Introduction
Heart failure (HF), commonly the end-stage of most
cardiovascular diseases, is a complex inflammatory neuroendocrine
syndrome not merely an exclusive problem of low cardiac output
(1,2). People older than 65 years with
chronic HF (CHF) represent the most with hospitalisation, and
approximately 50% of patients are dead within 5 years of the HF
diagnosis (3,4). Despite many advances in diagnosis and
treatment approaches, the condition continue to portend a high
morbidity and mortality (5). Early
identification of patients at high risk and accurate prognostic
evaluation is critical for physicians and patients. Although many
models of risk stratification and mortality prediction have been
developed (6), simple,
user-friendly and readily available methods are urgently needed in
the clinic. In this regard, the use of hematological factors is
becoming increasingly popular as a new approach.
Neutrophil-to-lymphocyte ratio (NLR), a new addition
to the list of systemic inflammatory biomarkers, has been
repeatedly reported as a prognostic marker and a risk
stratification index for various cardiovascular diseases including
HF by numerous studies (5,6). Like most commonly used markers for
cardiovascular diseases such as C-reactive protein (CRP) and
NT-proBNP levels, NLR has become emergingly popular and acceptable
as a biomarker for HF in clinical practice (7). NLR has been frequently found to have
an significant role in risk-stratification of patients with acute
decompensated HF and a obvious negative prognostic value for
long-term mortality by several studies (8–10).
Significatively, NLR was evaluated as an inexpensive and readily
available marker with similar independent prognostic power as
NT-proBNP in elderly patients with CHF (2). Additionally, NLR has also been
frequently reported to predict the clinical outcome with other
cardiovascular diseases and inflammatory diseases, even
malignancies (5,11).
Despite the remarkable significance of NLR in
clinical predictability and prognostication, NLR like most
inflammatory markers, is less specific to any disease (6), and the mechanism by which elevated
NLR is linked to outcomes with HF remains unknown. Therefore, it is
rational to propose that the identification of genes coupled with
altered NLR may contribute to the better understanding of the
predictive and prognostic value of NLR in HF and enhance the
interpretability of the pathobiology findings. Here, we analyzed
the gene signature in blood samples from 197 CHF patients in
attempt to screen for potential decisive factors of elevated
NLR.
Materials and methods
Microarrary analysis
The gene expression profile (GSE77343) based on the
GPL11532 platform {(HuGene-1_1-st) Affymetrix Human Gene 1.1 ST
Array [transcript (gene)]} for blood samples from 197 individuals
with CHF with corresponding hematological parameters and clinical
data were downloaded from the public GEO database (www.ncbi.nlm.gov/geo/), which was deposited by Shannon
et al (12). Total RNA
isolated from whole blood cells was used to determine the gene
expressions. The gene expression data were processed and normalized
by using the robust multi-array average method with the R software
(13). Missing data were imputed
by the KNN-based method (14).
Data were then filtered to include only probe sets with
annotations. Differentially expressed genes (DEGs) were identified
by significance analysis of the microarrays with the limma package
(15), following the criteria
‘fold-change >1.5 and P<0.05′. The heat map was plotted for
samples and DEGs by using the pheatmap package in R.
Protein-protein interaction (PPI)
network, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and
Genomes (KEGG) pathway analysis
The online database Search Tool for the Retrieval of
Interacting Genes (STRING, string-db.org)
was utilized to evaluate the PPI among the DEGs (16). To illustrate the comprehensive
information as possible, interactions with a combined score
>0.15 obtained from STRING were imported into the Cytoscape to
construct the PPI network, in which the upregulated DEGs were
indicated with red while downregulated genes were indicated with
wathet. GO analysis is a commonly used bioinformatics tool for
annotating genes by clustering DEGs into three aspects: Cellular
component, molecular function, and biological process (17). KEGG is a collection of manually
drawn pathway maps representing our knowledge of the molecular
interaction and reaction network f or genomes, biological pathways,
human diseases, drugs, and chemical substances (18). GO and KEGG pathway analysis were
performed with cluster Profiler in R (19), with P<0.05 as the cut-off.
Statistical analysis
NLR was calculated as neutrophil proportion divided
by lymphocyte proportion; the cut-off for NLR was defined as the
mean value for all samples at the time of evaluation. NLR greater
than the mean value was considered elevated.
Receiver operating characteristic (ROC) curves were
utilized to analyze the ability of DEGs in discriminating an
elevated NLR and to provide the optimal cut-off point between
sensitivity and specificity. The area under the curve (AUC) of ROC
was used to evaluate the performance of discrimination. AUC above
0.7 indicated satisfactory diagnostic accuracy, while AUC above 0.8
reflects excellent diagnostic accuracy. To guarantee a high
accuracy in the current study, only the DEGs with an AUC above 0.8
were considered for further analysis. The ROC curves were performed
using an R-based online-tool Cutoff Finder (20), which could provide the best cut-off
value, the AUC, sensitivity and specificity along with the plot.
Additionally, a Pearson linear correlation analysis between NLR and
the selected DEGs expression with high diagnostic accuracy was
performed with the normalized data.
Based on the cutoff values suggested by the ROC
curves, simple logistic regression analyses were performed to
further investigate the associations between selected DEGs and NLR
adjusted by possible confounders, age and sex. The median age of
the cohort was set as the cut-off for the logistic regression.
