FU treatment was not observed in the AGS cell line. This
discrepancy may be due to the cellular origins of these two
different cell types. AGS cells were derived from
well-differentiated gastric adenocarcinoma, whereas BGC-823 cells
originated from poorly-differentiated gastric adenocarcinoma. The
induction of autophagic processes following 5-FU chemotherapy in
poorly-differentiated gastric adenocarcinoma cells may be a
defensive response under stress conditions, whereas in
well-differentiated gastric adenocarcinoma cells, the lack of
autophagosome formation, usually as a guardian mechanism, may
promote tumor cell apoptosis under 5-FU treatment. The contribution
of autophagy to cell death or survival is controversial; however,
in the current experiments, autophagy inhibition was determined to
have enhanced 5-FU-mediated apoptosis. This suggests that
upregulation of autophagy by 5-FU does not support cell death.
Therefore, alteration in the phenotypic changes in BGC-823 cells
treated with 5-FU may be directly attributed to modulation of
apoptosis, as confirmed by the Annexin V-FITC/PI assay.
It is well-known that autophagy has a dual role in
cancer cells after chemotherapy. In some cases, it promotes
survival, whereas in other instances it facilitates cell death
(12). It is of note that in the
current study a specific autophagy inhibitor, 3-MA, enhanced
5-FU-induced apoptosis in BGC-823 cells. Additionally, it was also
confirmed that the augmentation of cell death by 5-FU, after
inhibiting autophagy through Beclin-1 ablation in BGC-823 cells.
These observations indicated that autophagy induction in BGC-823
cells delayed cell death; however, inhibition of this pathway
enhanced the 5-FU mediated cell death. Similar observations have
been made in our previous studies, where matrine-treated or
etoposide-treated hepatoma cells exhibited a delayed cell death
response due to autophagosome formation; however, autophagy
suppression augmented the cell death (30–32).
Autophagy is stimulated by stress conditions like nutrition
deprivation and starvation and is involved in the removal of
long-lived proteins and damaged organelles, in addition to
providing amino acids for maintaining the metabolism, essential for
survival in poor conditions (11).
Autophagy generally provides cancer cells with a rescue mechanism
(33,34). It has been previously reported that
in human skin squamous carcinoma cells, autophagy inhibition with
3-MA enhanced 5-FU mediated apoptosis (35). In another previous study, in the
absence of caspase activation, GAPDH-enhanced autophagy protected
cells from caspase-independent cell death (36). In apoptosis-defective cells,
autophagy has been identified to promote survival after metabolic
stress and autophagy inhibition induced necrotic cell death
(37). It is also possible that
autophagy sequesters and degrades proteins or organelles, such as
mitochondria damaged by 5-FU treatment and releases amino acids,
which in turn maintains gastric cancer cell viability.
Furthermore, when investigating the mechanistic role
of 5-FU in gastric cancer cells the present study determined that
SIRT1 and SIRT3-7 were upregulated, but not SIRT2, in 5-FU-treated
BGC-823 cells. It is of note that 3MA co-treatment with 5-FU
reduced the expression of four sirtuins, SIRT1, SIRT3, SIRT5 and
SIRT6, suggesting that sirtuins were important in the 5-FU-induced
autophagy in gastric cancer cells. SIRT3 and SIRT5 proteins were
primarily localized in the mitochondria, whereas SIRT1 and SIRT6
were in the nucleus. Previous studies have established the role of
these four sirtuins in autophagy induction and formation (38,39).
SIRT1 has predominantly been implicated in autophagosome formation
(16), with SIRT3, SIRT5, and
SIRT6 more recently revealed to be associated with autophagy
(18,19,40).
The SIRT1 mechanism of modulating autophagy occurs via
deacetylation of autophagy-related gene 5 (Atg5), Beclin-1, Atg7,
Atg8, and other autophagy mediators, which affects autophagosome
induction, maturation, and tumor growth (16,41,42).
Sirtuins appear to promote cellular proliferation and may also be
involved in 5-FU-induced autophagy, epithelial-mesenchymal
transition, cell invasion and chemoresistance in gastric cancer
cells (16). It is of note that
the present study identified SIRT inhibition by sirtinol reduced
5-FU-induced apoptotic cell death, suggesting that SIRT1 inhibition
may protect against cell death and function as a cell survival
mechanism under chemotherapy. However, this observation is
preliminary and further studies are warranted, for example using
SIRT siRNA, to elucidate the specific role of SIRT proteins in
5-FU-induced autophagy and cancer cell death.
In summary, the present study demonstrated that 5-FU
promoted apoptosis and autophagy in gastric cancer cells. However,
autophagy inhibition enhanced 5-FU-induced apoptosis. Furthermore,
the present study noted the probable involvement of four sirtuin
proteins, SIRT1, SIRT3, SIRT5, and SIRT6, in 5-FU mediated
autophagy induction. In conclusion, the current study revealed that
manipulation of autophagy for therapeutic purposes may be useful in
order to overcome chemoresistance and the use of autophagy
inhibitors along with 5-FU may enhance its therapeutic efficacy in
gastric carcinoma.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 81503437) and the
Natural Science Foundation of Jiangxi Province (grant no.
