Introduction
Glioma is one of the most common types of malignant
tumor in the central nervous system, which accounts for ~29% of all
types of brain tumor and contributes to cancer-associated mortality
rates (1). Characteristics of
glioma are high incidence and mortality rates, high recurrence and
low cure rates. It is difficult to diagnose and treat glioma at an
early stage due to the lack of auxiliary examination indexes.
Although advances have been made in the diagnostic and therapeutic
strategies for glioma, the prognoses of patients with glioma remain
poor, particularly for those with glioblastoma, with an estimated
2-year survival rate of 15% (2).
Epithelial-to-mesenchymal transition (EMT) is a
biological process in which epithelial cells are transformed to
mesenchymal-like cells. Throughout the EMT process, epithelial
cells lose their connectivity to basement membranes and acquire
improved migratory and invasive capabilities. EMT, which is
regulated by complex networks, is an important process in
development. It also serves important roles in the tumorigenic
process, resulting in improvement of the migration and invasion of
tumor cells (3–5).
Forkhead box (FOX) proteins are a class of conserved
transcription factors, which are implicated in various biological
and physiological processes. The dysregulation of FOX proteins is
also implicated in tumor progression; therefore, FOX proteins are
considered potential diagnostic markers or therapeutic targets for
cancer (6,7). As a member of the FOX protein family,
FOXC1 is located on chromosome 6p25. FOXC1 contains a conserved
forkhead domain, which binds upstream of target genes, thus
promoting gene activation. FOXC1 has critical roles in
physiological processes, including growth, development and
differentiation (8–11). FOXC1 also serves important roles in
pathological conditions and the dysregulation of FOXC1 additionally
contributes to tumorigenesis. Previous evidence has revealed that
FOXC1 is involved in tumor development (7). Notably, FOXC1 has been demonstrated
to be overexpressed in breast cancer and lung cancer, and is
correlated with the poor survival rates of these cancer types
(12–15). Furthermore, FOXC1 has been reported
to regulate the growth, metastasis and differentiation of several
types of cancer (16–20). Therefore, FOXC1 is regarded as a
potential biomarker for certain cancer types.
FOXC1 is associated with the proliferation and
migration of various types of cancer (15,21)
and is regarded as an EMT inducer. It has previously been reported
that FOXC1 is highly expressed in glioma (22); however, to the best of our
knowledge, there have been no previous studies on the association
between FOXC1 and glioma. The present study aimed to examine the
function of FOXC1 in glioma cells and to investigate the underlying
mechanism.
Materials and methods
Materials
FOXC1 small interfering (si)RNA1 (target sequence:
5′-CCACTGCAACCTGCAAGCCAT−3′), FOXC1 siRNA2 (target sequence:
5′-GCCGCACCATAGCCAGGGCTT-3′) and negative control (NC) siRNA
(5′-TTCTCCGAACGTGTCACGTTT-3′) were obtained from Shanghai
GenePharma Co., Ltd. (Shanghai, China). β-catenin overexpression
(OE) plasmid and empty vector were obtained from Addgene, Inc.
(cat. no. 19286; Cambridge, MA, USA). Antibodies against FOXC1
(cat. no. 55365-1-AP), N-cadherin (cat. no. 22018-1-AP), E-cadherin
(cat. no. 20874-1-AP), Vimentin (cat. no. 10366-1-AP), Snail (cat.
no. 13099-1-AP), Twist (cat. no. 25465-1-AP), β-catenin (cat. no.
17565-1-AP) and c-myc (cat. no. 10828-1-AP) were purchased from
Wuhan Sanying Biotechnology (Wuhan, China). Antibodies against
phosphorylated (p)-β-catenin (cat. no. 4176) and GAPDH (cat. no.
2118) were purchased from Cell Signaling Technology, Inc. (Danvers,
MA, USA).
Cell culture
U251 cells and SHG44 cells were obtained from
Procell Life Science and Technology Co., Ltd. (Wuhan, China). U251
cells were grown in Dulbecco's modified Eagle's medium (Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with
10% fetal bovine serum (FBS; Biological Industries, Kibbutz Beit
Haemek, Israel). SHG44 cells were grown in RPMI-1640 medium (Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 10% FBS. All
cells were cultured in a cell incubator at 37°C in an atmosphere
containing 5% CO2.
Transfection
Cells were seeded into a 6-well plate
(4×105 cells/well). A total of 1 h prior to
transfection, the medium was replaced with fresh serum-free medium.
A total of 100 pmol FOXC1 siRNA1, FOXC1 siRNA2 or negative control
(NC) siRNA were transfected into cells using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). After 4 h at 37°C, the cell medium was replaced
with fresh cell medium containing 10% FBS. Subsequently, the cells
were subjected to reverse transcription-quantitative polymerase
chain reaction (RT-qPCR) 24 h post-transfection or western blotting
48 h post-transfection. For co-transfection, cells were
co-transfected with 1 µg empty vector or β-catenin OE plasmid
alongside 50 pmol NC siRNA or FOXC1 siRNA2 using
Lipofectamine® 2000, as aforementioned.
RT-qPCR
TRIpure lysis buffer (BioTeke Corporation, Beijing,
China) was used to extract total RNA, according to the
manufacturer's protocol. Subsequently, total RNA was reverse
transcribed to cDNA using Super M-MLV reverse transcriptase
(BioTeke Corporation) according to the manufacturer's protocol.
FOXC1 mRNA expression levels were detected using the SYBR-Green
method. SYBR Green was obtained from Beijing Solarbio Science and
Technology Co., Ltd. (Beijing, China). The following primers were
used: FOXC1 forward, 5′-CAGAACAGCATCCGCCACA-3′ and reverse,
5′-TGTTGTAGGAGTCCGGGTC-3′; and GAPDH forward,
5′-GAAGGTCGGAGTCAACGGAT-3′ and reverse,
5′-CCTGGAAGATGGTGATGGGAT-3′. The thermocycling conditions were 94°C
for 5 min; 40 cycles of 94°C for 10 sec, 60°C for 20 sec, and 72°C
for 30 sec; then 72°C for 2.5 min and 40°C for 1.5 min; melting
from 60°C to 94°C, 1°C/sec. The mRNA expression levels of FOXC1
were calculated using the 2−ΔΔCq method (23).
Western blot analysis
Proteins were extracted using
radioimmunoprecipitation assay lysis buffer and protein
concentration was measured using a Bicinchoninic Acid Protein Assay
kit (both Beyotime Institute of Biotechnology, Shanghai, China).
Subsequently, 40 µg protein in each group were separated by 8, 10
or 12% SDS-PAGE and the separated proteins were transferred onto
polyvinylidene fluoride (PVDF) membranes (EMD Millipore, Billerica,
MA, USA). The PVDF membranes were blocked with 5% skim milk or 1%
bovine serum albumin (Biosharp, Hefei, China) at room temperature
for 1 h, and then incubated with FOXC1 (1:1,000), N-cadherin
(1:1,000), E-cadherin (1:1,000), Vimentin (1:1,000), Snail
(1:1,000), Twist (1:1,000), β-catenin (1:1,000), p-β-catenin
(1:1,000), c-myc (1:1,000) and GAPDH antibodies (1:1,000) overnight
at 4°C. The PVDF membranes were rinsed and then incubated with
corresponding horseradish peroxidase-labeled secondary antibodies
(cat. no. A0208; 1:5,000; Beyotime Institute of Biotechnology) for
45 min at 37°C. Subsequently, the PVDF membranes were visualized
using an enhanced chemiluminescent kit (Beyotime Institute of
Biotechnology). The target bands were scanned and analyzed by
Gel-Pro-Analyzer software version 4.0 (Media Cybernetics, Inc.,
Rockville, MD, USA).