Also, a comprehensive multivariate regression analysis was employed
to further identify the significant models including the selected
DEGs as well as age and sex. Pearson linear correlation and
logistic regression analysis involved use of SPSS 17.0 (SPSS, Inc.,
Chicago, IL, USA). P<0.05 was considered statistically
significant.
Results
Identification of DEGs
A total of 197 CHF patients with corresponding
neutrophil proportion and lymphocyte proportion were included in
the study. The mean of NLR was 3.96, which was set as the cut-off
for the further analysis. Based on the suggested cut-off for NLR,
31 genes were identified to be differentially expressed with the
criteria ‘fold-change >1.5 and P<0.05′, among which 21 genes
were upregulated and 10 genes were downregulated in patients with
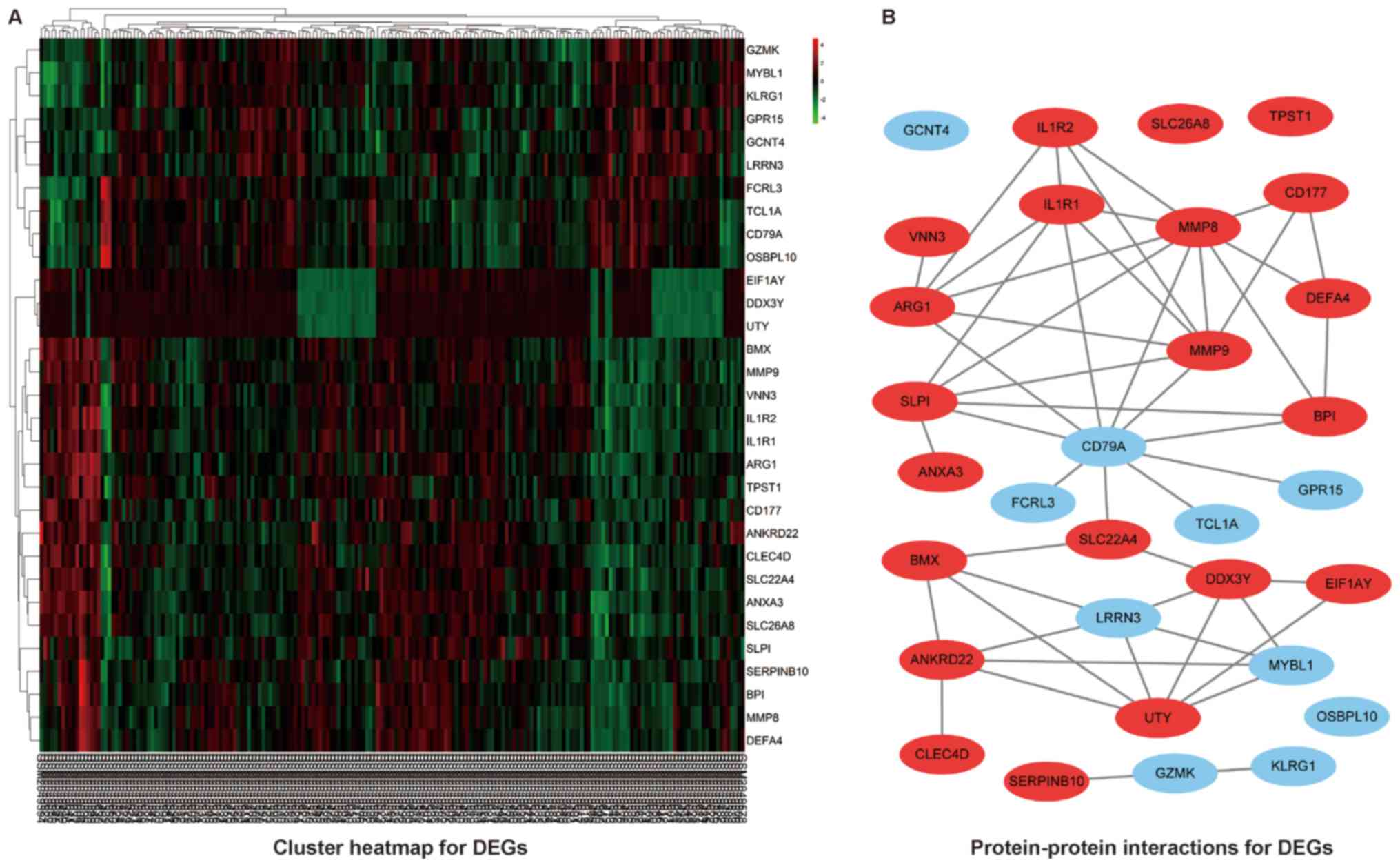
elevated NLR. The heat map for the DEGs was illustrated as Fig. 1A.
