Introduction
Prostate cancer (PCa) is the second most common
cancer among men worldwide (1).
Numerous factors are contribute to development of PCa, including
genetic mutations and the tumor microenvironment (2,3).
Therefore, understanding of the oncogenic pathologies in the
progression of PCa may contribute to improved clinical diagnosis
and treatment.
MicroRNAs (miRNA) are endogenous, non-coding short
chain RNAs, which alter gene expression at the post-transcriptional
level affecting angiogenesis, cell cycle, apoptosis, invasion and
migration of the cells and tumor-associated gene expression,
thereby affecting the tumor proliferation, invasion and metastasis
(4–6). Several studies have suggested that
miRNAs have been associated with PCa tumorigenesis and the clinical
outcome of patients (7–9). Previous studies confirmed that
microRNA-210 (miR-210) in renal cell carcinoma, pancreatic cancer,
colorectal cancer, and regulation of cancer cell proliferation,
migration and invasion in renal cell carcinoma (10–14).
Additionally, the present study investigated its function in the
regulation of proliferation and apoptosis and determined that the
regulator of differentiation 1 (ROD1), a protein also termed
polypyrimidine trace binding protein 3 (PTBP3), is a potential the
target of miR-210 in cancer cells (15). However, to the best of our
knowledge there is limited investigation of the correlation between
miR-210 and prostate cancer has been relatively limited.
In this study, we had detected the difference of
serum miR-210 expression between health and prostate cancer
patients at first, and further study the effect of miR-210 on
prostate cancer cells.
Materials and methods
Patients
A total of 30 prostate cancer patients that were
treated between June 2013 to May 2015 were selected for the
prostate cancer (PCa) group and 20 healthy people (all male; age
range, 47–83 years; median age, 58 years), which underwent physical
examination in our hospital at the same period were selected for
the healthy control group. Venous blood was collected from healthy
people and patients with PCa and serum was isolated immediately
using centrifugation in serum-gel tubes at 3,500 × g for 10 min at
4°C and stored at −80°C, until further experiments were
performed.
Cell line culture
The cells which have been collected from the PCa
patients and used for primary cell culture. The RPMI 1640 medium
containing 10% fetal bovine serum was used to culture the cells at
37°C and 5% CO2.
Materials
RPMI 1640 culture medium was purchased Gibco; Thermo
Fisher Scientific, Inc. (Waltham, MA, USA), fetal bovine serum was
purchased from ExCell Bio, Inc. (Shanghai, China), TRIzol Reagent,
PrimeScript RT Master Mix reagent kit and SYBR Premix Ex Taq kit
were purchased from Takara Bio, Inc. (Otsu, Japan), The miR-210
primer was constructed by the Shanghai GenePharma Co., Ltd.
(Shanghai, China), Opti-MEM medium and Lipofectamine™
2000 were purchased from Invitrogen; Thermo Fisher Scientific,
Inc., the miR-210 inhibitor was synthesized by the Invitrogen;
Thermo Fisher Scientific, Inc. (cat. no. 4464084), MTT was
purchased from Amresco LLC (Solon, OH, USA), fluorescence
quantitative PCR instrument purchased from ABI-7500 system
employing Primix Ex Taq II SYBR kit, the centrifuge was purchased
from Sigma-Aldrich; EMD Millipore (Billerica, MA, USA) and the flow
cytometer was purchased from BD Biosciences (Franklin Lakes, NJ,
USA).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the serum of healthy
and PCa groups and reverse transcribed to cDNA at 42°C for 60 min
and 70°C for 5 min. The following primer sequences were used:
miR-210 forward (F), 5′-CTGTGCGTGTGACAGCGGCTGA-3′, reverse (R)
primers for universal primer real-time PCR Uni-miR primers (Takara
Bio, Inc.); ROD1 F, 5′-AAGGAAATGAATGGGCAGCCGTTAG-3′ and R,
5′CATG-TAGTTGAGGTCAATGAAGGGGTC-3; GAPDH F,
5′-GCTCTCTGCTCCTCCTGTTC-3′ and R, 5′-GACTCCGACCTTCACCTTCC-3′. GAPDH
was used as the internal control, qPCR was performed according to
the manufacturer's instructions of the Primix Ex Taq II SYBR kit.