MTT assay
Cells were seeded in 96-well plates
(4×103 cells in each well) and were transfected with NC
siRNA, FOXC1 siRNA1 or FOXC1 siRNA2. MTT (Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany) at a final concentration of 0.5 mg/ml was
added to each well at 12, 24, 36, 48 and 72 h. After culturing for
4 h at 37°C, 150 µl dimethyl sulfoxide (Sigma-Aldrich; Merck KGaA)
was added to each well following the removal of cell medium.
Absorbance was measured at 570 nm using a microplate reader (BioTek
Instruments, Inc., Winooski, VT, USA).
Wound healing assay
Cells were seeded into a 6-well plate
(4×105 cells/well) and were transfected with negative
control siRNA, FOXC1 siRNA1 or FOXC1 siRNA2. After 24 h, when the
cell confluence reached 80%, cells were treated with mitomycin C (1
µg/ml; Sigma-Aldrich; Merck KGaA) for 1 h at 37°C. Subsequently,
wounds were created on the surface of the cell monolayer with a
200-µl pipette tip. Cells were cultured in a cell incubator and the
images were captured under an inverted microscope with
magnification, ×100 at 0 h and 24 h. The relative migration ratio
was calculated as follows: Relative migration ratio=(incipient gap
between two edges-migrated gap between two edges)/incipient gap
between the two edges.
Transwell assay
Transwell inserts were purchased from Corning
Incorporated (Corning, NY, USA). Matrigel was purchased from BD
Biosciences (San Jose, CA, USA). A total of 4×103 cells
in 200 µl cell medium were added into Transwell inserts (precoated
with Matrigel) and 800 µl medium supplemented with 30% FBS was
added into the lower chambers. Then the cells were transfected with
the negative control siRNA, FOXC1 siRNA1 or FOXC1 siRNA2, or
co-transfection with empty vector or β-catenin OE plasmid and
negative control siRNA or FOXC1 siRNA2. Thereafter, the cells were
cultured in a cell incubator and allowed to invade for 24 h. After
rinsing, cells on top of the membranes were removed. Cells that had
invaded through the membranes were fixed with 4% paraformaldehyde
at room temperature for 20 min and stained with 0.5% crystal violet
for 5 min. Images were captured under an inverted microscope (Motic
Instruments, Richmond, BC, Canada) with a magnification, ×200.
Immunofluorescence
Cells were seeded onto slides in 12-well plates
(5×104 cells/well) and were transfected with negative
control siRNA, FOXC1 siRNA1 or FOXC1 siRNA2. A total of 24 h
post-transfection, cells were fixed with 4% paraformaldehyde for 15
min at room temperature and permeabilized with 0.1% Triton X-100
for 30 min at room temperature. The cells were blocked with goat
serum (Beijing Solarbio Science and Technology Co., Ltd.) for 15
min at room temperature and were then incubated with a primary
antibody against N-cadherin (cat. no. 13116; 1:200; Cell Signaling
Technology, Inc.) overnight at 4°C. After rinsing, the cells were
incubated with a Cy3-labeled secondary antibody (cat. no. A0516;
1:400; Beyotime Institute of Biotechnology) for 60 min at room
temperature. Subsequently, the cells were rinsed, stained with DAPI
(Sigma-Aldrich; Merck KGaA), and observed under a fluorescence
microscope (Olympus Corporation, Tokyo, Japan) with magnification,
×400.
Statistical analysis
Each experiment was repeated three times and the
results are presented as the means ± standard deviation.
Differences between groups were analyzed using a one-way or two-way
analysis of variance, followed by Bonferroni's multiple comparison
as a post-hoc test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Silencing FOXC1 inhibits the
proliferation, migration and invasion of glioma cells
To examine the function of FOXC1 in glioma,
FOXC1-specific siRNAs were used in the present study, and the
expression levels of FOXC1 were detected in U251 and SHG44 cells.
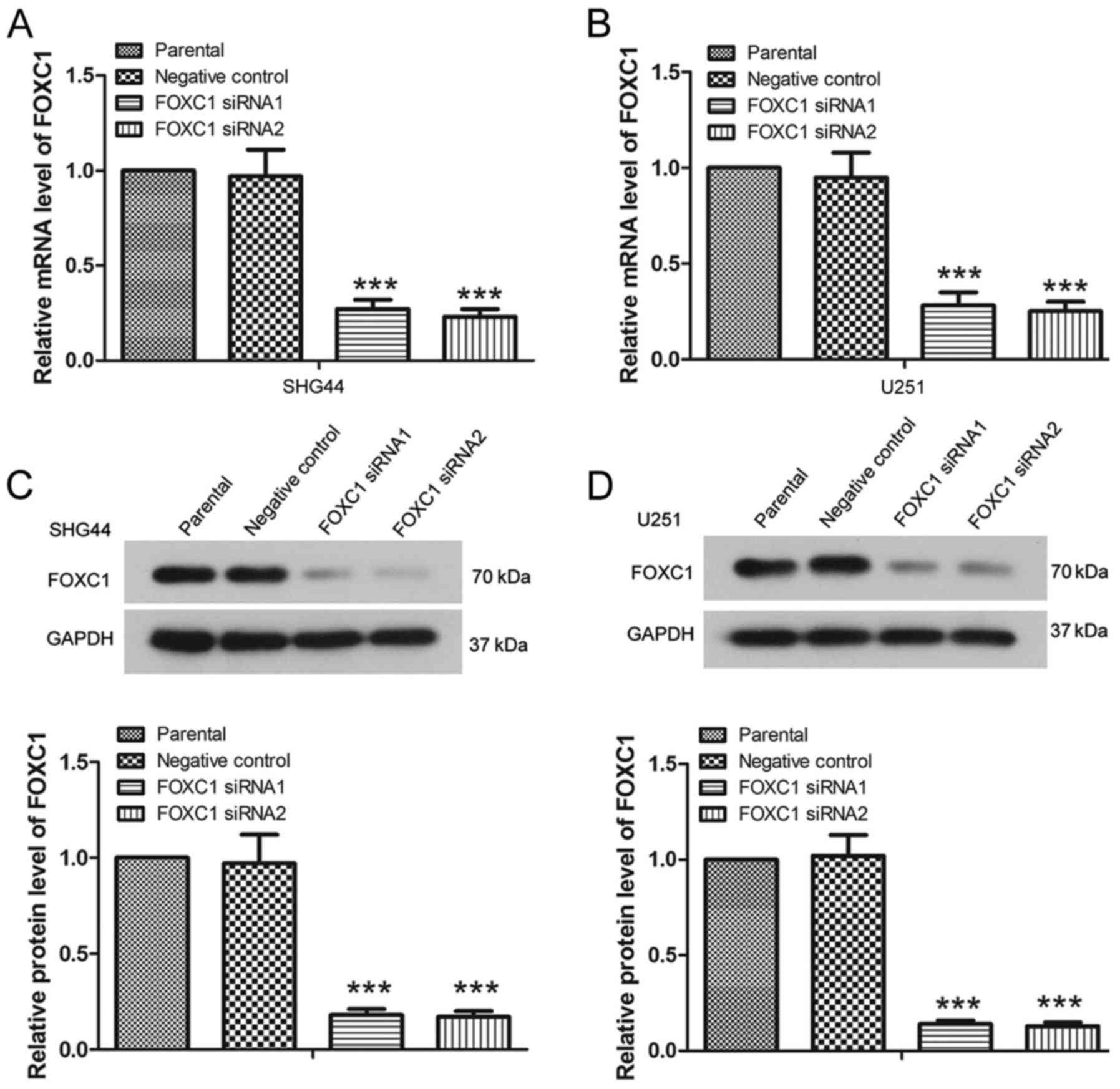
The RT-qPCR results revealed that, post-transfection with FOXC1
siRNA1 or FOXC1 siRNA2, the mRNA expression levels of FOXC1 were
significantly decreased (Fig. 1A and
B) (P<0.001). The results of western blot analysis also
revealed a significant decrease in the protein expression levels of
FOXC1 in U251 cells and SHG44 cells in response to FOXC1 siRNA
compared with in the NC siRNA group (Fig. 1C and D) (P<0.001). These results
indicated that FOXC1 siRNA1 and FOXC1 siRNA2 effectively decreased
FOXC1 expression.