PPI network, GO and KEGG pathway
analysis
A basic PPI network between DEGs was constructed by
using the STRING database, with potential links displayed by
Cytoscape (Fig. 1B). To gain the
cellular function, process and signal pathway associated with
elevated NLR, GO and KEGG pathway enrichment analyses were
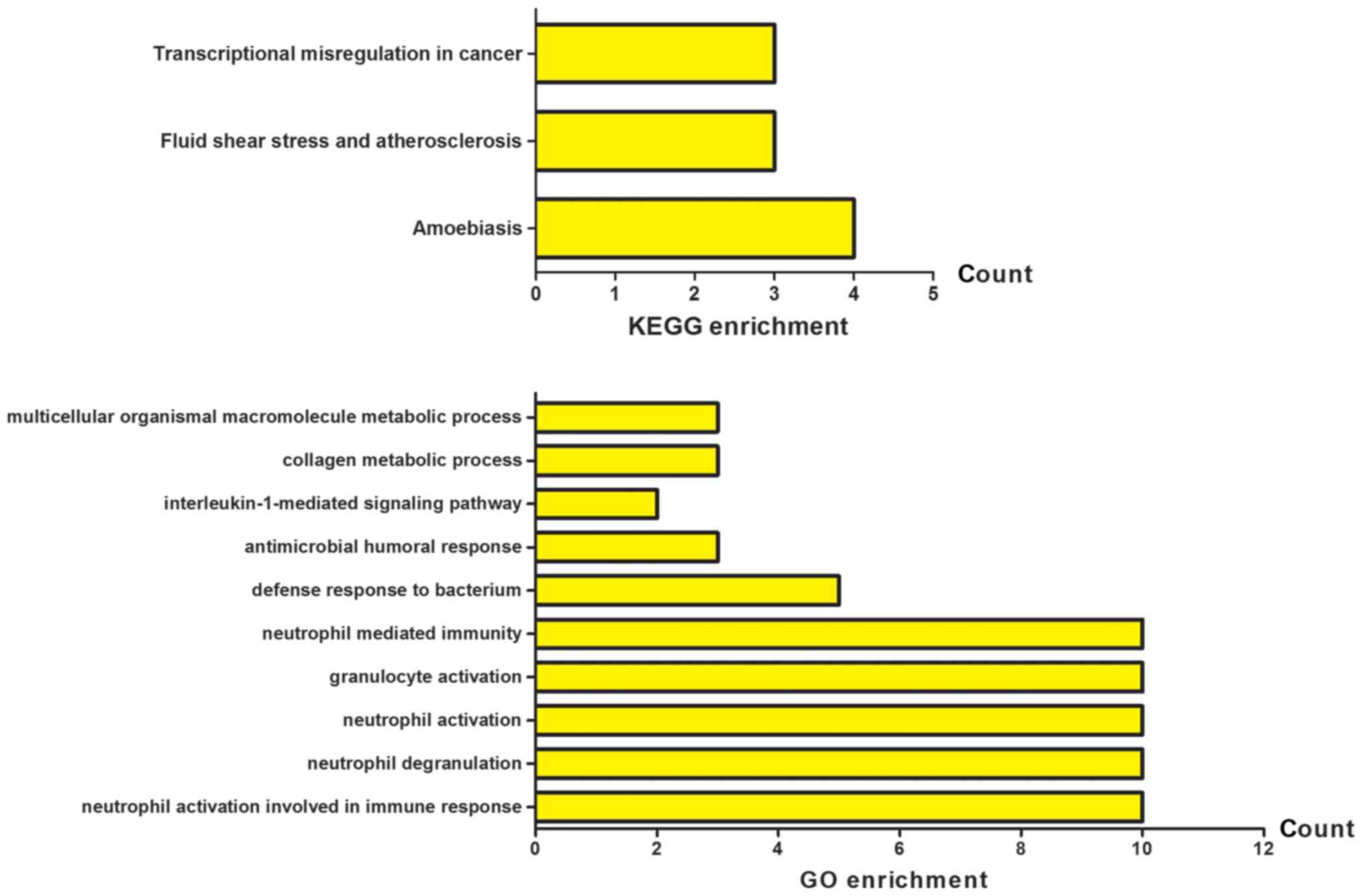
performed using cluster Profiler. As shown in Fig. 2, the results of GO analysis showed
that the DEGs were mainly enriched in neutrophil activation
involved in immune response, neutrophil degranulation, neutrophil
activation, granulocyte activation and neutrophil mediated
immunity. The KEGG pathway analysis revealed that the DEGs were
significantly enriched in fluid shear stress and atherosclerosis,
transcriptional misregulation in cancer and amoebiasis. These
significantly enriched GO terms and pathways will provide further
insight into the relation of decisive factors to the elevated NLR
in CHF patients for future researches, thereby potentially
proposing promising biomarkers for the prognostication and rational
interpretability of pathobiology for HF.