Each sample was analyzed in triplicate. The following thermocycling
conditions were used: 95°C for 30 sec, 95°C for 5 sec, 60°C for 31
sec and for 40 cycles. The 2−∆∆Cq method (16) was used to calculate the expression
levels of miR-210 and ROD1.
Transfection
The transfection was performed according to the
manufacturer's protocol using Lipofectamine™ 2000 for a
96-well (1×104 cells/per well) transfection. A total of
48 h following transfection, cells divided into the following
treatment groups: i) NC group, untreated; ii) BL group, empty
vector; and iii) anti-miR-210 group, miR-210
inhibitor-transfected.
Cell apoptosis detection
Apoptosis of cells was detected using Annexin
V-fluorescein isothiocyanate/propidium iodide (Nanjing Keygen
Biotech, Nanjing, China) double staining kit. Following
transfection for 48 h, 1.0×105 cells/well, washed with
PBS, 50 µl Annexin buffer, 5 µl 7-aminoactinomycin D, 5 µl Annexin
V-phycoerythrin dye were added, following a reaction of 15 min, 200
µl buffer was added to detect apoptosis by flow cytometry using the
BD FACSuite™ software (version 1.0; BD Biosciences). The
experiment was repeated four times.
Western blot analysis
Cells were lysed in cold
radioimmunoprecipitationlysis buffer (Thermo Fisher Scientific,
Inc.). Protein concentration was measured using a bicinchoninic
acid protein assay kit. Proteins (30 µg per lane) were separated on
10% SDS-PAGE and subsequently transferred to nitrocellulose
membranes and then incubated with 5% skimmed milk for 2 h at room