20161BAB205246).
References
|
1
|
Xiao S and Zhou L: Gastric cancer:
Metabolic and metabolomics perspectives (Review). Int J Oncol.
51:5–17. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hartgrink HH, Jansen EP, van Grieken NC
and van de Velde CJ: Gastric cancer. Lancet. 374:477–490. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Greenspan EM: Adjuvant chemotherapy for
stomach cancer. Lancet. 1:1459–1460. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
MacDonald JS, Schein PS, Woolley PV,
Smythe T, Ueno W, Hoth D, Smith F, Boiron M, Gisselbrecht C, Brunet
R and Lagarde C: 5-Fluorouracil, doxorubicin, and mitomycin (FAM)
combination chemotherapy for advanced gastric cancer. Ann Intern
Med. 93:533–536. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Longley DB, Harkin DP and Johnston PG:
5-fluorouracil: Mechanisms of action and clinical strategies. Nat
Rev Cancer. 3:330–338. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wohlhueter RM, McIvor RS and Plagemann PG:
Facilitated transport of uracil and 5-fluorouracil, and permeation
of orotic acid into cultured mammalian cells. J Cell Physiol.
104:309–319. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rani I, Sharma B, Kumar S, Kaur S and
Agnihotri N: Apoptosis mediated chemosensitization of tumor cells
to 5-fluorouracil on supplementation of fish oil in experimental
colon carcinoma. Tumour Biol. 39:10104283176950192017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu J, Hu D and Zhang R: Depletion of Bmi-1
enhances 5-fluorouracil-induced apoptosis and autophagy in
hepatocellular carcinoma cells. Oncol Lett. 4:723–726. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yao CW, Kang KA, Piao MJ, Ryu YS, Fernando
PMDJ, Oh MC, Park JE, Shilnikova K, Na SY, Jeong SU, et al: Reduced
autophagy in 5-fluorouracil resistant colon cancer cells. Biomol
Ther. 25:315–320. 2017. View Article : Google Scholar
|
|
10
|
Suzuki R, Kang Y, Li X, Roife D, Zhang R
and Fleming JB: Genistein potentiates the antitumor effect of
5-Fluorouracil by inducing apoptosis and autophagy in human
pancreatic cancer cells. Anticancer Res. 34:4685–4692.
2014.PubMed/NCBI
|
|
11
|
Mizushima N: Autophagy: Process and
function. Genes Dev. 21:2861–2873. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mathew R, Karantza-Wadsworth V and White
E: Role of autophagy in cancer. Nat Rev Cancer. 7:961–967. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Singh SS, Vats S, Chia AY, Tan TZ, Deng S,
Ong MS, Arfuso F, Yap CT, Goh BC, Sethi G, et al: Dual role of
autophagy in hallmarks of cancer. Oncogene. 2017.Doi:
10.1038/s41388-017-0046-6.
|
|
14
|
Chen HY and White E: Role of autophagy in
cancer prevention. Cancer Prev Res. 4:973–983. 2011. View Article : Google Scholar
|
|
15
|
Guarente L: Introduction: Sirtuins in
aging and diseases. Methods Mol Biol. 1077:3–10. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Qiu G, Li X, Wei C, Che X, He S, Lu J,
Wang S, Pang K and Fan L: The prognostic role of SIRT1-autophagy
axis in gastric cancer. Dis Markers. 2016:68694152016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gal J, Bang Y and Choi HJ: SIRT2
interferes with autophagy-mediated degradation of protein
aggregates in neuronal cells under proteasome inhibition. Neurochem
Int. 61:992–1000. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cho CS, Lombard DB and Lee JH: SIRT3 as a
regulator of hepatic autophagy. Hepatology. 66:700–702. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Polletta L, Vernucci E, Carnevale I,
Arcangeli T, Rotili D, Palmerio S, Steegborn C, Nowak T,
Schutkowski M, Pellegrini L, et al: SIRT5 regulation of
ammonia-induced autophagy and mitophagy. Autophagy. 11:253–270.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Takasaka N, Araya J, Hara H, Ito S,
Kobayashi K, Kurita Y, Wakui H, Yoshii Y, Yumino Y, Fujii S, et al:
Autophagy induction by SIRT6 through attenuation of insulin-like
growth factor signaling is involved in the regulation of human
bronchial epithelial cell senescence. J Immunol. 192:958–968. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tang JC, Feng YL, Liang X and Cai XJ:
Autophagy in 5-fluorouracil therapy in gastrointestinal cancer:
Trends and challenges. Chin Med J. 129:456–463. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kimura S, Fujita N, Noda T and Yoshimori
T: Monitoring autophagy in mammalian cultured cells through the
dynamics of LC3. Methods Enzymol. 452:1–12. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hu Y, Liu J, Wu YF, Lou J, Mao YY, Shen HH
and Chen ZH: mTOR and autophagy in regulation of acute lung injury:
A review and perspective. Microbes Infect. 16:727–734. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Petibone DM, Majeed W and Casciano DA:
Autophagy function and its relationship to pathology, clinical
applications, drug metabolism and toxicity. J Appl Toxicol.