The effects of FOXC1 silencing were also detected on
the proliferation, migration and invasion of glioma cells. An MTT
assay revealed that, post-transfection with FOXC1 siRNA1 or FOXC1
siRNA2, the proliferation of SHG44 and U251 cells was reduced
compared with those transfected with the negative control siRNA
(Fig. 2A and B). These results
revealed that silencing FOXC1 inhibited the proliferation of glioma
cells. A wound healing assay demonstrated that, compared with the
negative control siRNA group, the relative migration ratio of
glioma cells was decreased by FOXC1 siRNA1 and FOXC1 siRNA2
(Fig. 2C). In addition, a
Transwell assay revealed that the ratio of invasive cells was
decreased by FOXC1 siRNA1 and FOXC1 siRNA2 compared with the
negative control siRNA (Fig. 2D).
These results revealed that silencing FOXC1 inhibited the migration
and invasion of glioma cells.
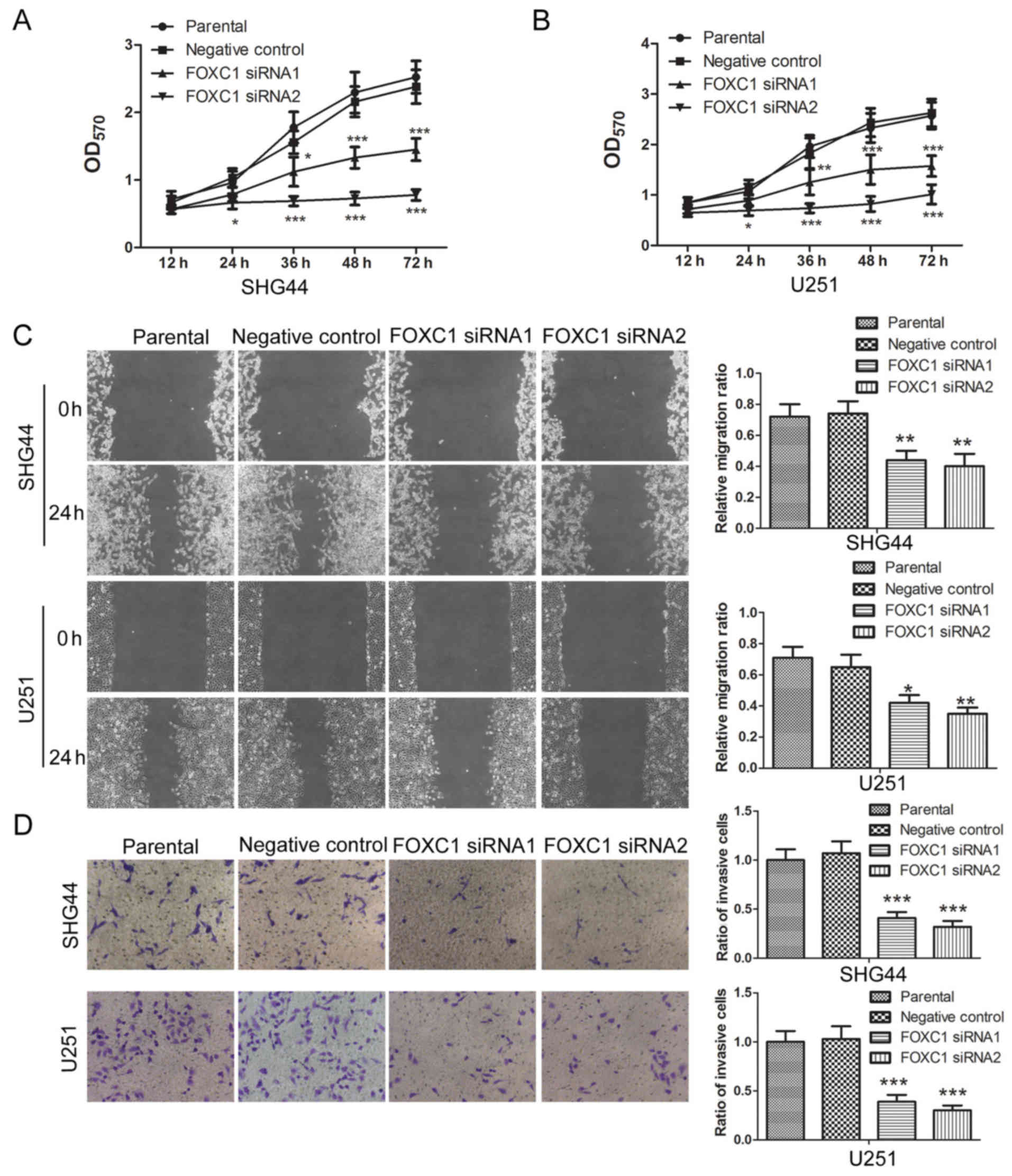 | Figure 2.Silencing of FOXC1 inhibits the
proliferation, migration and invasion of glioma cells. (A and B)
Post-transfection with FOXC1 siRNA1 or FOXC1 siRNA2, the
proliferation of SHG44 and U251 cells was assessed by an MTT assay.
(C) Migratory capability of SHG44 and U251 cells was assessed by a
wound healing assay, after which, the relative migration ratio was
calculated. (D) Invasive capability of glioma cells was assessed
using a Transwell assay, after which, the ratio of invasive cells
was calculated. All experiments were repeated three times and the
results are presented as the means ± standard deviation.