temperature. Subsequently, the membranes were probed with
antibodies specific to ROD1 (cat. no. WH0009991M1; 1:200;
Sigma-Aldrich; Merck Millipore) and GAPDH (cat. no. G8795; 1:1,000;
Sigma-Aldrich; Merck Millipore) overnight at 4°C. Membranes were
then incubated with anti-mouse horseradish peroxidase-conjugated
secondary antibodies for 1 h at room temperature (cat. no. A9044;
1:5,000; Sigma-Aldrich; Merck KGaA). The membranes were washed
three times in PBS and analyzed using enhanced chemiluminesence
film system (GE Healthcare, Chicago, IL, USA).
Statistical analysis
Analysis was performed using SPSS version 19.0 (IBM
Corporation, Armonk, NY, USA). Data were expressed as the mean ±
standard deviation. One-way analysis of variance followed by a
least significant difference post hoc test were used to identify
statistical differences groups. P<0.05 was considered to
indicate a statistically significant difference.
Results
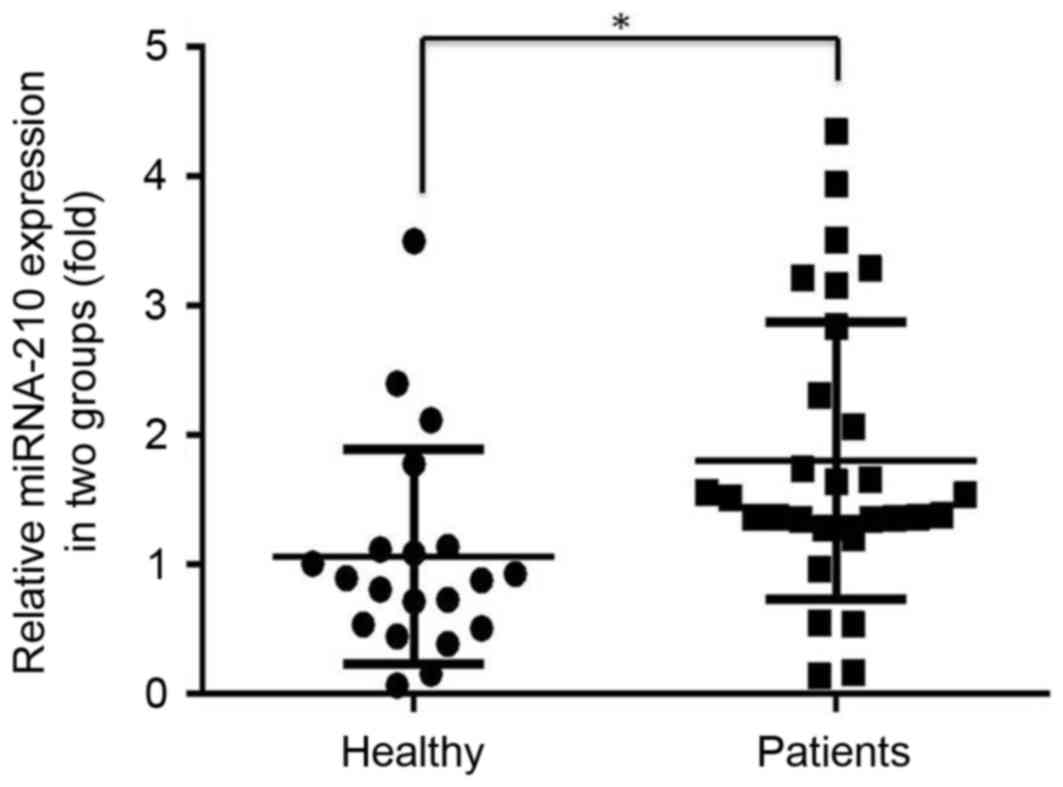
Expression levels of miR-210 in
healthy individuals and patients with PCa
The expression level of miR-210 in the serum of
prostate cancer patients was significantly higher compared with
that in healthy individuals (P<0.05; Fig. 1).
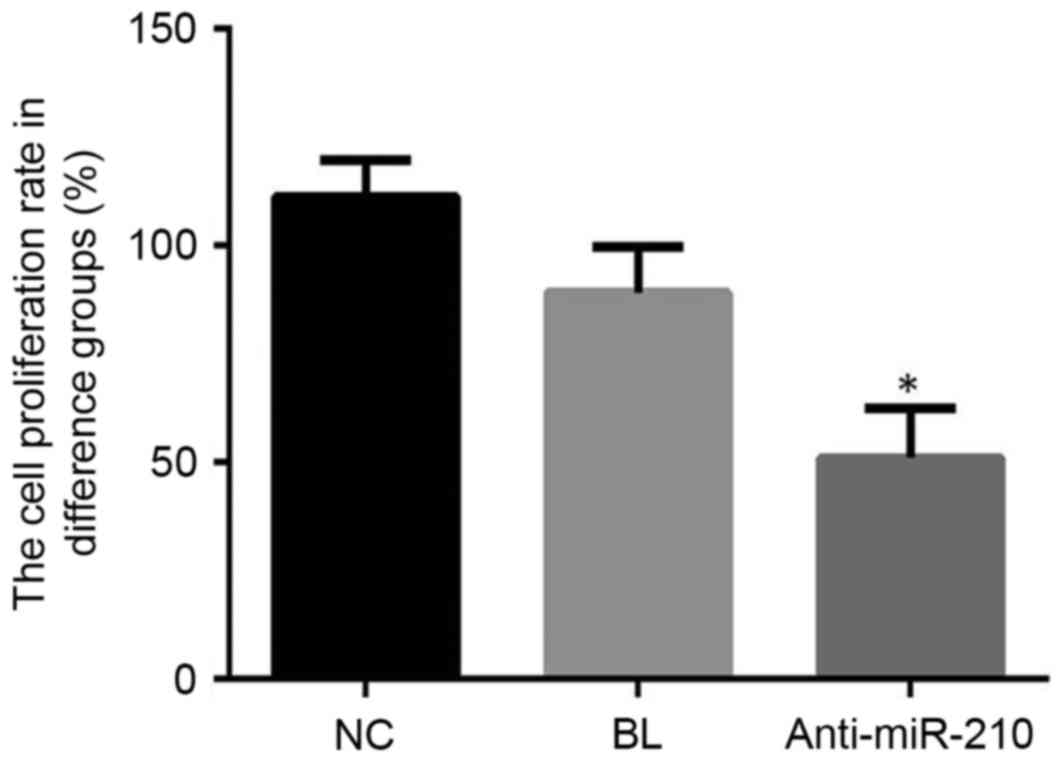
Cell proliferation
Cell proliferation in the anti-miR-210 group was
significantly lower when compared with the NC group (P<0.05;
Fig. 2). These findings suggested
that a miR-210 inhibitor may effectively inhibit the proliferation
of primary PCa cells.
