Introduction
Macrophages are the primary effector cells of the
immune system and are involved in inflammatory and anti-infective
responses (1). They additionally
serve an important role in tissue homeostasis, promote the growth
of cells in tissues and are involved in the repair of tissue damage
(2). Macrophages undergo different
forms of polarization under the action of different inducers and
are polarized to M1 or M2 macrophages (3). M1 macrophages are additionally known
as classical pathway-activated macrophages. They are primarily
induced by bacterial products and T helper (Th)1-type cytokines,
including interferon (IFN)-γ. Their characteristic effects include
eliminating intracellular microorganisms and producing large
quantities of pro-inflammatory mediators (4). M2 macrophages are additionally termed
alternative pathway-activated macrophages (5). The primary inducers of M2 macrophages
are Th2-type cytokines [interleukin (IL)-4, IL-13 and IL-10],
glucocorticoids and immunoglobulin complexes and Toll-like receptor
(TLR) ligands (6). The principal
effect of M2 macrophages is the suppression of inflammatory
responses (7). Macrophage
polarization commonly occurs during the pathogenesis and
progression of inflammatory diseases, including cancer, obesity and
cardiovascular diseases, and has a guiding significance for the
prognosis of specific tumors (8–11).
In the present study, a protein-protein interaction
(PPI) network was constructed based on differentially expressed
genes in macrophages of different polarization types from the Gene
Expression Omnibus (GEO) database. The aim of the present study was
to identify important signaling pathways and gene groups that are
involved in the regulation of macrophage polarization through
modular analysis and functional enrichment analysis, which may
provide specific novel insight for the treatment of human
immune-associated diseases.
Materials and methods
Gene expression profiles of human M1
and M2 macrophages
Gene expression profiles of human M1 and M2
macrophages were downloaded from the GEO database [https://www.ncbi.nlm.nih.gov/geo/; accession nos.
GSE18686 (12) and GSE35449
(13)]. Expression profile data of
GSE18686 and GSE35449 were obtained from M1 and M2 macrophages
cultured with inducer or without inducer, respectively (12,13).
The GSE18686 and GSE35449 data sets were tested using the Illumina
HumanHT-12 v3.0 Gene Expression BeadChip platform (Illumina, Inc.,
San Diego, CA, USA).
The data of M1 macrophages treated with
lipopolysaccharide (LPS) and IFN-γ, M2 macrophages cultured with
IL-4 and the corresponding control group (M0 macrophages) were
selected from GSE18686, with six biological replicates included in
each group. The data of the M1 and M2 macrophage groups in addition
to the control group were selected from GSE35449, and seven
biological replicates were included in each group.
Pretreatment of expression profile
data
GSE18686 and GSE35449 expression matrix data sets
were downloaded and pretreated using the quantile method in the
lumi software package v2.32.0 (14,15).
The mean value of the different probes that mapped to the same gene
(GeneSymbol) was calculated. The GSE18686 data set consists of
48,802 probes, and a final total of 19,489 genes remained following
the treatment. The GSE35449 data set consists of 48,797 probes, and
19,487 genes remained following the treatment. The gene expression
data sets were extracted for M1 and M2 macrophages, according to
the aforementioned methods.
Detection of differentially expressed
genes
The limma package in Bioconductor (v3.36.2)
(16) was employed to analyze
genes that were differentially expressed between M1 and M0 or
between M2 and M0 in GSE18686 and GSE35449. This method is
comprised of the following steps: i) Constructing a design matrix
for the preprocessed data; ii) estimating the number of folds in
differential gene expression using a linear model; iii) followed by
using a Bayesian approach for smoothing the standard deviation
(17); and iv) finally utilizing
different parameters for output of the differentially expressed
genes. The log2 of fold change (log2FC) and P-values were used as
parameters for the selection of differentially expressed genes;
P<0.05 was considered to indicate a statistically significant