*P<0.05, **P<0.01, ***P<0.001 vs. the negative control
siRNA group. FOXC1, forkhead box C1; OD, optical density; siRNA,
small interfering RNA. |
FOXC1 modulates the expression of
EMT-associated proteins
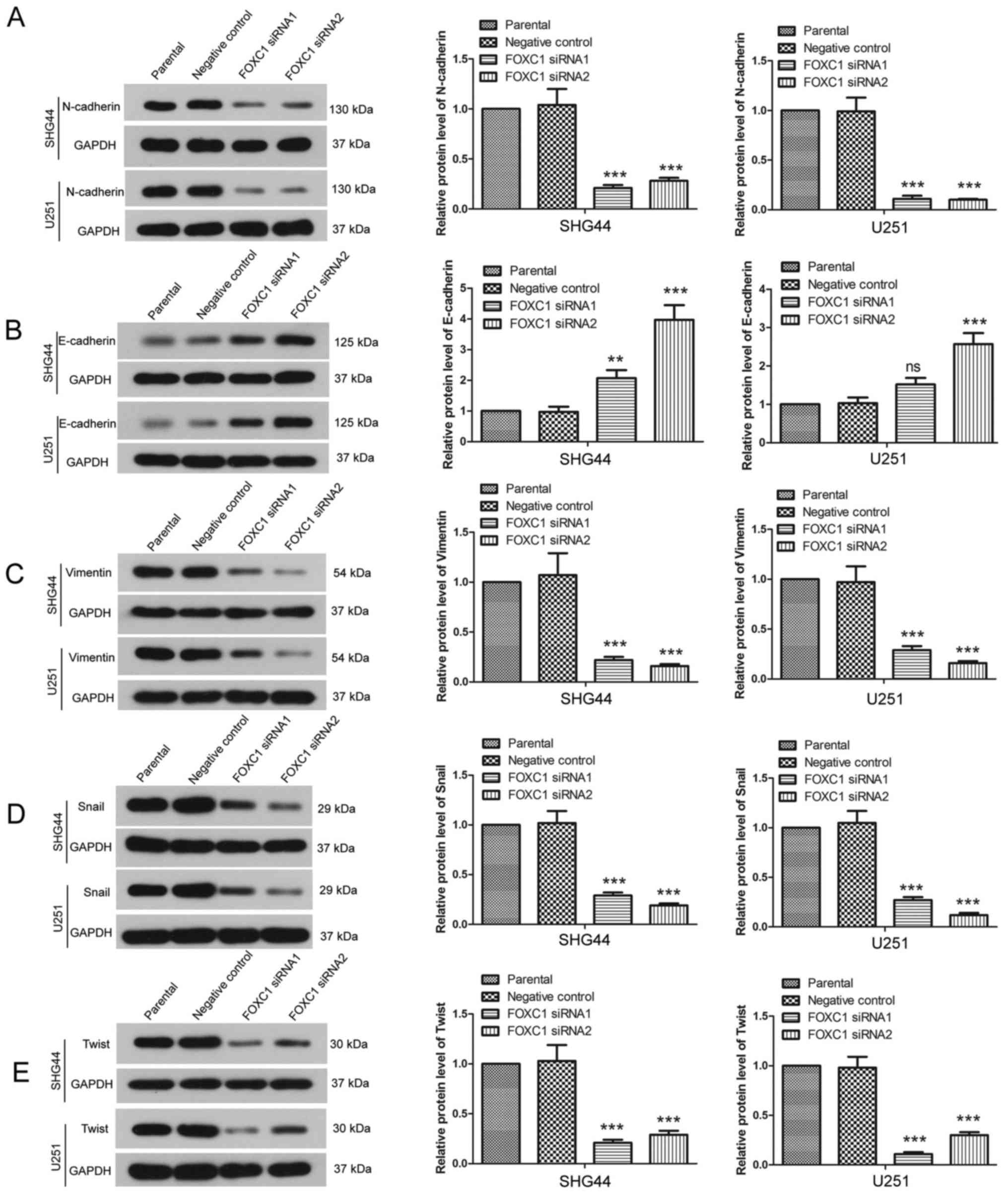
Since EMT contributes to the migration and invasion
of cancer cells, the effects of FOXC1 silencing on the expression
of EMT-associated proteins were detected by western blot analysis.
As presented in Fig. 3A and B, in
SHG44 and U251 cells, the expression levels of N-cadherin were
decreased by FOXC1 siRNAs, whereas the expression levels of
E-cadherin were increased by FOXC1 silencing. In addition, the
expression levels of Vimentin were decreased post-transfection with
FOXC1 siRNAs (Fig. 3C), and the
protein expression levels of Snail and Twist were also decreased
(Fig. 3D and E). Furthermore, the
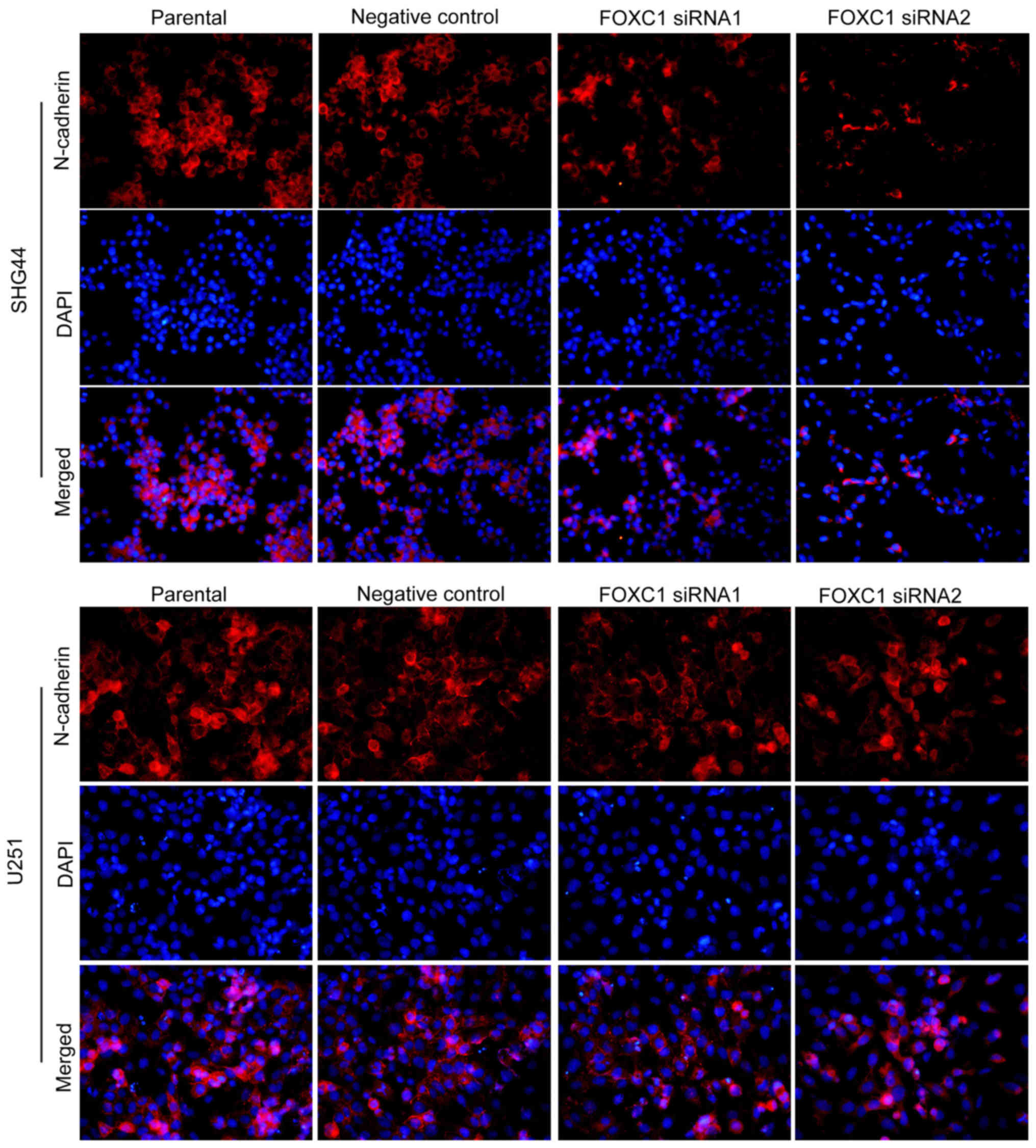
expression and distribution of N-cadherin were detected by
immunofluorescence. Consistent with the results of western blot
analysis, immunofluorescence analysis revealed that, following
silencing of FOXC1, N-cadherin expression was reduced, particularly
in the cell membranes (Fig. 4).