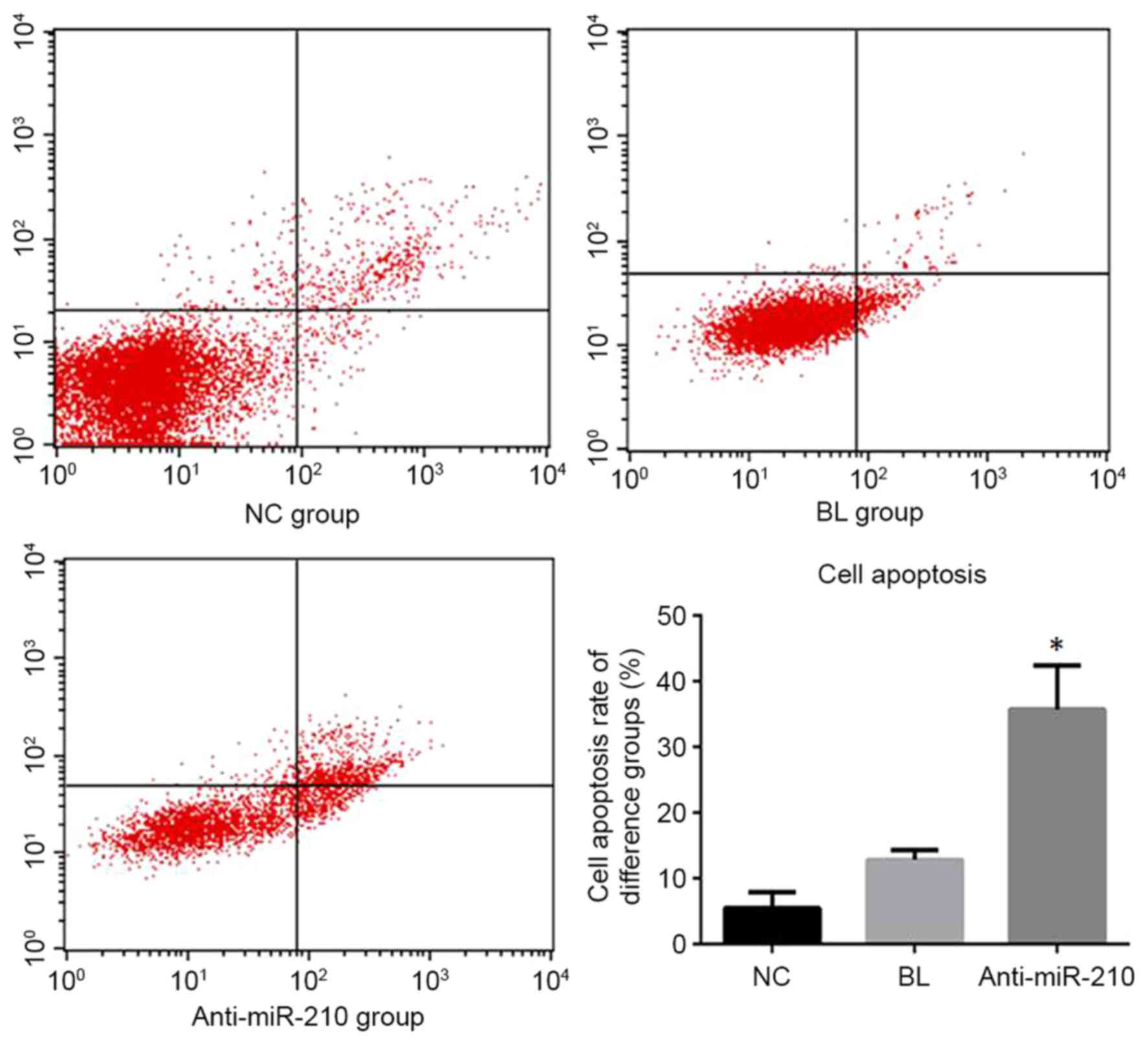
Cell apoptosis detection
The anti-miR-210 group exhibited an apoptotic rate
of 35.75+6.71%, which was significantly higher than the NC group
5.48+2.47% and BL group 12.83+1.47% (P<0.05; Fig. 3).