difference, determined using a Student's t-test, and an absolute
value of log2FC≥1 (the differential expression coefficient was 2).
Subsequently, the differentially expressed genes in the
intersection of GSE18686 and GSE35449 were selected.
Cluster analysis of differentially
expressed genes in intersection
The gplots software package (v2.17.0) (18) was employed to calculate and
construct thermal graphs from the cluster analysis of the
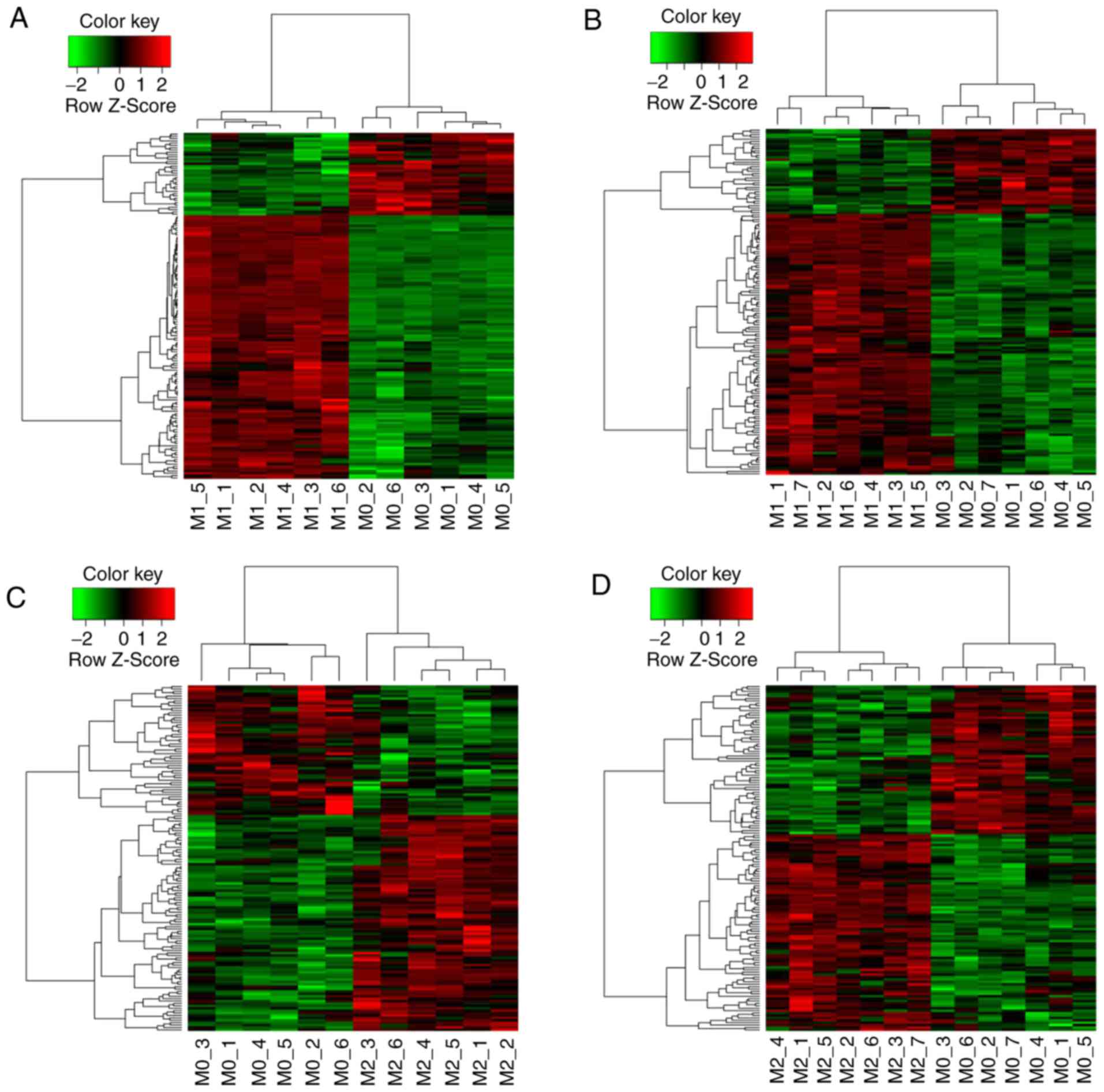
differentially expressed genes in M1 (Fig. 1A and B) and M2 (Fig. 1C and D) in the intersection of
GSE18686 and GSE35449. The expression values of these
differentially expressed genes were analyzed for hierarchical
clustering.
Modular analysis of differentially
expressed genes in the intersection
To further analyze the differentially expressed
genes at the molecular level, Search Tool for the Retrieval of
Interacting Genes/Proteins v9.1 (https://string-db.org/) was used to obtain information
regarding PPI pairs based on the differentially expressed genes in
the GSE18686 and GSE35449 intersection and Cytoscape (v3.4.0)
(19) to construct PPI networks.
To obtain functional modules in the PPI networks, communities in
the PPI networks were extracted using CFinder (v2.0.6) (20).
Functional enrichment analysis of each
community
The candidate genes of each community were submitted
to the Database for Annotation, Visualization and Integrated
Discovery database (http://david.abcc.ncifcrf.gov/), and the complete
genome of Homo sapiens was used for assessment and
comparison as background genes. The ‘Functional Annotation Tool’
was used to obtain the Kyoto Encyclopedia of Genes and Genomes
(KEGG; http://www.genome.jp/kegg/) pathway
enrichment analysis results (the P-value cutoff was 0.05).
Results
Identification of differentially
expressed genes in gene expression profiles
A total of 338 genes that were differentially
expressed between the M1 test group and the M0 control group were
obtained from GSE18686 when P<0.05 and the absolute log2FC value
was ≥0.58; whereas, 636 genes that were differentially expressed
between the M1 test group and the M0 control group were obtained
from GSE35449 (Table I). A total
of 151 differentially expressed genes were obtained from the
intersection of differentially expressed genes from GSE18686 and
GSE35449 between the M1 test group and the M0 control group (data
not shown). Within the same threshold range as above, 273 genes
that were differentially expressed between the M2 test group and
the M0 control group were obtained in GSE18686; whereas, 1,171
genes that were differentially expressed between the M2 test group
and the M0 control group were identified in GSE35449 (Table I). A total of 144 differentially
expressed genes were obtained from the intersection of
differentially expressed genes from GSE18686 and GSE35449 between
the M2 test group and the M0 control group.
 | Table I.Differentially expressed genes
between M1 experimental group and M0 control group. |
Table I.
Differentially expressed genes
between M1 experimental group and M0 control group.
| A, GSE18686 |
|---|
|
|---|
| Contrast group | Number of
differentially expressed genes | Number of
upregulated genes | Number of
downregulated genes |
|---|
| M1 vs. M0 | 338 | 249 | 89 |
| M2 vs. M0 | 273 | 181 | 92 |
|
| B,
GSE35449 |
|
| Contrast
group | Number of
differentially expressed genes | Number of
upregulated genes | Number of
downregulated genes |
|
| M1 vs. M0 | 636 | 298 | 338 |
| M2 vs. M0 | 1,171 | 399 | 772 |
Cluster analysis of differentially
expressed genes in intersection
The thermal graphs from the cluster analysis of the
151 differentially expressed genes between the M1 and M0 groups in
the intersection of GSE18686 and GSE35449 are demonstrated in
Fig. 1A and B. The majority of the
differentially expressed genes in the intersection were upregulated
in the M1 group when compared with their expression levels in the
M0 samples. The thermal graphs from the cluster analysis of the 144
differentially expressed genes between the M2 and M0 groups in the
intersection of GSE18686 and GSE35449 are demonstrated in Fig. 1C and D. The majority of the
differentially expressed genes in the intersection were upregulated
in the M2 group when compared with their expression levels in the
M0 samples.