These results revealed that FOXC1 silencing modulated the
expression of EMT-associated proteins.
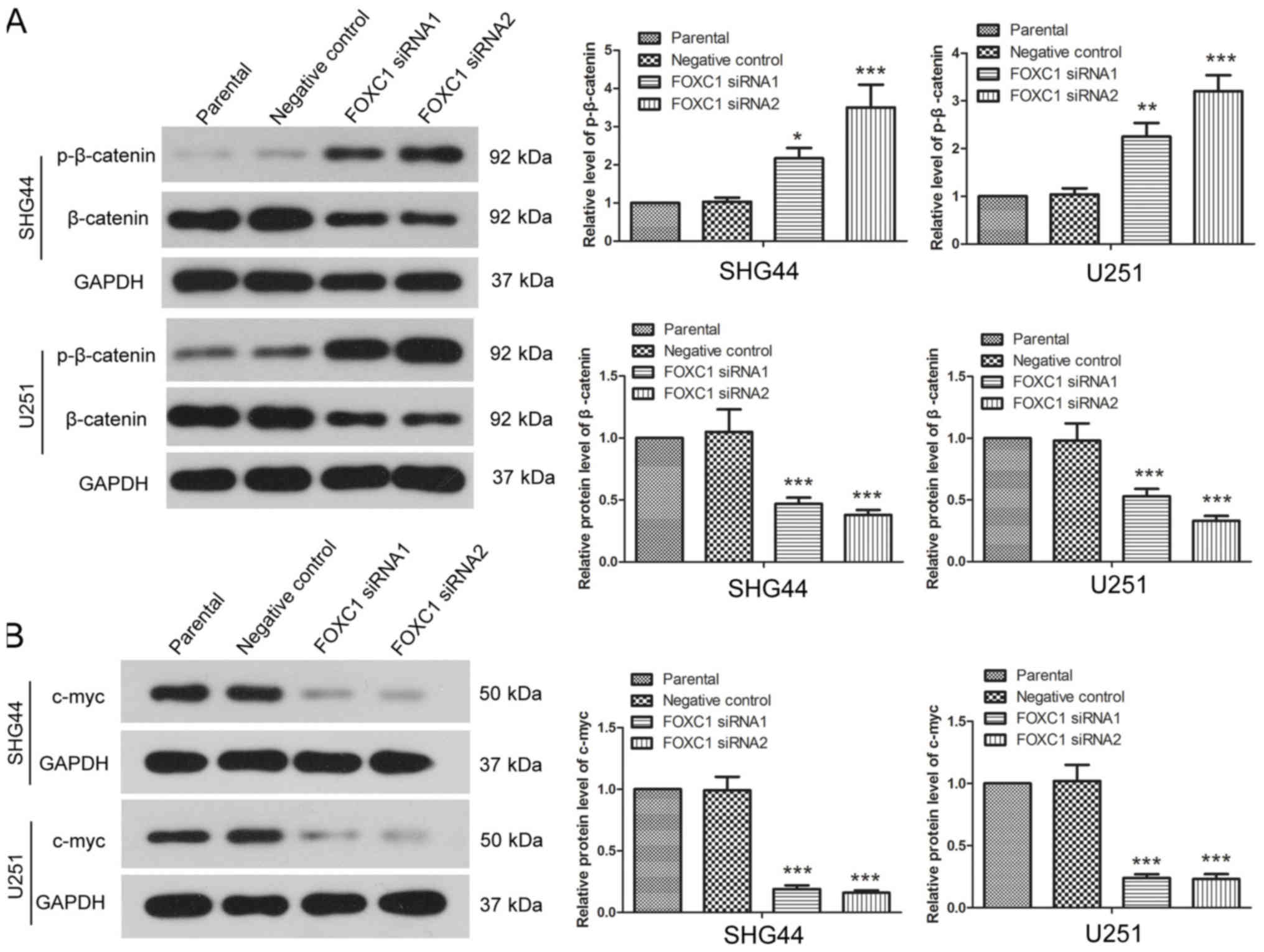
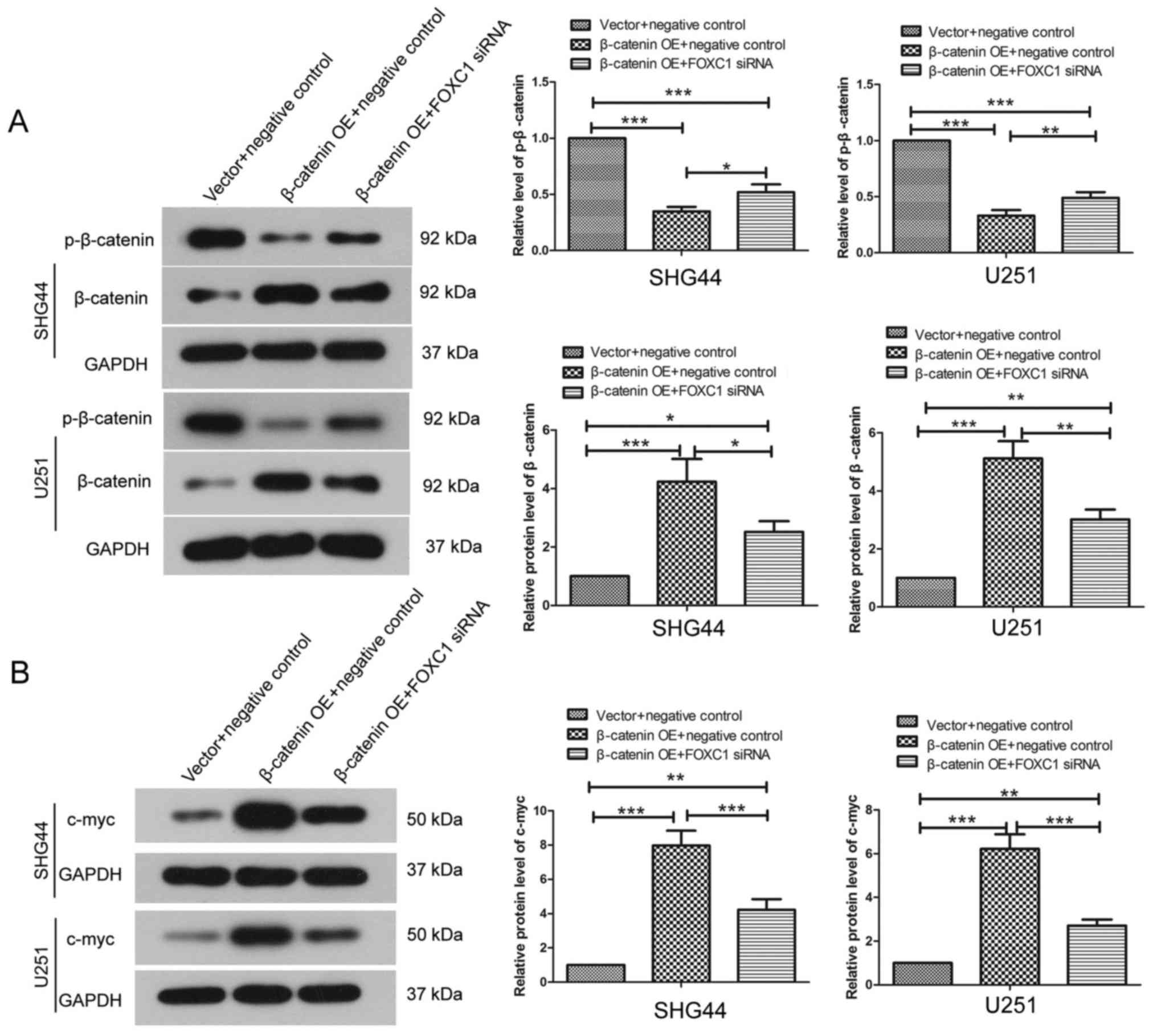
FOXC1 silencing affects β-catenin
signaling
The expression and phosphorylation levels of
β-catenin were detected by western blot analysis. Post-transfection
with FOXC1 siRNA1 and FOXC1 siRNA2, the expression levels of
p-β-catenin were increased in SHG44 and U251 cells, whereas the
protein expression levels of total β-catenin were significantly
decreased compared with the negative control siRNA group (Fig. 5A). The protein expression levels of
c-myc, which is a downstream target of β-catenin signaling, were
also detected by western blotting. After silencing FOXC1, the
expression levels of c-myc in SHG44 and U251 cells were decreased
compared with the negative control siRNA group (Fig. 5B). These results revealed that
silencing FOXC1 affected β-catenin signaling.