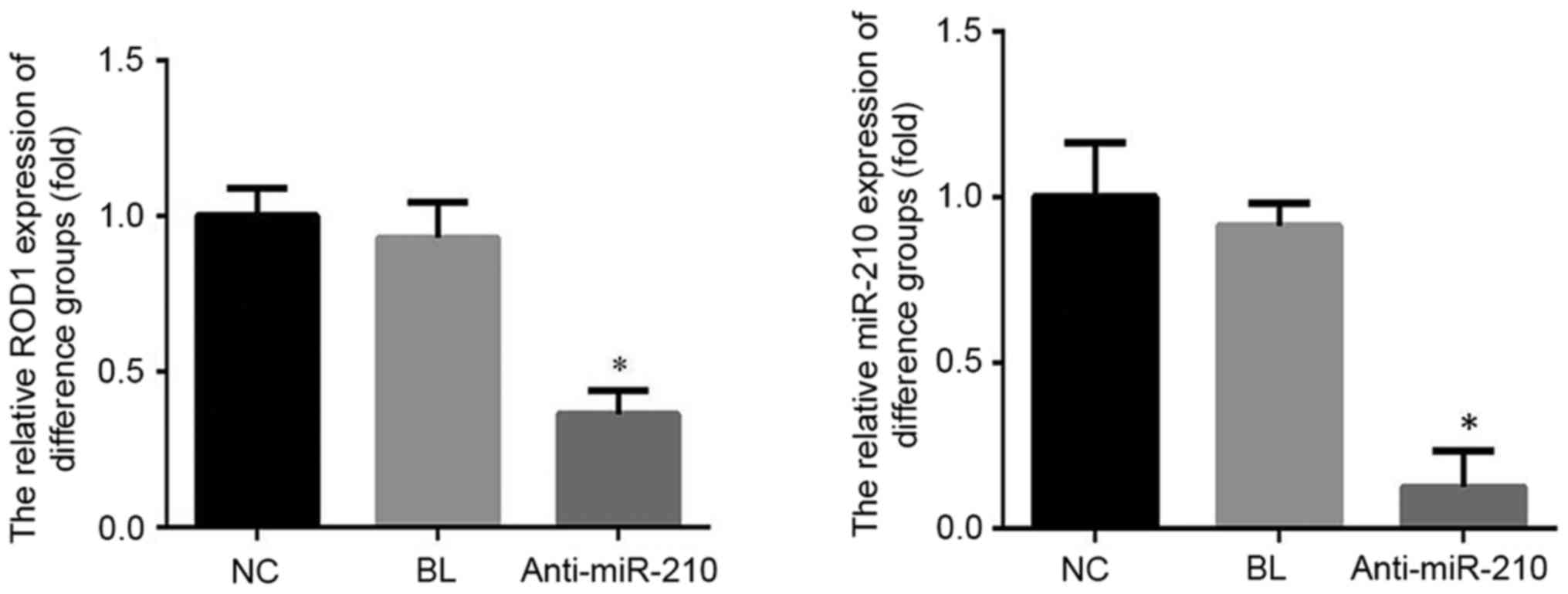
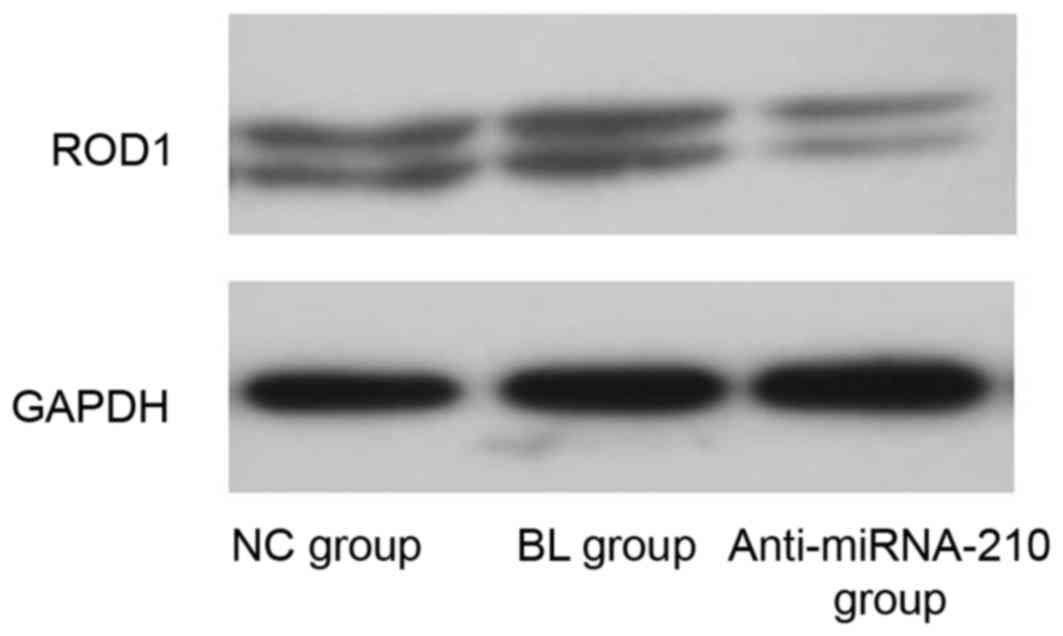
ROD1 mRNA and protein expression
level
The expression level of miR-210 and ROD1 in the
anti-miR-210 group was significantly reduced when compared with the
NC and BL groups (P<0.05; Fig.
4). The expression level of ROD1 protein in the anti-miR-210
group was reduced compared with the NC group (Fig. 5).
Discussion
PCa is a common malignant tumor in men (1). It is the second most common malignant
tumor in the United States. Additionally, it is the sixth most
common mortality due to cancer (17). Recently, the incidence and
mortality of PCa in China has significantly increased and is
gradually approaching that of European and American countries, and
has become a serious threat to the health of the elderly men
(18). Molecular-targeted therapy
is a novel therapeutic method, developed in recent years (19). It has become an important area of
research for effective therapeutic targets, and miR-210 appears to
provide a new target for the diagnosis and treatment of prostate
cancer (20). A previous study
revealed that more than half of the miRNAs are located in the
region of tumor-associated genes and fragile sites, loss of
heterozygosity, amplification area or fault zone, suggesting that
miRNAs may be used as oncogenes or tumor suppressor genes (21). At present, miRNAs may regulate gene
expression at the post-transcriptional level, which is primarily
used as a negative regulator of gene expression. A previous study
confirmed that miRNA affected angiogenesis, cell cycle, apoptosis,
cell invasion and migration (19).
A previous study confirmed that many tumors, including prostate
cancer, have many miRNA expression abnormalities, and have a