ROC curve and Pearson correlation
analysis
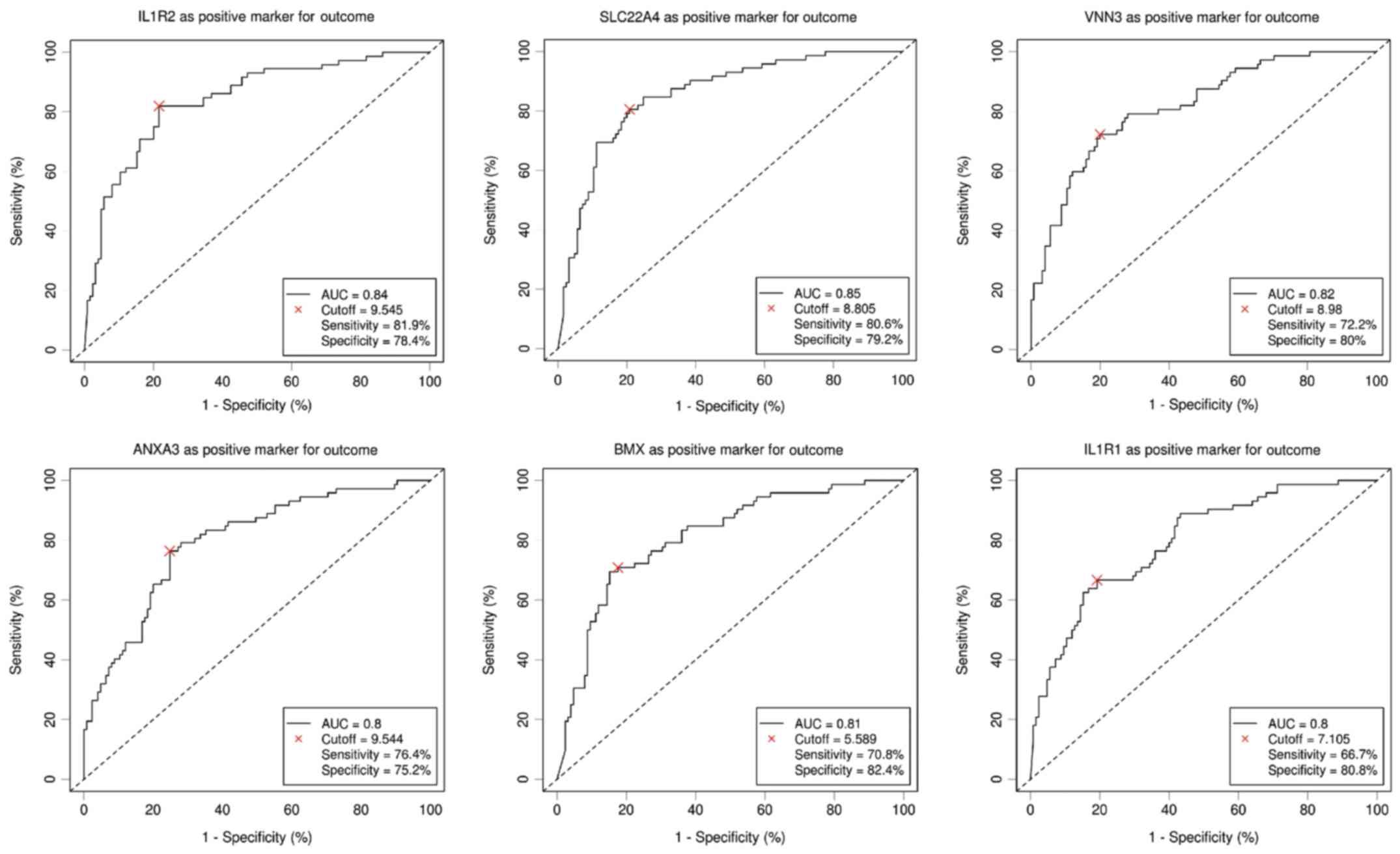
Six DEGs with an AUC above 0.8 were selected to be
potentially related to elevated NLR by the ROC curve analysis. As
shown in Fig. 3, ROC curve
demonstrated the predicting power with the optimal cut-off on
elevated NLR with an AUC value of 0.84, sensitivity 81.9%,
specificity 78.4% for interleukin-1 receptor 2 (IL1R2), an AUC
value of 0.85, sensitivity 80.6%, specificity 79.2% for solute
carrier family 22 member 4 (SLC22A4), an AUC value of 0.82,
sensitivity 72.2%, specificity 80% for vanin 3 (VNN3), an AUC value
of 0.8, sensitivity 76.4%, specificity 75.2% for Annexin A3
(ANXA3), an AUC value of 0.81, sensitivity 70.8%, specificity 82.4%
for BMX non-receptor tyrosine kinase (BMX) and an AUC value of 0.8,
sensitivity 66.7%, specificity 80.8% for interleukin-1 receptor
type 1 (IL1R1), respectively. To further study the relationships
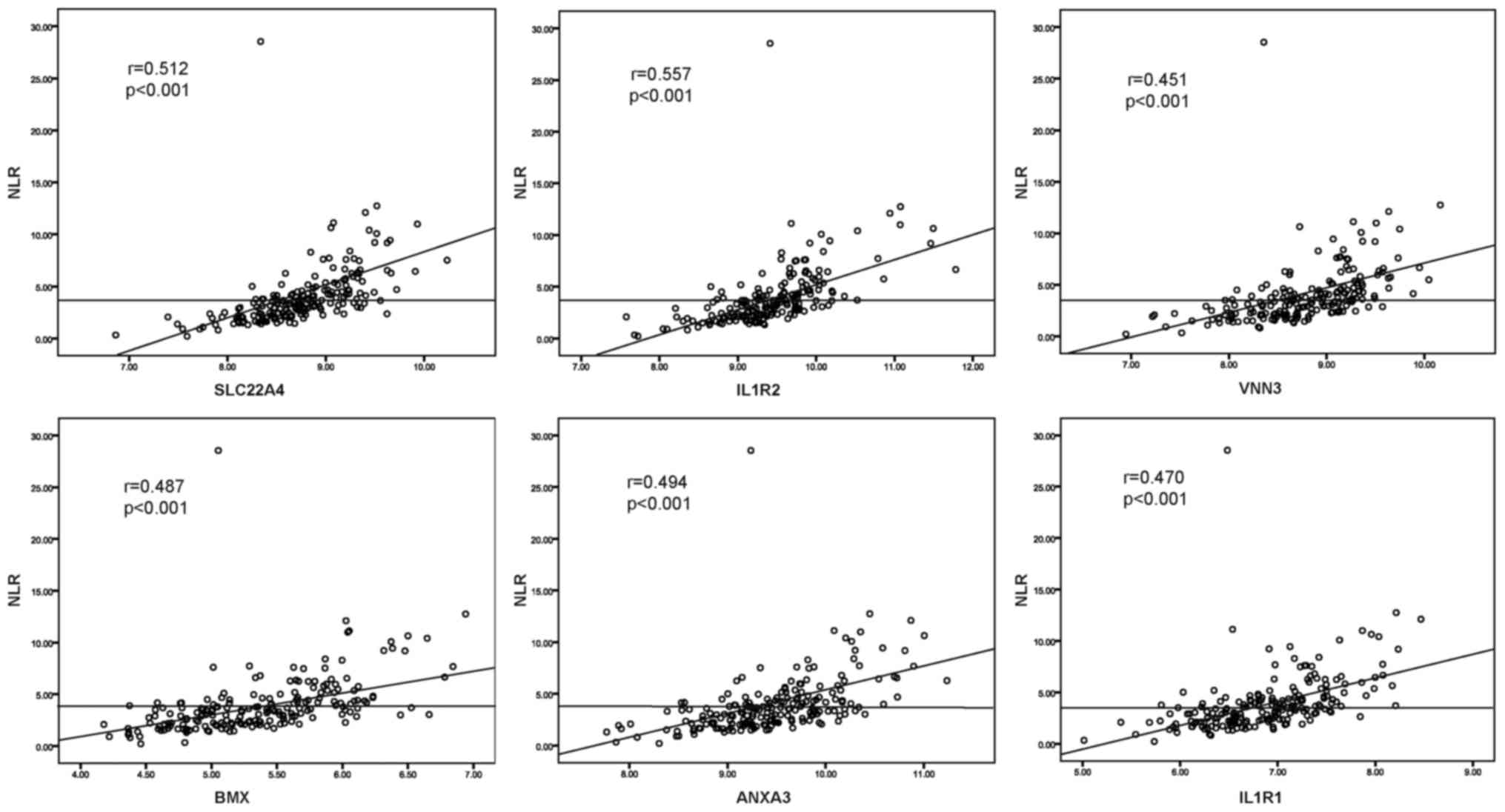
between selected DEGs and NLR, the Pearson relation analysis was
performed using the normalized expression data, and the results
revealed significant associations of all six genes (r=0.512,
P<0.001 for SLC22A4; r=0.557, P<0.001 for IL1R2; r=0.451,
P<0.001 for VNN3; r=0.487, P<0.001 for BMX; r=0.494,
P<0.001 for ANXA3; r=0.470, P<0.001 for IL1R1) with elevated
NLR (Fig. 4).
Logistic regression analyses
To further study the association of the selected six
DEGs with the alteration of NLR, a simple logistic regression
adjusted for age and gender was performed. As shown in Table I, significant associations of
elevated NLR with SLC22A4, IL1R2, VNN3, ANXA3, BMX, IL1R1 and
gender were revealed (all P<0.05). Considering the potential
links between the DEGs proposed by the PPI network, a multivariate