β-catenin signaling is involved in the
effects of FOXC1 silencing
Since FOXC1 silencing inhibited β-catenin signaling,
a β-catenin OE plasmid was used in the present study. β-catenin OE
plasmid and FOXC1 siRNA were co-transfected into cells, and the
protein expression levels of β-catenin, p-β-catenin and c-myc were
detected by western blot analysis. As presented in Fig. 6, following OE of β-catenin, the
protein expression levels of β-catenin were increased, whereas the
expression levels of p-β-catenin were decreased compared with the
negative control group. Co-transfection with β-catenin OE plasmid
and FOXC1 siRNA increased the expression levels of β-catenin, but
decreased the levels of p-β-catenin compared with the empty vector
and negative control siRNA group (Fig.
6A). In addition, co-transfection with β-catenin OE plasmid and
FOXC1 siRNA increased the protein expression levels of c-myc
compared with the empty vector and negative control siRNA group
(Fig. 6B), indicating that
co-transfection with β-catenin OE and FOXC1 siRNA enhanced the
activation of β-catenin signaling. However, our previous data
(Fig. 5) demonstrated that FOXC1
silencing alone suppressed β-catenin signaling, which was
distinctly different from the effects of co-transfection with
β-catenin OE and FOXC1 siRNA, suggesting that β-catenin OE may
reverse the effects of FOXC1 silencing on β-catenin signaling.
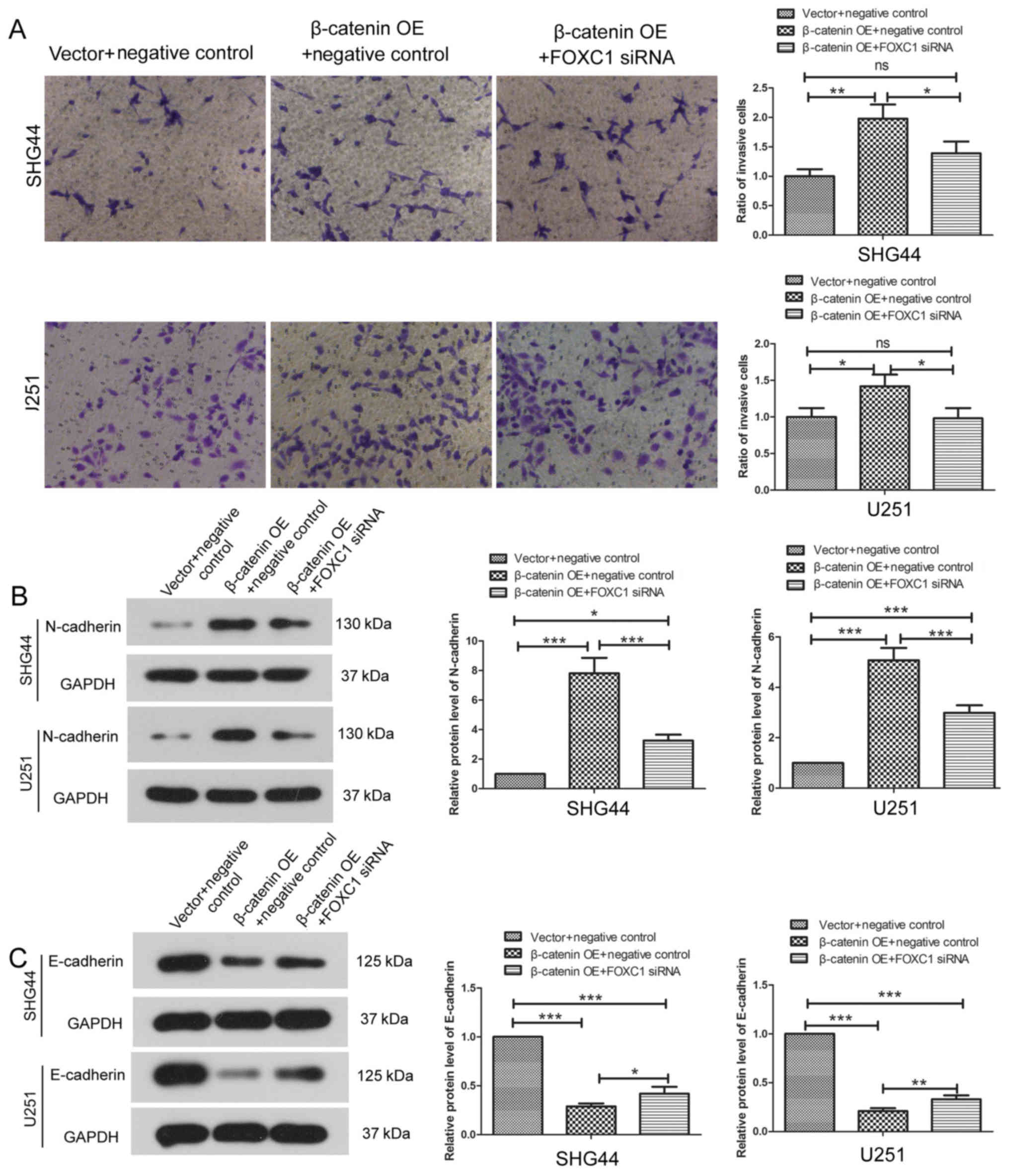
Following co-transfection with β-catenin OE plasmid
and FOXC1 siRNA, the invasion of SHG44 cells and U251 cells was
assessed using a Transwell assay. β-catenin OE enhanced the
invasive capability of SHG44 and U251 cells. However, following
co-transfection with β-catenin OE and FOXC1 siRNA, the invasive
capability of SHG44 and U251 cells demonstrated no significant
difference compared with cells co-transfected with the empty vector
and negative control siRNA (Fig.
7A). Our previous data in Fig.
2 showed that FOXC1 silencing alone inhibited the invasion of
glioma cells. These results indicated that β-catenin OE may abolish
the effects of FOXC1 silencing on the invasion of glioma cells.