different role in different stages of tumor cell migration,
invasion and angiogenesis (20).
Volinia et al determined that miR-20a was
overexpressed in prostate cancer tissues by using miRNA microarrays
(22). Sylvestre et al
identified that after transfection of an miR-20a inhibitor into PCa
cells, the apoptotic rate was significantly increased, which
indicated that miR-20a had an anti-apoptotic role in PCa cells
(23). Previous studies have
confirmed that expression level of miR-210 is increased in a number
of solid tumors; however, no uniform trend has been identified, to
the best of our knowledge (24–29).
The present study detected the expression of miR-210
in the serum of patients with PCa and healthy individuals. The
current findings revealed that miR-210 in the serum of patients
with PCa was overexpressed. In order to further clarify the role of
mir-210 in PCa, an miR-210 antagonist transfection was used in
primary PCa cells. The present findings revealed that the
anti-miR-210 group cell proliferation rate was significantly
reduced compared with that of the NC and BL groups, and the
apoptotic was significantly greater. In order to determine the
mechanism of action of miR-210 in prostate cancer, the expression
levels of the associated genes and proteins were also
determined.
ROD1, a protein also termed PTBP3, is the potential
target of miRNA-210 in various types of cancer, including
glioblastoma multiforme (30,31).
The present study determined that the ROD1 mRNA and protein
expression levels in the anti-miR-210 group were significantly
reduced and it may be possible that miR-210 inhibitors reduced cell
proliferation and increased apoptotic rate of PCa cells by
inhibiting ROD1 expression level.