regression analysis including the six DEGs, age and gender was also
conducted. As shown in Table II,
the results suggested that a higher expression of SLC22A4
(OR=5.219, P=0.003), IL1R2 (OR=5.228, P=0.007), VNN3 (OR=3.478,
P=0.012) and female (OR=0.208, P=0.005) might be independently
associated with elevated NLR in CHF patients.
 | Table I.Results of logistic regression
analysis adjusted by age and sex. |
Table I.
Results of logistic regression
analysis adjusted by age and sex.
|
| Logistic
regression |
|
|---|
|
|
|
|
|---|
| Variables | OR | 95% CI | P-value |
|---|
| SLC22A4 |
|
|
|
| Age,
years (≥66 vs. <66) | 1.702 | 0.805–3.599 | 0.164 |
| Sex
(female vs. male) | 0.311 | 0.122–0.789 | 0.014a |
|
Expression (high vs. low) | 16.844 | 7.910–35.869 |
<0.001c |
| IL1R2 |
|
|
|
| Age,
years (≥66 vs. <66) | 1.054 | 0.496–2.239 | 0.891 |
| Sex
(female vs. male) | 0.252 | 0.100–0.636 | 0.004b |
|
Expression (high vs. low) | 18.799 | 8.617–41.013 |
<0.001c |
| VNN3 |
|
|
|
| Age,
years (≥66 vs. <66) | 1.434 | 0.710–2.898 | 0.315 |
| Sex
(female vs. male) | 0.294 | 0.120–0.720 | 0.007b |
|
Expression (high vs. low) | 11.136 | 5.501–22.534 |
<0.001c |
| BMX |
|
|
|
| Age,
years (≥66 vs. <66) | 1.076 | 0.526–2.202 | 0.841 |
| Sex
(female vs. male) | 0.325 | 0.132–0.800 | 0.014a |
|
Expression (high vs. low) | 11.716 | 5.742–23.908 |
<0.001c |
| ANXA3 |
|
|
|
| Age,
years (≥66 vs. <66) | 1.461 | 0.732–2.918 | 0.282 |
| Sex
(female vs. male) | 0.390 | 0.163–0.931 | 0.034a |
|
Expression (high vs. low) | 9.562 | 4.796–19.063 |
<0.001c |
| IL1R1 |
|
|
|
| Age,
years (≥66 vs. <66) | 1.228 | 0.616–2.448 | 0.559 |
| Sex
(female vs. male) | 0.263 | 0.108–0.636 | 0.003b |
|
Expression (high vs. low) | 9.585 | 4.736–19.400 |
<0.001c |
 | Table II.Result of multiple logistic
regression analysis. |
Table II.