Construction of PPI networks based on
differentially expressed intersection genes
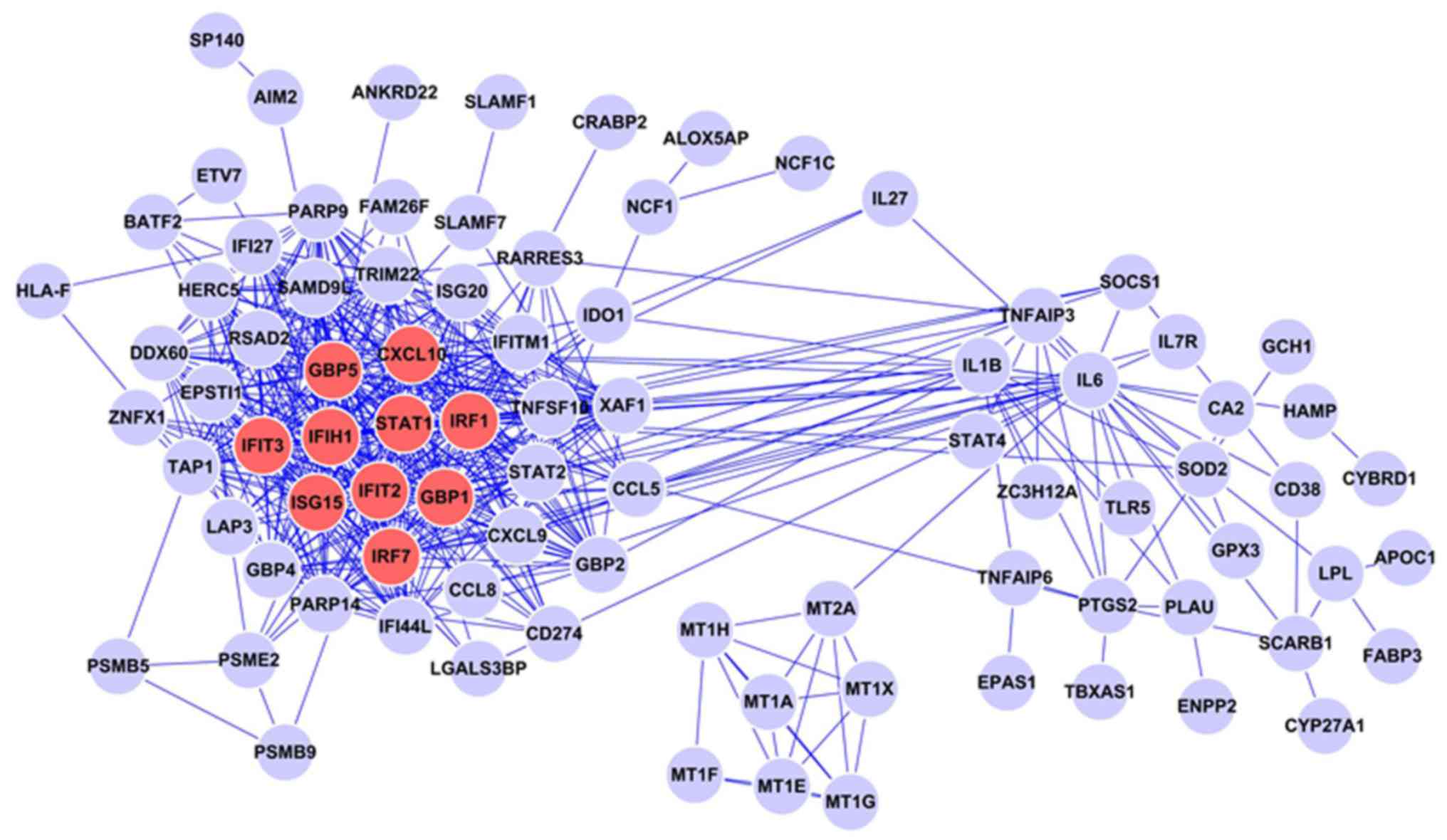
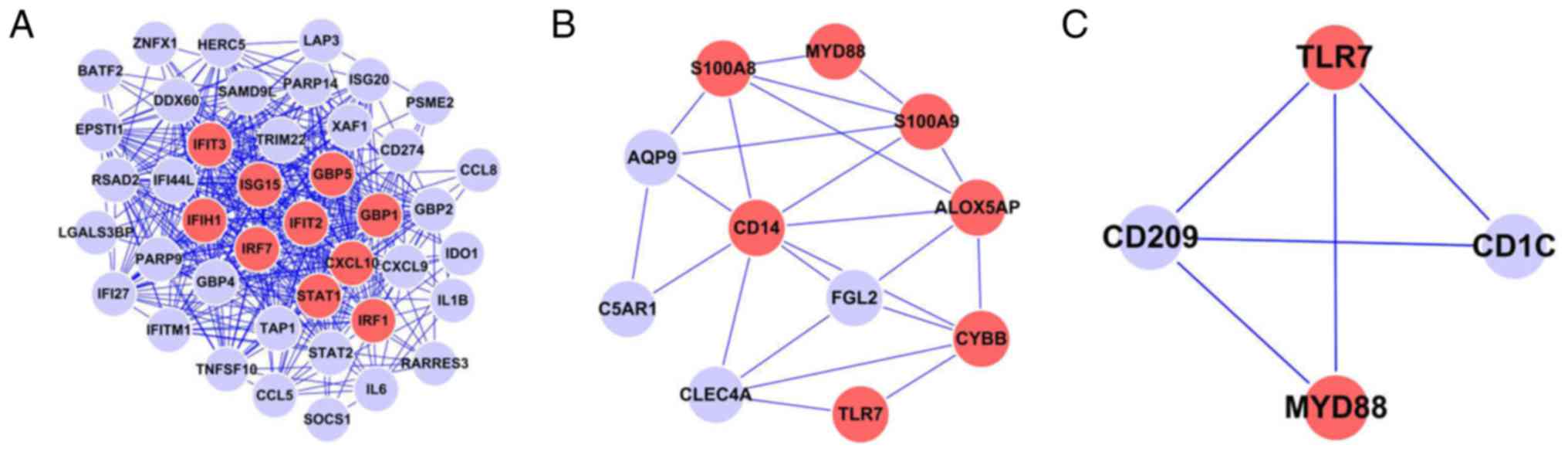
The PPI network based on the intersection of the
differentially expressed genes between the M1 and M0 groups was
constructed. This network consists of 94 protein nodes and 523
PPIs. As demonstrated in the network diagram, the proteins encoded
by these intersecting differentially expressed genes exhibit a
complex association. In this PPI network, the 10 highest degree
proteins were signal transducer and activator of transcription
(STAT)1 (degree=41), guanylate-binding protein (GBP)1 (degree=35),
GBP5 (degree=35), C-X-C motif chemokine 10 (CXCL10; degree=35),
interferon-induced protein with tetratricopeptide (IFIT)2
(degree=32), interferon regulator factor (IRF)7 (degree=32), IFIT3
(degree=32), ubiquitin-like protein ISG15 (ISG15; degree=31), IRF1
(degree=31) and interferon-induced helicase C domain-containing
protein 1 (IFIH1; degree=2; Fig.
2). These 10 proteins are significant nodes in the network. A
previous study demonstrated that IFN signaling activates the
IRF/STAT signaling pathways through STAT1, leading to the
transformation of macrophages to the M1 type (21).