Additionally, compared with the empty vector and negative control
siRNA group, the expression levels of N-cadherin were increased and
the expression levels of E-cadherin were decreased following
co-transfection with β-catenin OE and FOXC1 siRNA (Fig. 7B and C). However, our previous data
(Fig. 3) demonstrated that FOXC1
silencing alone decreased that levels of N-cadherin and increased
the levels of E-cadherin. These results provide further evidence
supporting the hypothesis that β-catenin signaling is involved in
the inhibitory effects of FOXC1 silencing on the EMT of glioma
cells.
Discussion
Glioma is a type of malignant brain cancer, which is
difficult to diagnose and cure at an early stage. FOXC1 has been
revealed to be highly expressed in glioma (22,24).
In the present study, it was demonstrated that silencing FOXC1
inhibited the proliferation, migration and invasion of glioma
cells. Further experiments revealed that β-catenin signaling was
involved in the effects of FOXC1 silencing.
Growth of cancer cells is necessary for
tumorigenesis. In numerous cancer types, FOXC1 functions as an
oncogene; silencing FOXC1 inhibits the proliferation of cancer
cells (15,21,25,26),
whereas OE of FOXC1 promotes the proliferation of cancer cells
(19,27). In the present study, silencing
FOXC1 was revealed to suppress the proliferation of glioma cells,
indicating that FOXC1 may function as an oncogene and promote the
growth of glioma. The cell cycle is a crucial factor impacting
cancer cell growth. FOXC1 silencing has been revealed to arrest the
cell cycle of non-small-cell lung carcinoma cells at the
G0/G1 phase and regulate cyclin D1 expression
levels (15). FOXC1 also exerts
effects on cancer cell apoptosis; knockdown of FOXC1 induces
apoptosis in cervical cancer and endometrial cancer (25,26).
These findings indicated that the effects of FOXC1 silencing on
glioma cell growth may be associated with its effects on the cell
cycle and apoptosis of glioma. However, future studies
investigating this implication are required.
Migration and invasion additionally contribute to
tumor development. The invasion of glioblastoma into adjacent
normal tissues often results in incomplete resection. Liu et
al (22) revealed that,
through targeting FOXC1, microRNA-133 is able to inhibit the
proliferation and invasion of glioma, indicating that FOXC1 may
function as an oncogene in glioma. In the present study, silencing
FOXC1 was revealed to inhibit the migration and invasion of glioma
cells, indicating that FOXC1 may regulate the metastasis of glioma.
The present study provided direct evidence to suggest that FOXC1
performs as an oncogene in glioma cells. Consistently, the
expression of FOXC1 is positively correlated with lymph node
metastasis and the distant metastasis of nasopharyngeal cancer
(18). FOXC1 also affects the
migration and invasion of cervical cancer, endometrial cancer,
osteosarcoma and melanoma (19,21,25–27).
The results of the present study verified the hypothesis that FOXC1
functions as an oncogene in glioma at the cellular level; however,
a lack of in vivo data is a limitation of this study.
EMT is a process that enables epithelial cells to
lose their cell-cell adhesion, and gain migratory and invasive
properties. EMT also serves a crucial role in tumor metastasis and
is regarded as a crucial function for cancer cells to escape from
primary sites (28). N-cadherin is
a cell-cell adhesion glycoprotein. It is able to facilitate
transendothelial migration and is regarded as a marker of
mesenchymal cells. E-cadherin is a component of an adhesion complex
located in adherens junctions, and is regarded as a marker of
epithelial cells. Dysregulation of E-cadherin results in the
disintegration of adherens junctions (29). Vimentin is responsible for
stabilization of the cytoskeleton, and Snail and Twist are
regulators of E-cadherin and N-cadherin. All of these proteins
serve important roles in the EMT process. The results of the
present study revealed that these proteins, which are closely
associated with EMT, were modulated by FOXC1 silencing, indicating
that FOXC1 exerts effects on the EMT of glioma cells, which may
contribute to its effects on the metastasis of glioma. Consistent
with the present study, in other types of cancer, FOXC1 also
contributes to the process of EMT (17,20,21).
In addition, throughout the EMT process, FOXC1 regulates the
microvascular invasion of hepatocellular carcinoma (30).
The Wnt/β-catenin pathway is involved in the
regulation of numerous biological processes (31) and has been implicated in various
malignancies (32,33). The accumulation and nuclear
translocation of β-catenin is a vital event in the activation of
Wnt signaling. Nuclear β-catenin serves an important role in
tumorigenesis; it has been reported that the expression levels of
nuclear β-catenin, and its downstream targets cyclin D1 and c-Myc,
are increased in glioma tissues and cell lines (34), and are associated with the
proliferation and apoptosis of glioma cells (35,36).
In the present study, it was revealed that FOXC1 silencing alone
suppressed β-catenin signaling, however, co-transfection of
β-catenin OE and FOXC1 siNRA enhanced the activation of β-catenin
signaling, indicating that the OE of β-catenin abolished the
effects of FOXC1 silencing, thus it was concluded that β-catenin
signaling may be implicated in the effects of FOXC1 on glioma.
FOXC1 mutations may also result in a reduction of endothelial Wnt
signaling (37). Furthermore, it
has been demonstrated that there is a β-catenin binding site near
the transcriptional start site of FOXC1, and β-catenin is able to
directly regulate the transcription of FOXC1 (38). Therefore, the regulatory
association between FOXC1 and β-catenin requires further
exploration. In addition to β-catenin signaling, alternative
signaling pathways, including phosphoinositide 3-kinase/protein
kinase B and nuclear factor-κB, are also involved in the effects of
FOXC1 in cancer. Whether these signaling pathways are additionally
implicated in the effects of FOXC1 on glioma remains unclear and
requires further investigation (16,19,21).
In conclusion, silencing FOXC1 was demonstrated to
suppress the proliferation, migration and invasion of glioma cells.
Further study revealed that β-catenin signaling was implicated in
the function of FOXC1. The present study is the first, to the best
of our knowledge, to provide direct evidence to suggest that FOXC1
functions as an oncogene in glioma cells. The present study
indicated that silencing FOXC1 may be considered a potential
therapeutic method for glioma; however, further investigation is
required.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81570203) and the
Scientific Research Initiation Foundation for Youth of the First
Affiliated Hospital of Zhengzhou University (grant no. 161032;
Zhengzhou, China).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
QC, MZ and YS contributed to the study design,
experiment performance and manuscript writing. XW, JY, MD, YM, ZZ,
KL, LJ, NW and PW contributed to the performance of the
experiments.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ostrom QT, Gittleman H, Fulop J, Liu M,
Blanda R, Kromer C, Wolinsky Y, Kruchko C and Barnholtz-Sloan JS:
CBTRUS statistical report: Primary brain and central nervous system
tumors diagnosed in the united states in 2008-2012. Neuro Oncol. 4
Suppl 17:iv1–iv62. 2015. View Article : Google Scholar
|
|
2
|
Ho VK, Reijneveld JC, Enting RH, Bienfait
HP, Robe P, Baumert BG and Visser O; Dutch Society for
Neuro-Oncology (LWNO), : Changing incidence and improved survival
of gliomas. Eur J Cancer. 50:2309–2318. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Brabletz T, Kalluri R, Nieto MA and
Weinberg RA: EMT in cancer. Nat Rev Cancer. 18:128–134. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Myatt SS and Lam EW: The emerging roles of
forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 7:847–859.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Han B, Bhowmick N, Qu Y, Chung S, Giuliano
AE and Cui X: FOXC1: An emerging marker and therapeutic target for
cancer. Oncogene. 36:3957–3963. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Seo S, Fujita H, Nakano A, Kang M, Duarte
A and Kume T: The forkhead transcription factors, foxc1 and foxc2,
are required for arterial specification and lymphatic sprouting
during vascular development. Dev Biol. 294:458–470. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sun J, Ishii M, Ting MC and Maxson R:
Foxc1 controls the growth of the murine frontal bone rudiment by
direct regulation of a Bmp response threshold of Msx2. Development.