Result of multiple logistic
regression analysis.
|
| Logistic
regression |
|
|---|
|
|
|
|
|---|
| Variables | OR | 95% CI | P-value |
|---|
| Age, years (≥66 vs.
<66) | 1.278 | 0.532–3.071 | 0.584 |
| Gender (female vs.
male) | 0.208 | 0.069–0.629 | 0.005b |
| SLC22A4 (high vs.
low expression) | 5.219 | 1.762–15.458 | 0.003b |
| IL1R2 (high vs. low
expression) | 5.228 | 1.560–17.515 | 0.007b |
| VNN3 (high vs. low
expression) | 3.478 | 1.322–9.195 | 0.012a |
| BMX (high vs. low
expression) | 2.012 | 0.775–5.221 | 0.151 |
| ANXA3 (high vs. low
expression) | 1.870 | 0.681–5.133 | 0.225 |
| IL1R1 (high vs. low
expression) | 0.625 | 0.180–2.173 | 0.460 |
Discussion
The current study is the first to investigate the
potential decisive factors associated with elevated NLR in CHF by
microarray assay of genes, demonstrating that several genes
including SLC22A4, IL1R2 and VNN3 may be independently associated
with the elevation of NLR.
NLR reflecting the systemic inflammatory response
has been consistently confirmed to affect the severity and
prognosis of many cardiovascular diseases (6), which implies its role as a reliable
prognostic index. NLR has also been found with similar independent
prognostic power as NT-proBNP for older people with HF but with
inexpensive and readily available testing properties (2). To be biologically reasonable, the
capability of NLR by integrating two different yet complementary
immune pathways may account for its predictive superiority of NLR.
Neutrophils represent a subclinical inflammatory condition whereby
endothelial damage and platelet aggregation are initiated by the
release of pro-oxidants and prothrombotic substances (21). By contrast, reduced lymphocytes
proportion represents a physiological stress and poor general
health (22). Therefore, NLR
actually acts as a balance of inflammation and stress response
(5). Clinically, NLR has been
found predictive for cardiac arrhythmias and mortality in patients
with acute coronary syndromes. High NLR has also been found to be
associated with arterial stiffness and high coronary calcium score.
Moreover, elevated NLR has been repeatedly found a powerful
predictor of worse clinical outcome with HF. Despite the
significance of NLR as a prognostic index in the prognostication
and risk stratification of cardiovascular disease including HF
verified by many studies, NLR, like most inflammatory markers, is
trapped in the lack of specificity to a certain disease. Therefore,
several decisive factors associated with the elevation of NLR in
CHF may aid in the prediction of prognosis of CHF in combination
with NLR.
In the present study involving 197 CHF patients,
increased expressions of SLC22A4, IL1R2 and VNN3 were found in
patients with elevated NLR and independent significant associations
of SLC22A4, IL1R2 and VNN3 with elevated NLR in CHF patients were
also revealed through a further comprehensive analysis including
ROC, simple and multivariate regression analysis, which suggests
the possible predictive role of SLC22A4, IL1R2 and VNN3 in NLR.
Considering the significance of NLR as a prognostic index in HF,
the suggested genes as potential decisive factors for NLR may be
hypothesized for the prediction of prognostication in CHF.
Biologically, the detrimental effect of over-expressed SLC22A4
(also known as OCTN1) which acts as an organic cation transporter
in cardiomyocytes has been demonstrated by a previous study
(23). SLC22A4 was suggested to be
a potential risk factor for torsade de pointes by intensifying
quinidine-induced HERG block (23). In addition, OCTN1 was also found to
be possibly associated with rheumatoid arthritis (24). Although modest, it is plausible to
propose a deleterious effect of dysregulated SLC22A4 on CHF
patients. Similarly, the inflammatory cytokine IL1R2 acting as an
important component of IL1 signal pathway was found associated with
several common cardiovascular diseases including coronary artery
disease (25), hypertension
(26), cardiac allograft rejection
and coronary heart disease (27,28).
The inflammation-related function of IL1R2 may also support a role
in the pathogenesis and prognosis of CHF. Moreover, the vanin
family gene VNN3 is a secreted and membrane-bound ectoenzyme which
converts pantetheine into pantothenic acid and cysteamine. A
previous study screening a blood transcriptome-based molecular
signature demonstrated that VNN3 may serve as a more specific and
sensitive diagnostic biomarker for ST-segment-elevation myocardial
infarction than traditional creatine kinase-MB or troponin
(29). Additionally, the
expression of VNN3 was also found to be upregulated in mesangial
proliferative nephritis and skin psoriasis lesions, suggesting its
proinflammatory activity in diseases (30,31).
Collectively, the findings of the present study may hint at the
potential for SLC22A4, IL1R2 and VNN3 associated with altered NLR
to predict the prognosis with CHF patients owing to their evidenced
biological properties in cardiovascular and inflammatory diseases
to some extent. However, their prognostic value remains undefined
to date. Furthermore, the present study also indicated that female
gender tended to present a lower NLR, potentially suggesting a
favorable prognosis for female CHF patients, which was supported by
a large-scale cohort study which demonstrated that women had a
lower risk of death irrespective of cause of HF or left-ventricular
ejection fraction (32).