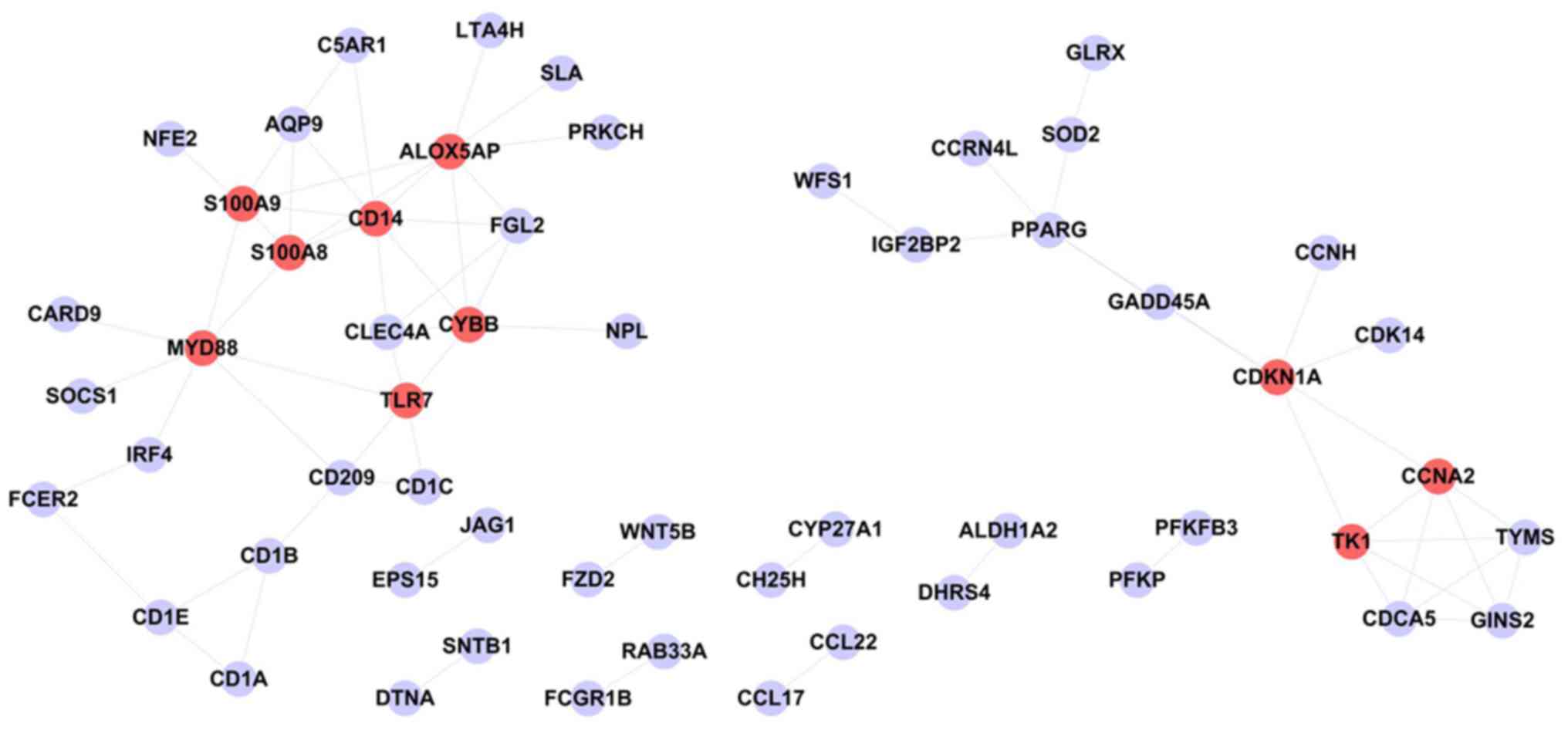
The PPI network based on the intersection of the
differentially expressed genes between the M2 and M0 groups is
demonstrated in Fig. 3. This
network consists of 56 protein nodes and 72 PPIs. The 10 highest
degree proteins in this PPI network were monocyte differentiation
antigen CD14 (CD14; degree=8), arachidonate
5-lipoxygenase-activating protein (ALOX5AP; degree=8), myeloid
differentiation primary response gene 88 (MYD88; degree=7),
cyclin-dependent kinase inhibitor 1A (CDKN1A; degree=6), protein
S100-A9 (S100A9; degree=32), cytochrome b-245 heavy chain (CYBB;
degree=6), cyclin-A2 (CCNA2; degree=5), protein S100-A8 (S100A8;
degree=5), thymidine kinase, cytosolic (TK1; degree=5) and TLR7
(degree=5).
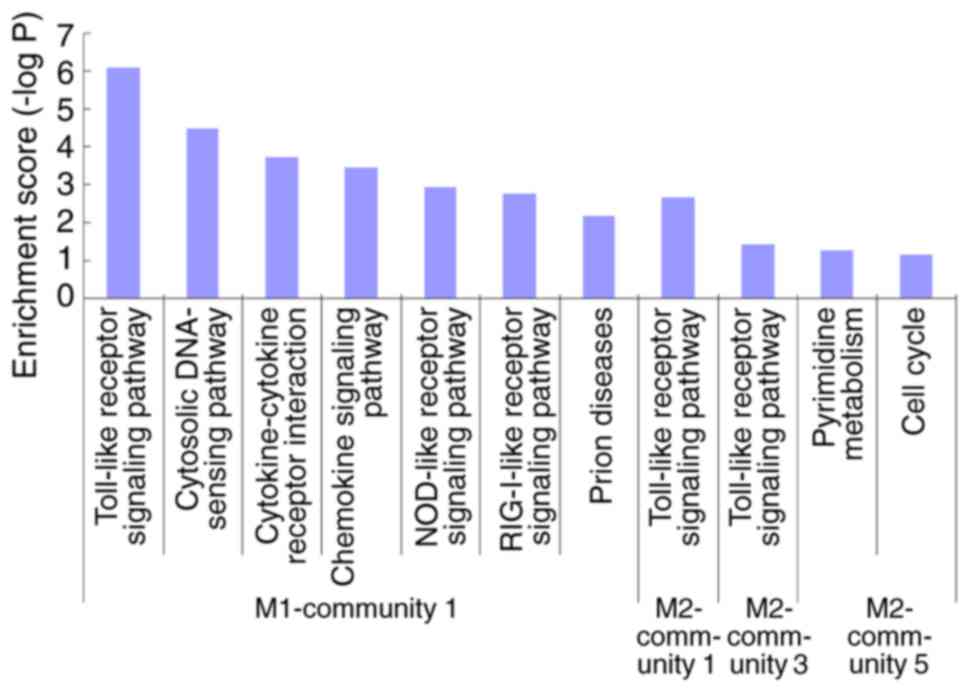
Modular analysis and KEGG functional
analysis of each module
In the M1 vs. M0 group, communities in the PPI
network were identified using CFinder software. When K=5, three
communities were identified, and there were 432, 15 and 14 pairs of
interacting genes in the first, second and third communities,
respectively.