In summary, the present study demonstrated that
miR-210 was overexpressed in patients with PCa and that miR-210 may
function as an oncogene. Based on the present study, it is possible
that miR-210 may be a novel biomarker for prostatic cancer, and it
may regulate tumor progression through by targeting ROD1. As PCa is
a form of cancer that progresses and spreads quickly, traditional
therapies, such as surgery and chemo and radiotherapy have only
limited effectiveness in treating this aggressive disease.
Therefore, novel approaches, such as molecular targeting would be
beneficial in the therapeutic treatment of PCa. Targeting miR-210
may provide a novel therapeutic option in the treatment of this
disease. In conclusion, miR-210 may have a key role in PCa
diagnosis and treatment by regulating ROD1 miRNA and protein
expression levels.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The analyzed data sets generated during the study
are available from the corresponding author on reasonable
request.
Authors' contributions
YQ and WH contributed to study design, data
collection, data interpretation, preparation of manuscript and
literature analysis.
Ethics approval and consent to
participate
The present study was approved by the Human Ethics
Committee Review Board at the Medical Group of Ping Mei Shenma
General Hospital. All participants signed written informed consent
forms.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Santanam U, Banach-Petrosky W, Abate-Shen
C, Shen MM, White E and DiPaola RS: Atg7 cooperates with Pten loss
to drive prostate cancer tumor growth. Genes Dev. 30:399–407. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Miftakhova R, Hedblom A, Semenas J,
Robinson B, Simoulis A, Malm J, Rizvanov A, Heery DM, Mongan NP,
Maitland NJ, et al: Cyclin A1 and P450 aromatase promote metastatic
homing and growth of stem-like prostate cancer cells in the bone
marrow. Cancer Res. 76:2453–2464. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Negrini M and Calin GA: Breast cancer
metastasis: A microRNA story. Breast Cancer Res. 10:2032008.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Redova M, Poprach A, Besse A, Iliev R,
Nekvindova J, Lakomy R, Radova L, Svoboda M, Dolezel J, Vyzula R
and Slaby O: MiR-210 expression in tumor tissue and in vitro
effects of its silencing in renal cell carcinoma. Tumour Biol.
34:481–491. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bakirzi K, Law IK, Xue X, Iliopoulos D,
Shah YM and Pothoulakis C: Neurotensin promotes the development of
colitis and intestiinal angiogenesis via Hif-1α-miR-210 signaling.
J Immunol. 196:4311–4321. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rane JK, Scaravilli M, Ylipää A, Pellacani
D, Mann VM, Simms MS, Nykter M, Collins AT, Visakorpi T and
Maitland NJ: MicroRNA expression profile of primary prostate cancer
stem cells as a source of biomarkers and therapeutic targets. Eur
Urol. 67:7–10. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Alhasan AH, Scott AW, Wu JJ, Feng G, Meeks
JJ, Thaxton CS and Mirkin CA: Circulating microRNA signature for
the diagnosis of very high-risk prostate cancer. Proc Natl Acad Sci
USA. 113:10655–10660. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li X, Wan X, Chen H, Yang S, Liu Y, Mo W,
Meng D, Du W, Huang Y, Wu H, et al: Identification of miR-133b and
RB1CC1 as independent predictors for biochemical recurrence and
potential therapeutic targets for prostate cancer. Clin Cancer Res.
20:2312–2325. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Toyama T, Kondo N, Endo Y, Sugiura H,
Yoshimoto N, Iwasa M, Takahashi S, Fujii Y and Yamashita H: High
expression of microRNA-210 is an independent factor indicating a
poor prognosis in Japanese triple-negative breast cancer patients.