37:23–37. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liang XH, Jackson S, Seaman M, Brown K,
Kempkes B, Hibshoosh H and Levine B: Induction of autophagy and
inhibition of tumorigenesis by beclin 1. Nature. 402:672–676. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hu F, Guo XL, Zhang SS, Zhao QD, Li R, Xu
Q and Wei LX: Suppression of p53 potentiates chemosensitivity in
nutrient-deprived cholangiocarcinoma cells via inhibition of
autophagy. Oncol Lett. 14:1959–1966. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Inada T, Ichikawa A, Igarashi S, Kubota T
and Ogata Y: Effect of preoperative 5-fluorouracil on apoptosis of
advanced gastric cancer. J Surg Oncol. 65:106–110. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Klionsky DJ, Abdelmohsen K, Abe A, Abedin
MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD,
Adeli K, et al: Guidelines for the use and interpretation of assays
for monitoring autophagy (3rd edition). Autophagy. 12:1–222. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang L, Gao C, Yao S and Xie B: Blocking
autophagic flux enhances matrine-induced apoptosis in human
hepatoma cells. Int J Mol Sci. 14:23212–23230. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xie BS, Zhao HC, Yao SK, Zhuo DX, Jin B,
Lv DC, Wu CL, Ma DL, Gao C, Shu XM and Ai ZL: Autophagy inhibition
enhances etoposide-induced cell death in human hepatoma G2 cells.
Int J Mol Med. 27:599–606. 2011.PubMed/NCBI
|
|
32
|
Xie SB, He XX and Yao SK: Matrine-induced
autophagy regulated by p53 through AMP-activated protein kinase in
human hepatoma cells. Int J Oncol. 47:517–526. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tylichová Z, Straková N, Vondráček J,
Vaculová AH, Kozubík A and Hofmanová J: Activation of autophagy and
PPARγ protect colon cancer cells against apoptosis induced by
interactive effects of butyrate and DHA in a cell type-dependent
manner: The role of cell differentiation. J Nutr Biochem.
39:145–155. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu Z, Xu J, He J, Liu H, Lin P, Wan X,
Navone NM, Tong Q, Kwak LW, Orlowski RZ and Yang J: Mature
adipocytes in bone marrow protect myeloma cells against
chemotherapy through autophagy activation. Oncotarget.
6:34329–34341. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang L, Zhang J, Chen L and Wang J:
Autophagy in human skin squamous cell carcinoma: Inhibition by 3-MA
enhances the effect of 5-FU-induced chemotherapy sensitivity. Oncol
Rep. 34:3147–3155. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Colell A, Ricci JE, Tait S, Milasta S,
Maurer U, Bouchier-Hayes L, Fitzgerald P, Guio-Carrion A,
Waterhouse NJ, Li CW, et al: GAPDH and autophagy preserve survival
after apoptotic cytochrome c release in the absence of
caspase activation. Cell. 129:983–997. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Degenhardt K, Mathew R, Beaudoin B, Bray
K, Anderson D, Chen G, Mukherjee C, Shi Y, Gélinas C, Fan Y, et al:
Autophagy promotes tumor cell survival and restricts necrosis,
inflammation, and tumorigenesis. Cancer Cell. 10:51–64. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Dai Y and Faller DV: Transcription
regulation by class III histone deacetylases (HDACs)-sirtuins.
Transl Oncogenomics. 3:53–65. 2008.PubMed/NCBI
|
|
39
|
Chalkiadaki A and Guarente L: The
multifaceted functions of sirtuins in cancer. Nat Rev Cancer.
15:608–624. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
40
|
He J, Zhang G, Pang Q, Yu C, Xiong J, Zhu
J and Chen F: SIRT6 reduces macrophage foam cell formation by
inducing autophagy and cholesterol efflux under ox-LDL condition.
FEBS J. 284:1324–1337. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Qiu G, Li X, Che X, Wei C, He S, Lu J, Jia
Z, Pang K and Fan L: SIRT1 is a regulator of autophagy:
Implications in gastric cancer progression and treatment. FEBS
Lett. 589:2034–2042. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sun T, Li X, Zhang P, Chen WD, Zhang HL,
Li DD, Deng R, Qian XJ, Jiao L, Ji J, et al: Acetylation of Beclin
1 inhibits autophagosome maturation and promotes tumour growth. Nat
Commun. 6:72152015. View Article : Google Scholar : PubMed/NCBI
|