In the M2 vs. M0 group, communities in the PPI
network were identified using CFinder software. When K=3, five
communities were identified, and there were 23, 3, 5, 3 and 12
pairs of interacting genes in the first, second, third, fourth and
fifth communities, respectively (data not shown).
The results from KEGG functional analysis of each
module are demonstrated in Fig. 4.
Enrichment was obtained in community 1 of the M1 group and in
communities 1, 3 and 5 of the M2 group. Community 1 of the M1 group
and communities 1 and 3 of the M2 group demonstrated functions
primarily enriched in the ‘toll-like receptor signaling pathway’.
Detailed information regarding gene-interaction pairs and the
degree of gene nodes in these three communities is presented in
Fig. 5.
Discussion
Macrophages are multifunctional cells that perform
different functions depending on the type and state of the tissues
where they reside. Dysregulation of macrophage functions may lead
to numerous diseases, including infectious diseases and immune
disorders. In addition, macrophages serve a role in the destruction
of endocrine pancreatic-cells in the autoimmune response of type 1
diabetes (21), metabolic diseases
(22) and malignancies (23). The transition between macrophage
polarization types serves a significant and pivotal role in the
progression of these diseases (24). Therefore, the identification of the
molecules and molecular groups associated with the dynamic
processes of macrophages is crucial for the elucidation of the
molecular basis of disease progression and the design of novel
macrophage-based therapeutic strategies.
The concept of macrophage polarization types has
been described as a dynamic, stepwise and continuous process of
alteration from M1 to M2 (25–27).
Macrophage polarization is classified into M1 (classical) and M2
(alternative) types, which is currently an intense area of research
in the study of macrophage function (3,28).
The present study employed gene expression profiling data of M1 and
M2 macrophages in the GEO database to evaluate protein-protein
interrelationships and construct PPI networks through a more
efficient method based on protein semantic similarity (29). However, it is critical to identify
functional modules in the PPI network for analysis with the aim of
identifying cell functions (30).
The subsequent modular and KEGG enrichment analyses of each module
demonstrated that the majority of the M1 and M2 modules were
primarily involved in the TLR signaling pathway, suggesting that
this pathway is involved in the regulation of macrophage M1 and M2
polarization.
Macrophages are the sensing cells of the immune
system and are crucial mediators of the inflammatory response.
TLRs, which serve an important role in the body's defense against
specific pathogenic microorganisms, are the best-characterized
inducers of acute inflammation (31). The initial characterization of
enhancers involved in LPS-inducible gene expression in macrophages
is based on the ability of stimulus-activated translational
factors, including NF-κB and IRFs (32). The TLR/NF-κB and Janus kinase/STAT
signaling pathways (33) are
involved in the regulation of macrophage polarization. In the
majority of cases, the TLR/NF-κB signaling pathway promotes M1-type
macrophage polarization when external microorganisms invade.
However, this signaling pathway may additionally select the type of
macrophage polarization based on the subunit composition of NF-κB.
When NF-κB is activated in the form of p65/p50, macrophages
increase pro-inflammatory cytokine production (34), and M1 macrophages are formed. When
NF-κB is activated in the form of p50/p50, M2 macrophages are
formed, which occurs in tumor-associated and LPS-tolerant
macrophages (35). In addition,
TLR serves an important role in the regulation of
post-transcriptional polarization of macrophages.
In the present study, the macrophage polarization
regulatory gene profile was efficiently extracted from the
accessible online database and a PPI network map of M1 and M2
macrophages, based on genes demonstrating significantly upregulated
expression identified by cluster analysis of differentially
expressed genes was constructed. The network map demonstrated that
the first 10 high-degree proteins in the PPI network of the M1-type
polarized macrophages were STAT1, GBP1, GBP5, CXCL10, IFIT2, IRF7,
IFIT3, ISG15, IRF1 and IFIH1. A PPI network map of M2-type
polarized macrophages was constructed using the same method, and
the first 10 high-degree proteins were CD14, ALOX5AP, MYD88,
CDKN1A, S100A9, CYBB, CCNA2, S100A8, TK1 and TLR7. The majority of
the high-degree proteins in the PPI network that are involved in
the molecular regulation of macrophage polarization are closely
associated with proteins involved in the TLR signaling pathway. It
was identified that the TLR7 signaling pathway serves an important
role in the regulation of macrophage polarization. These results
are consistent with a previous study, which demonstrated that the
TLR family regulates M1 and M2 differentiation, in addition to
demonstrating the potential importance of TLR7 in virus-induced M2
polarization (36). Furthermore,
other high-degree proteins identified during comparison not only
help suggest the modulating role of the TLR pathway during
macrophage polarization; however, may additionally function
individually in the program and link macrophage polarization to
crucial biological programs, including immune response and
malignancy progression.
In a previous study of mouse bone marrow-derived
macrophages, macrophages were mock transfected and their
transcriptional products analyzed (37). IFN responses were the primary
reactions identified at 5 and 24 h after transfection, and the
expression levels of IFN-mediated proteins (IFIH1, IFIT2 and
IFIT3), the GBP protein family (GBP1 and GBP5), the transcription
factor STAT1 and cytokine CXCL10 were detected. Increased
expression of IRF1 was additionally detected in the
transcriptional products of macrophages stimulated by IFN-γ
(37).