140:1034–1044. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bin L, Deng L, Yang H, Zhu L, Wang X,
Edwards MG, Richers B and Leung DY: Forkhead box C1 regulates human
primary keratinocyte terminal differentiation. PLoS One.
11:e01673922016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mirzayans F, Lavy R, Penner-Chea J and
Berry FB: Initiation of early osteoblast differentiation events
through the direct transcriptional regulation of Msx2 by FOXC1.
PLoS One. 7:e490952012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang J, Xu Y, Li L, Wang L, Yao R, Sun Q
and Du G: FOXC1 is associated with estrogen receptor alpha and
affects sensitivity of tamoxifen treatment in breast cancer. Cancer
Med. 6:275–287. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sizemore ST and Keri RA: The forkhead box
transcription factor FOXC1 promotes breast cancer invasion by
inducing matrix metalloprotease 7 (MMP7) expression. J Biol Chem.
287:24631–24640. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wei LX, Zhou RS, Xu HF, Wang JY and Yuan
MH: High expression of FOXC1 is associated with poor clinical
outcome in non-small cell lung cancer patients. Tumour Biol.
34:941–946. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen S, Jiao S, Jia Y and Li Y: Effects of
targeted silencing of FOXC1 gene on proliferation and in vitro
migration of human non-small-cell lung carcinoma cells. Am J Transl
Res. 8:3309–3318. 2016.PubMed/NCBI
|
|
16
|
Wang J, Ray PS, Sim MS, Zhou XZ, Lu KP,
Lee AV, Lin X, Bagaria SP, Giuliano AE and Cui X: FOXC1 regulates
the functions of human basal-like breast cancer cells by activating
NF-kB signaling. Oncogene. 31:4798–4802. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xia L, Huang W, Tian D, Zhu H, Qi X, Chen
Z, Zhang Y, Hu H, Fan D, Nie Y and Wu K: Overexpression of forkhead
box C1 promotes tumor metastasis and indicates poor prognosis in
hepatocellular carcinoma. Hepatology. 57:610–624. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ou-Yang L, Xiao SJ, Liu P, Yi SJ, Zhang
XL, Ou-Yang S, Tan SK and Lei X: Forkhead box C1 induces
epithelialmesenchymal transition and is a potential therapeutic
target in nasopharyngeal carcinoma. Mol Med Rep. 12:8003–8009.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang J, Li L, Liu S, Zhao Y, Wang L and Du
G: FOXC1 promotes melanoma by activating MST1R/PI3K/AKT.
Oncotarget. 7:84375–84387. 2016.PubMed/NCBI
|
|
20
|
Zhu X, Wei L, Bai Y, Wu S and Han S: FoxC1
promotes epithelial-mesenchymal transition through PBX1 dependent
transactivation of ZEB2 in esophageal cancer. Am J Cancer Res.
7:1642–1653. 2017.PubMed/NCBI
|
|
21
|
Huang L, Huang Z, Fan Y, He L, Ye M, Shi
K, Ji B, Huang J, Wang Y and Li Q: FOXC1 promotes proliferation and
epithelial-mesenchymal transition in cervical carcinoma through the
PI3K-AKT signal pathway. Am J Transl Res. 9:1297–1306.
2017.PubMed/NCBI
|
|
22
|
Liu Y, Han L, Bai Y, Du W and Yang B:
Down-regulation of MicroRNA-133 predicts poor overall survival and
regulates the growth and invasive abilities in glioma. Artif Cells
Nanomed Biotechnol. 46:206–210. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yu H, Xue Y, Wang P, Liu X, Ma J, Zheng J,
Li Z, Cai H and Liu Y: Knockdown of long non-coding RNA XIST
increases blood-tumor barrier permeability and inhibits glioma
angiogenesis by targeting miR-137. Oncogenesis. 6:e3032017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang L, Chai L, Ji Q, Cheng R, Wang J and
Han S: Forkhead box protein C1 promotes cell proliferation and
invasion in human cervical cancer. Mol Med Rep. 17:4392–4398.
2018.PubMed/NCBI
|
|
26
|
Xu YY, Tian J, Hao Q and Yin LR:
MicroRNA-495 downregulates FOXC1 expression to suppress cell growth
and migration in endometrial cancer. Tumour Biol. 37:239–251. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Deng L, Liu T, Zhang B, Wu H, Zhao J and
Chen J: Forkhead box C1 is targeted by microRNA-133b and promotes
cell proliferation and migration in osteosarcoma. Exp Ther Med.
14:2823–2830. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kalluri R and Weinberg RA: The basics of
epithelial-mesenchymal transition. J Clin Invest. 119:1420–1428.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yilmaz M and Christofori G: EMT, the
cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev.
28:15–33. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xu ZY, Ding SM, Zhou L, Xie HY, Chen KJ,
Zhang W, Xing CY, Guo HJ and Zheng SS: FOXC1 contributes to
microvascular invasion in primary hepatocellular carcinoma via
regulating epithelial-mesenchymal transition. Int J Biol Sci.
8:1130–1141. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Clevers H and Nusse R: Wnt/β-catenin
signaling and disease. Cell. 149:1192–1205. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Saito-Diaz K, Chen TW, Wang X, Thorne CA,
Wallace HA, Page-McCaw A and Lee E: The way Wnt works: Components
and mechanism. Growth Factors. 31:1–31. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Anastas JN and Moon RT: WNT signalling
pathways as therapeutic targets in cancer. Nat Rev Cancer.
13:11–26. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li Y, Zhu G, Zeng W, Wang J, Li Z, Wang B,
Tian B, Lu D, Zhang X, Gao G and Li L: Long noncoding RNA AB073614
promotes the malignance of glioma by activating Wnt/β-catenin
signaling through downregulating SOX7. Oncotarget. 8:65577–65587.
2017.PubMed/NCBI
|
|
35
|
Liu X, Wang L, Zhao S, Ji X, Luo Y and
Ling F: β-catenin overexpression in malignant glioma and its role
in proliferation and apoptosis in glioblastma cells. Med Oncol.
28:608–614. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang K, Zhang J, Han L, Pu P and Kang C:
Wnt/beta-catenin signaling in glioma. J Neuroimmune Pharmacol.
7:740–749. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mishra S, Choe Y, Pleasure SJ and
Siegenthaler JA: Cerebrovascular defects in Foxc1 mutants correlate
with aberrant WNT and VEGF-A pathways downstream of retinoic acid
from the meninges. Dev Biol. 420:148–165. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Savage J, Voronova A, Mehta V,
Sendi-Mukasa F and Skerjanc IS: Canonical Wnt signaling regulates
Foxc1/2 expression in P19 cells. Differentiation. 79:31–40. 2010.
View Article : Google Scholar : PubMed/NCBI
|