Jpn J Clin Oncol. 42:256–263. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Eilertsen M, Andersen S, Al-Saad S,
Richardsen E, Stenvold H, Hald SM, Al-Shibli K, Donnem T, Busund LT
and Bremnes RM: Positive prognostic impact of miR-210 in non-small
cell lung cancer. Lung Cancer. 83:272–278. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Neal CS, Michael MZ, Rawlings LH, Van der
Hoek MB and Gleadle JM: The VHL-dependent regulation of microRNAs
in renal cancer. BMC Med. 8:642010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lai NS, Dong QS, Ding H, Miao ZL and Lin
YC: MicroRNA-210 overexpression predicts poorer prognosis in glioma
patients. J Clin Neurosci. 21:755–760. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cai H, Lin L, Cai H, Tang M and Wang Z:
Prognostic evaluation of microRNA-210 expression in pediatric
osteosarcoma. Med Oncol. 30:4992013.Andersen S, Richardsen E, Moi
L, Donnem T, Nordby Y, Ness N, Holman ME, Bremnes RM and Busund LT:
Fibroblast miR-210 overexpression is independently associated with
clinical failure in prostate cancer-a multicenter (in situ
hybridization) study. Sci Rep 6: 36573, 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bray F and Soerjomataram I: The changing
global burden of cancer: Transitions in human development and
implications for cancer prevention and controlCancer: Disease
Control Priorities. Gelband H, Jha P, Sankaranarayanan R and Horton
S: 3. 3rd edition. The International Bank for Reconstruction and
Development/The World Bank; Washington, DC: 2015
|
|
17
|
Zhang K, Bangma CH and Roobol MJ: Prostate
cancer screening in Europe and Asia. Asian J Urol. 4:86–95. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Riquelme I, Saavedra K, Espinoza JA, Weber
H, García P, Nervi B, Garrido M, Corvalán AH, Roa JC and Bizama C:
Molecular classification of gastric cancer: Towards a
pathway-driven targeted therapy. Oncotarget. 6:24750–24779. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Taylor DD and Gecel-Taylor C: MicroRNA
signatures of tumor-derived exosomes as diagnostic biomarkers of
ovarian cancer. Gynecol Oncol. 110:13–21. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K,
Guo J, Zhang Y, Chen J, Guo X, et al: Characterization of microRNAs
in serum: A novel class of biomarkers for diagnosis of cancer and
other diseases. Cell Res. 18:997–1006. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dumont N and Tlsty TD: Reflections on
miR-ing effects in metastais. Cancer Cell. 16:3–4. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sylvestre Y, De Guire V, Querido E,
Mukhopadhyay UK, Bourdeau V, Major F, Ferbeyre G and Chartrand P:
An E2F/miR-20a autoregulatory feedback loop. J Biol Chem.
282:2135–2143. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
McCormick R, Buffa FM, Ragoussis J and
Harris AL: The role of hypoxia regulated microRNAs in cancer. Curr
Top Microbiol Immunol. 345:47–70. 2010.PubMed/NCBI
|
|
25
|
Tsuchiya S, Fujiwara T, Sato F, Shimada Y,
Tanaka E, Sakai Y, Shimizu K and Tsujimoto G: MicroRNA-210
regulates cancer cell proliferation through targeting fibroblast
growth factor receptor-like 1 (FGFRL1). J Biol Chem. 286:420–428.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
White NM, Bao TT, Grigull J, Youssef YM,
Girgis A, Diamandis M, Fatoohi E, Metias M, Honey RJ, Stewart R, et
al: miRNA profiling for clear cell renal cell carcinoma: Biomarker
discovery and identification of potential controls and consequences
of miRNA dysregulation. J Urol. 186:1077–1083. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhao A, Li G, Péoc'h M, Genin C and
Gigante M: Serum miR-210 as a biomarker for molecular diagnosis of
clear cell renal cell carcinoma. Exp Mol Pathol. 94:115–120. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shen J, Liu Z, Todd NW, Zhang H, Liao J,
Yu L, Guarnera MA, Li R, Cai L, Zhan M and Jiang F: Diagnosis of
lung cancer in individuals with solitary pulmonary nodules by
plasma microRNA biomarkers. BMC Cancer. 11:3742011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ho AS, Huang X, Cao H, Christman-Skieller
C, Bennewith K, Le QT and Koong AC: Circulating miR-210 as a novel
hypoxia marker in pancreatic cancer. Transl Oncol. 3:109–113. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang S, Lai N, Liao K, Sun J and Lin Y:
MicroRNA-210 regulates cell proliferation and apoptosis by
targeting regulator of differentiation 1 in glioblastoma cells.
Folia Neuropathol. 53:236–244. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fasanaro P, Romani S, Voellenkie C,
Maimone B, Capogrossi MC and Martelli F: ROD1 is a seedless target
gene of hypoxia-induced miR-210. PLoS One. 7:e446512012. View Article : Google Scholar : PubMed/NCBI
|