LPS, as well as IFN-γ and TNF Th1-type cytokines,
mediate the classical macrophage activation pathway (33). It has been demonstrated that LPS
mediates the expression of IFIH1, GBP5, IRF1, IRF7, IFIT2,
IFIT3 and CXCL10 and that their expression is due to
pro-inflammatory responses of M1 macrophages. The expression of a
number of genes stimulated by LPS is mediated by IFN-γ and not IL-4
or IL-10. These genes include GBP5, IFIH1, IFIT2, IFIT3 and
STAT1 (38). During the
replication of Orientia tsutsugamushi in macrophages, the
expression of genes involved in M1 macrophage polarization is
upregulated. When macrophages are infected by Orientia
tsutsugamushi, the expression of IFN-stimulated genes,
including CXCL10, IRF7 and ISG15, is increased
(39). Exposure to LPS increases
blood TNF levels via the canonical TLR4-associated NF-κB signaling
pathway, resulting in inflammation. TLR4 recognizes LPS in the
canonical NF-κB signaling pathway and initiates a signaling
cascade. This leads to the activation of NF-κB and the expression
of pro-inflammatory cytokines. Therefore, the complex
transcriptional programme induced in macrophages following LPS
stimulation is a product of the coordinated action of the
transcription factors (40), and
inhibition of the TLR4-NF-κB signaling pathway may shift M1
macrophage polarization toward the M2 phenotype.
The TLR signaling pathway upregulates the expression
of pro-inflammatory gene products by activating STAT1 and NF-κB
(41), thereby regulating M1
macrophage polarization. STAT1 has been demonstrated to be an
important regulator of the biological responses of different TLRs.
The expression of inflammatory cytokines mediated by TLR2, TLR4,
TLR7, TLR8 or TLR9 is suppressed in STAT1-deficient macrophages
(42). In addition, activation of
IRF3 is selectively coupled to TLR3 and TLR4 (43). Previous studies have demonstrated
that IFN is involved in the regulation of TLR-triggered gene
expression, including the TLR4 upregulation of IRF7
expression through TLR-, activator- and interferon-mediated
signaling pathways and the upregulation of IRF1 expression
through TLR9 (44,45). The present results further
suggested that TLR7 serves as a mediator of M2 macrophage
polarization. In contrast to the well-known macrophage
polarization-associated TLR transmembrane family members, including
TLR4, TLR7 is localized in the endosomal compartment, along with
TLR3/9, and is associated with the viral-induced immune response
(46). A recent study demonstrated
that Hepatitis C virus-induced TLR7 stimulation results in monocyte
differentiation and M2 macrophage polarization (36), which is consistent with the present
study and further suggests that TLR7 stimulated macrophage
polarization associated with chronic liver disease
pathogenesis.
A previous study demonstrated that CDKN1A, or p21,
serves an important role in the production of IL-1β and the
progression of inflammatory diseases (47). CDKN1A-deficient mice are more prone
to endotoxic shock, and CDKN1A-deficient macrophages produce more
IL-1β. CDKN1A was additionally demonstrated to suppress the
stimulatory effect of macrophages on inflammatory stimulation
factors, and CDKN1A-deficient macrophages produce more inflammatory
response factors, which promote M1 macrophage polarization
(47). In contrast, the presence
of CDKN1A regulates M2 macrophage polarization. It was identified
that CDKN1A inhibits the activation of macrophages by TLR (47). The carboxyl-terminal domain of
CDKN1A inhibits macrophage function by enhancing protein kinase B
phosphorylation, which suppresses p38 activation, thereby
inhibiting the production of inflammatory cytokines in macrophages
activated by TLR (48).
Consequently, CDKN1A limits macrophage activation in inflammatory
reactive diseases, including rheumatoid arthritis. Previous studies
demonstrated that CDKN1A-deficient macrophages exhibit enhanced
activation by TLR agonists compared with control macrophages,
regardless of the study background (47,49),
which confirms the inhibitory effect of CDKN1A on TLR.
CCNA2 is essential for the initiation of DNA
replication, transcription and regulation of the cell cycle and has
been reported to be a key regulator of cell differentiation
(50). It was reported that the
expression of microRNA (miR)-125b is downregulated during
macrophage TLR4 signaling (51).
TLR4 activates macrophages to produce pro-inflammatory cytokines;
whereas, mmu-miR-125b reduces the production of nitric oxide in
activated macrophages, promotes the growth of tumor cells, and, at
least partially, inhibits the expression of CCNA2 (52).
MYD88 is an adapter protein that transduces
intracellular signals induced by TLRs and IL-1 receptors (IL-1Rs)
and serves a critical role in TLR/IL-1R-mediated immune responses
(53). It has been demonstrated
that MYD88 is essential for most TLR signaling pathways (54) and serves an important role in the
activation of the signaling pathways induced by all TLRs/IL-1Rs
(except TLR3) (55). Therefore,
MYD88 is a suitable target for abnormal regulation of the TLR/IL-1R
signaling pathways under pathological conditions (56,57).
The absence of MYD88 in macrophages leads to a reduction in
pro-inflammatory cytokine production mediated by TLR2, TLR5, TLR7
and TLR9 (58–61).
In conclusion, the present study used bioinformatics
analysis to demonstrate that the TLR signaling pathway serves an
important role in the regulation of macrophage polarization and
that the high-degree proteins in the PPI network involved in
molecular regulation of macrophage polarization are closely
associated with proteins of the TLR signaling pathway, suggesting
that the TLR signaling pathway will be an important direction for
future studies of macrophage function in a systematic view. The
aforementioned proteins that were identified may serve as a primary
focus and may provide a useful reference for the intervention and
regulation of macrophage polarization.
Acknowledgements
Not applicable.
Funding
The present study was supported by National Natural
Science Foundation of China (grant nos. 81570579, 81602337 and
81702564).
Availability of data and materials
The datasets used and analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
BM, YY, ZL, DZ and WZ analyzed the Gene Expression
Omnibus database, conducted the cluster analysis of differentially
expressed genes and constructed the protein-protein interaction
networks. BM and YY were primary contributors in writing the
manuscript. YJ and DX conceived the study. All authors read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Murray PJ: Macrophage polarization. Annu
Rev Physiol. 79:541–566. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wynn TA, Chawla A and Pollard JW:
Macrophage biology in development, homeostasis and disease. Nature.
496:445–455. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Murray PJ and Wynn TA: Protective and
pathogenic functions of macrophage subsets. Nat Rev Immunol.