To be noteworthy, the uniform cut-off of NLR remains
undefined in the scientific community, so we used the mean 3.96 of
NLR as the cut-off to define the elevation of NLR, while the
cut-off utilized in several studies reporting a significant
association of elevated NLR with a worse clinical outcome of CHF
varied from 2.74 to 4.1 (2,7,10,33).
Therefore, it may indicate a comparatively high risk of mortality
for CHF patients with NLR greater than 3.96 in this study.
Accordingly, combined with the biological inherent property
discussed previously, the genes SLC22A4, IL1R2 and VNN3 associated
with the useful prognostic index NLR may be potential predictors of
CHF prognosis in combination with NLR. However, further perspective
studies are needed to confirm the findings.
Our study has certain limitations. First, the NLR
cut-off based on mean, median or quantile was previously used
widely to explore the diagnostic and prognostic role of NLR in most
cardiovascular diseases due to the lack of a uniform cut-off of NLR
defined by the scientific community, we therefore used the mean
3.96 of NLR as the cut-off to define the elevation of NLR, which
may have resulted in misclassification of patients. Second, several
well known pro-inflammatory and prognostic markers such as
NT-proBNP, interleukin-6, high-sensitivity CRP and cardiac troponin
were not analyzed and compared with the NLR due to the lack of
available relevant information. Nevertheless, the present study
also has unoverlooked redeeming feature. Although measurements of
serum products were more common and convenient in the clinics, some
genes with prognostic potential may be undetectable in serum.
Moreover, it may suffer from limitations in providing a complete
and unbiased genetic profile of the circulating peripheral blood
compartment of an individual, which may be affected by factors such
as preprocessing of samples, cellular contamination. Collectively,
the detection of gene expression from peripheral blood may be a
significant supplement to routine measurement of serum products
when necessary in the clinic.
In summary, the current study was designed to
identify DEGs associated with elevated NLR, which may be candidate
predictors for CHF prognosis. A total of 31 DEGs were revealed, of
which three potential decisive factors SLC22A4, IL1R2 and VNN3
independently associated with elevated NLR were focused.
Nevertheless, the present study was devised using a bioinformatics
strategy, further clinical studies are warranted to verify the
findings.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
GXW conceived and designed the study, and drafted
the manuscript. LHJ, WBX and LC performed the statistical analysis,
data collation and interpretation. YGZ designed the study, and
reviewed and edited the manuscript. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Tasal A, Erturk M, Uyarel H, Karakurt H,
Bacaksiz A, Vatankulu MA, Turfan M, Sonmez O, Erdogan E and Ergelen
M: Utility of the neutrophil to lymphocyte ratio for predicting
in-hospital mortality after levosimendan infusion in patients with
acute decompensated heart failure. J Cardiol. 63:418–423. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yan W, Li RJ, Jia Q, Mu Y, Liu CL and He
KL: Neutrophil-to-lymphocyte ratio compared to N-terminal pro-brain
natriuretic peptide as a prognostic marker of adverse events in
elderly patients with chronic heart failure. J Geriatr Cardiol.
14:127–134. 2017.PubMed/NCBI
|
|
3
|
Ostrowska M, Ostrowski A, Łuczak M,
Jaguszewski M, Adamski P, Bellwon J, Rynkiewicz A and Gruchała M:
Basic laboratory parameters as predictors of in-hospital death in
patients with acute decompensated heart failure: Data from a large
single-centre cohort. Kardiol Pol. 75:157–163. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ho KK, Anderson KM, Kannel WB, Grossman W
and Levy D: Survival after the onset of congestive heart failure in
Framingham Heart Study subjects. Circulation. 88:107–115. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Afari ME and Bhat T: Neutrophil to
lymphocyte ratio (NLR) and cardiovascular diseases: An update.
Expert Rev Cardiovasc Ther. 14:573–577. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bhat T, Teli S, Rijal J, Bhat H, Raza M,
Khoueiry G, Meghani M, Akhtar M and Costantino T: Neutrophil to
lymphocyte ratio and cardiovascular diseases: A review. Expert Rev
Cardiovasc Ther. 11:55–59. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wasilewski J, Pyka Ł, Hawranek M, Osadnik
T, Kurek A, Skrzypek M, Niedziela J, Desperak P, Kułaczkowska Z,
Brzezina M, et al: Prognostic value of neutrophil-to-lymphocyte
ratio in predicting long-term mortality in patients with ischemic
and nonischemic heart failure. Pol Arch Med Wewn. 126:166–173.
2016.PubMed/NCBI
|
|
8
|
Uthamalingam S, Patvardhan EA, Subramanian
S, Ahmed W, Martin W, Daley M and Capodilupo R: Utility of the
neutrophil to lymphocyte ratio in predicting long-term outcomes in
acute decompensated heart failure. Am J Cardiol. 107:433–438. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Benites-Zapata VA, Hernandez AV, Nagarajan
V, Cauthen CA, Starling RC and Tang WH: Usefulness of
neutrophil-to-lymphocyte ratio in risk stratification of patients
with advanced heart failure. Am J Cardiol. 115:57–61. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yan W, Liu C, Li R, Mu Y, Jia Q and He K:
Usefulness of the neutrophil-to-lymphocyte ratio in predicting
adverse events in elderly patients with chronic heart failure. Int
Heart J. 57:615–621. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mei Z, Shi L, Wang B, Yang J, Xiao Z, Du
P, Wang Q and Yang W: Prognostic role of pretreatment blood
neutrophil-to-lymphocyte ratio in advanced cancer survivors: A
systematic review and meta-analysis of 66 cohort studies. Cancer
Treat Rev. 58:1–13. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shannon CP, Balshaw R, Chen V, Hollander
Z, Toma M, McManus BM, Fitzgerald JM, Sin DD, Ng RT and Tebbutt SJ:
A novel approach to identifying marker genes and estimating the
cellular composition of whole blood from gene expression profiles.