11:723–737. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
O'Shea JJ and Murray PJ: Cytokine
signaling modules in inflammatory responses. Immunity. 28:477–487.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Vergadi E, Ieronymaki E, Lyroni K,
Vaporidi K and Tsatsanis C: Akt signaling pathway in macrophage
activation and M1/M2 polarization. J Immunol. 198:1006–1014. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mantovani A, Biswas SK, Galdiero MR, Sica
A and Locati M: Macrophage plasticity and polarization in tissue
repair and remodelling. J Pathol. 229:176–185. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Grailer JJ, Haggadone MD, Sarma JV,
Zetoune FS and Ward PA: Induction of M2 regulatory macrophages
through the β2-adrenergic receptor with protection during
endotoxemia and acute lung injury. J Innate Immun. 6:607–618. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Labonte AC, Tosello-Trampont AC and Hahn
YS: The role of macrophage polarization in infectious and
inflammatory diseases. Mol Cells. 37:275–285. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Incio J, Tam J, Rahbari NN, Suboj P,
McManus DT, Chin SM, Vardam TD, Batista A, Babykutty S, Jung K, et
al: PlGF/VEGFR-1 signaling promotes macrophage polarization and
accelerated tumor progression in obesity. Clin Cancer Res.
22:2993–3004. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Castoldi A, Naffah de Souza C, Câmara NO
and Moraes-Vieira PM: The macrophage switch in obesity development.
Front Immunol. 6:6372016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kralova Lesna I, Petras M, Cejkova S,
Kralova A, Fronek J, Janousek L, Thieme F, Tyll T and Poledne R:
Cardiovascular disease predictors and adipose tissue macrophage
polarization: Is there a link? Eur J Prev Cardiol. 25:328–334.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fuentes-Duculan J, Suarez-Fariñas M, Zaba
LC, Nograles KE, Pierson KC, Mitsui H, Pensabene CA, Kzhyshkowska
J, Krueger JG and Lowes MA: A subpopulation of CD163-positive
macrophages is classically activated in psoriasis. J Invest
Dermatol. 130:2412–2422. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Beyer M, Mallmann MR, Xue J,
Staratschek-Jox A, Vorholt D, Krebs W, Sommer D, Sander J, Mertens
C, Nino-Castro A, et al: High-resolution transcriptome of human
macrophages. PLoS One. 7:e454662012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bolstad BM, Irizarry RA, Astrand M and
Speed TP: A comparison of normalization methods for high density
oligonucleotide array data based on variance and bias.
Bioinformatics. 19:185–193. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Marabita F, Almgren M, Lindholm ME,
Ruhrmann S, Fagerström-Billai F, Jagodic M, Sundberg CJ, Ekström
TJ, Teschendorff AE, Tegnér J and Gomez-Cabrero D: An evaluation of
analysis pipelines for DNA methylation profiling using the Illumina
HumanMethylation450 BeadChip platform. Epigenetics. 8:333–346.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Smyth GK, Michaud J and Scott HS: Use of
within-array replicate spots for assessing differential expression
in microarray experiments. Bioinformatics. 21:2067–2075. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Vallejos CA, Marioni JC and Richardson S:
BASiCS: Bayesian analysis of single-cell sequencing data. PLoS
Comput Biol. 11:e10043332015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Warnes GR: Gplots: Various R Programming
Tools for Plotting Data. Version 3.0.1. https://cran.r-project.org/web/packages/gplots/index.htmlMarch
30–2016
|
|
19
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Palla G, Derényi I, Farkas I and Vicsek T:
Uncovering the overlapping community structure of complex networks
in nature and society. Nature. 435:814–818. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sica A and Mantovani A: Macrophage
plasticity and polarization: In vivo veritas. J Clin Invest.
122:787–795. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Komohara Y, Fujiwara Y, Ohnishi K,
Shiraishi D and Takeya M: Contribution of macrophage polarization
to metabolic diseases. J Atheroscler Thromb. 23:10–17. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tariq M, Zhang J, Liang G, Ding L, He Q
and Yang B: Macrophage polarization: Anti-cancer strategies to
target tumor-associated macrophage in breast cancer. J Cell
Biochem. 118:2484–2501. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shapouri-Moghaddam A, Mohammadian S,
Vazini H, Taghadosi M, Esmaeili SA, Mardani F, Seifi B, Mohammadi
A, Afshari JT and Sahebkar A: Macrophage plasticity, polarization,
and function in health and disease. J Cell Physiol. 233:6425–6440.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mosser DM and Edwards JP: Exploring the
full spectrum of macrophage activation. Nat Rev Immunol. 8:958–969.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Geissmann F, Manz MG, Jung S, Sieweke MH,
Merad M and Ley K: Development of monocytes, macrophages, and
dendritic cells. Science. 327:656–661. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Noel W, Raes G, Hassanzadeh Ghassabeh G,
De Baetselier P and Beschin A: Alternatively activated macrophages
during parasite infections. Trends Parasitol. 20:126–133. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Davies LC, Jenkins SJ, Allen JE and Taylor
PR: Tissue-resident macrophages. Nat Immunol. 14:986–995. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cui G, Kim B, Alguwaizani S and Han K:
Assessing protein-protein interactions based on the semantic
similarity of interacting proteins. Int J Data Min Bioinform.
13:75–83. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yu L, Gao L and Sun PG: A hybrid
clustering algorithm for identifying modules in Protein-Protein
Interaction networks. Int J Data Min Bioinform. 4:600–615. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ullah MO, Sweet MJ, Mansell A, Kellie S
and Kobe B: TRIF-dependent TLR signaling, its functions in host
defense and inflammation, and its potential as a therapeutic
target. J Leukoc Biol. 100:27–45. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tong AJ, Liu X, Thomas BJ, Lissner MM,
Baker MR, Senagolage MD, Allred AL, Barish GD and Smale ST: A
stringent systems approach uncovers gene-specific mechanisms
regulating inflammation. Cell. 165:165–179. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhou D, Huang C, Lin Z, Zhan S, Kong L,
Fang C and Li J: Macrophage polarization and function with emphasis
on the evolving roles of coordinated regulation of cellular
signaling pathways. Cell Signal. 26:192–197. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bonizzi G and Karin M: The two NF-kappaB
activation pathways and their role in innate and adaptive immunity.
Trends Immunol. 25:280–288. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Porta C, Rimoldi M, Raes G, Brys L, Ghezzi
P, Di Liberto D, Dieli F, Ghisletti S, Natoli G, De Baetselier P,
et al: Tolerance and M2 (alternative) macrophage polarization are
related processes orchestrated by p50 nuclear factor kappaB. Proc
Natl Acad Sci USA. 106:14978–14983. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Saha B, Kodys K, Adejumo A and Szabo G:
Circulating and exosome-packaged hepatitis C single-stranded RNA
induce monocyte differentiation via TLR7/8 to polarized macrophages
and fibrocytes. J Immunol. 198:1974–1984. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lacaze P, Raza S, Sing G, Page D, Forster
T, Storm P, Craigon M, Awad T, Ghazal P and Freeman TC: Combined
genome-wide expression profiling and targeted RNA interference in
primary mouse macrophages reveals perturbation of transcriptional
networks associated with interferon signalling. BMC Genomics.
10:3722009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang S, Kim CC, Batra S, McKerrow JH and
Loke P: Delineation of diverse macrophage activation programs in
response to intracellular parasites and cytokines. PLoS Negl Trop
Dis. 4:e6482010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tantibhedhyangkul W, Prachason T, Waywa D,
El Filali A, Ghigo E, Thongnoppakhun W, Raoult D, Suputtamongkol Y,
Capo C, Limwongse C and Mege JL: Orientia tsutsugamushi stimulates
an original gene expression program in monocytes: Relationship with
gene expression in patients with scrub typhus. PLoS Negl Trop Dis.
5:e10282011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Medzhitov R and Horng T: Transcriptional
control of the inflammatory response. Nat Rev Immunol. 9:692–703.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Smale ST: Selective transcription in
response to an inflammatory stimulus. Cell. 140:833–844. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kim HS, Kim DC, Kim HM, Kwon HJ, Kwon SJ,
Kang SJ, Kim SC and Choi GE: STAT1 deficiency redirects IFN
signalling toward suppression of TLR response through a feedback
activation of STAT3. Sci Rep. 5:134142015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Smale ST and Natoli G: Transcriptional
control of inflammatory responses. Cold Spring Harb Perspect Biol.
6:a0162612014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Battistini A: Interferon regulatory
factors in hematopoietic cell differentiation and immune
regulation. J Interferon Cytokine Res. 29:765–780. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Aly S, Mages J, Reiling N, Kalinke U,
Decker T, Lang R and Ehlers S: Mycobacteria-induced granuloma
necrosis depends on IRF-1. J Cell Mol Med. 13:2069–2082. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Petes C, Odoardi N and Gee K: The toll for
trafficking: Toll-like receptor 7 delivery to the endosome. Front
Immunol. 8:10752017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Scatizzi JC, Mavers M, Hutcheson J, Young
B, Shi B, Pope RM, Ruderman EM, Samways DS, Corbett JA, Egan TM and
Perlman H: The CDK domain of p21 is a suppressor of
IL-1beta-mediated inflammation in activated macrophages. Eur J
Immunol. 39:820–825. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Mavers M, Cuda CM, Misharin AV, Gierut AK,
Agrawal H, Weber E, Novack DV, Haines GK III, Balomenos D and
Perlman H: Cyclin-dependent kinase inhibitor p21, via its
C-terminal domain, is essential for resolution of murine
inflammatory arthritis. Arthritis Rheum. 64:141–152. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Trakala M, Arias CF, Garcia MI,
Moreno-Ortiz MC, Tsilingiri K, Fernández PJ, Mellado M, Díaz-Meco
MT, Moscat J, Serrano M, et al: Regulation of macrophage activation
and septic shock susceptibility via p21(WAF1/CIP1). Eur J Immunol.
39:810–819. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Anger M, Bryja V, Jirmanova L, Hampl A,
Carrington M, Motlik J, Dvorak P and Kubelka M: The appearance of
truncated cyclin A2 correlates with differentiation of mouse
embryonic stem cells. Biochem Biophys Res Commun. 302:825–830.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Tili E, Michaille JJ, Cimino A, Costinean
S, Dumitru CD, Adair B, Fabbri M, Alder H, Liu CG, Calin GA and
Croce CM: Modulation of miR-155 and miR-125b levels following
lipopolysaccharide/TNF-alpha stimulation and their possible roles
in regulating the response to endotoxin shock. J Immunol.
179:5082–5089. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Xu Z, Zhao L, Yang X, Ma S, Ge Y, Liu Y,
Liu S, Shi J and Zheng D: Mmu-miR-125b overexpression suppresses NO
production in activated macrophages by targeting eEF2K and CCNA2.
BMC Cancer. 16:2522016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Deguine J and Barton GM: MyD88: A central
player in innate immune signaling. F1000Prime Rep. 6:972014.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
O'Neill LA and Bowie AG: The family of
five: TIR-domain-containing adaptors in Toll-like receptor
signalling. Nat Rev Immunol. 7:353–364. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Alexopoulou L, Holt AC, Medzhitov R and
Flavell RA: Recognition of double-stranded RNA and activation of
NF-kappaB by Toll-like receptor 3. Nature. 413:732–738. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Loiarro M, Ruggiero V and Sette C:
Targeting TLR/IL-1R signalling in human diseases. Mediators
Inflamm. 2010:6743632010. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Loiarro M, Ruggiero V and Sette C:
Targeting the Toll-like receptor/interleukin 1 receptor pathway in
human diseases: Rational design of MyD88 inhibitors. Clin Lymphoma
Myeloma Leuk. 13:222–226. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Hayashi F, Smith KD, Ozinsky A, Hawn TR,
Yi EC, Goodlett DR, Eng JK, Akira S, Underhill DM and Aderem A: The
innate immune response to bacterial flagellin is mediated by
Toll-like receptor 5. Nature. 410:1099–1103. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Hemmi H, Kaisho T, Takeuchi O, Sato S,
Sanjo H, Hoshino K, Horiuchi T, Tomizawa H, Takeda K and Akira S:
Small anti-viral compounds activate immune cells via the TLR7
MyD88-dependent signaling pathway. Nat Immunol. 3:196–200. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Schnare M, Holt AC, Takeda K, Akira S and
Medzhitov R: Recognition of CpG DNA is mediated by signaling
pathways dependent on the adaptor protein MyD88. Curr Biol.
10:1139–1142. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Takeuchi O, Kaufmann A, Grote K, Kawai T,
Hoshino K, Morr M, Mühlradt PF and Akira S: Cutting edge:
Preferentially the R-stereoisomer of the mycoplasmal lipopeptide
macrophage-activating lipopeptide-2 activates immune cells through
a toll-like receptor 2- and MyD88-dependent signaling pathway. J
Immunol. 164:554–557. 2000. View Article : Google Scholar : PubMed/NCBI
|