bioRxiv. 2:387942016.
|
|
13
|
Irizarry RA, Hobbs B, Collin F,
Beazer-Barclay YD, Antonellis KJ, Scherf U and Speed TP:
Exploration, normalization, and summaries of high density
oligonucleotide array probe level data. Biostatistics. 4:249–264.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Troyanskaya O, Cantor M, Sherlock G, Brown
P, Hastie T, Tibshirani R, Botstein D and Altman RB: Missing value
estimation methods for DNA microarrays. Bioinformatics. 17:520–525.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:(Database Issue). D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gene Ontology Consortium, . The Gene
Ontology (GO) project in 2006. Nucleic Acids Res. 34:(Database
Issue). D322–D326. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Budczies J, Klauschen F, Sinn BV, Győrffy
B, Schmitt WD, Darb-Esfahani S and Denkert C: Cutoff Finder: A
comprehensive and straightforward Web application enabling rapid
biomarker cutoff optimization. PLoS One. 7:e518622012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Weber C, Zernecke A and Libby P: The
multifaceted contributions of leukocyte subsets to atherosclerosis:
Lessons from mouse models. Nat Rev Immunol. 8:802–815. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Azab B, Zaher M, Weiserbs KF, Torbey E,
Lacossiere K, Gaddam S, Gobunsuy R, Jadonath S, Baldari D, McCord D
and Lafferty J: Usefulness of neutrophil to lymphocyte ratio in
predicting short- and long-term mortality after non-ST-elevation
myocardial infarction. Am J Cardiol. 106:470–476. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
McBride BF, Yang T, Liu K, Urban TJ,
Giacomini KM, Kim RB and Roden DM: The organic cation transporter,
OCTN1, expressed in the human heart, potentiates antagonism of the
HERG potassium channel. J Cardiovasc Pharmacol. 54:63–71. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Taubert D, Lazar A, Grimberg G, Jung N,
Rubbert A, Delank KS, Perniok A, Erdmann E and Schömig E:
Association of rheumatoid arthritis with ergothioneine levels in
red blood cells: A case control study. J Rheumatol. 33:2139–2145.
2006.PubMed/NCBI
|
|
25
|
Long F, Wang L, Yang L, Ji Z and Hu Y:
Screening hub genes in coronary artery disease based on integrated
analysis. Cardiol J. Oct 5–2017.(Epub ahead of print). PubMed/NCBI
|
|
26
|
Stoynev N, Dimova I, Rukova B, Hadjidekova
S, Nikolova D, Toncheva D and Tankova T: Gene expression in
peripheral blood of patients with hypertension and patients with
type 2 diabetes. J Cardiovasc Med (Hagerstown). 15:702–709. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mehra MR, Uber PA and Benitez RM:
Gene-based bio-signature patterns and cardiac allograft rejection.
Heart Fail Clin. 6:87–92. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen X, Chen X, Xu Y, Yang W, Wu N, Ye H,
Yang JY, Hong Q, Xin Y, Yang MQ, et al: Association of six CpG-SNPs
in the inflammation-related genes with coronary heart disease. Hum
Genomics. 10 Suppl 2:S212016. View Article : Google Scholar
|
|
29
|
Park HJ, Noh JH, Eun JW, Koh YS, Seo SM,
Park WS, Lee JY, Chang K, Seung KB, Kim PJ and Nam SW: Assessment
and diagnostic relevance of novel serum biomarkers for early
decision of ST-elevation myocardial infarction. Oncotarget.
6:12970–12983. 2015.PubMed/NCBI
|
|
30
|
Lu Y and Chen X, Yin Z, Zhu S, Wu D and
Chen X: Screening for potential serum biomarkers in rat mesangial
proliferative nephritis. Proteomics. 16:1015–1022. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jansen PA, Kamsteeg M, Rodijk-Olthuis D,
van Vlijmen-Willems IM, de Jongh GJ, Bergers M, Tjabringa GS,
Zeeuwen PL and Schalkwijk J: Expression of the vanin gene family in
normal and inflamed human skin: Induction by proinflammatory
cytokines. J Invest Dermatol. 129:2167–2174. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
O'Meara E, Clayton T, McEntegart MB,
McMurray JJ, Piña IL, Granger CB, Ostergren J, Michelson EL,
Solomon SD, Pocock S, et al: Sex differences in clinical
characteristics and prognosis in a broad spectrum of patients with
heart failure: Results of the Candesartan in Heart failure:
Assessment of Reduction in Mortality and morbidity (CHARM) program.
Circulation. 115:3111–3120. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cakici M, Cetin M, Doğan A, Oylumlu M,
Aktürk E, Polat M, Suner A and Abuş S: Neutrophil to lymphocyte
ratio predicts poor functional capacity in patients with heart
failure. Turk Kardiyol Dern Ars. 42:612–620. 2014. View Article : Google Scholar : PubMed/NCBI
|