Introduction
Cutaneous T-cell lymphoma (CTCL) represents serial
diseases, which mainly involve malignant clonal T-lymphocytes of
the CD4 phenotype due to a heterogeneous population of
lymphoproliferative disorders (1).
The annual incidence of primary cutaneous lymphomas is estimated to
be 1:100,000, of which CTCL accounts for ~75% of cases; therefore,
CTCL is the most common type of primary cutaneous lymphoma
(2,3). Due to the clonal proliferation of
skin-invasive mature T lymphocytes, CTCL is characterized as a type
of non-Hodgkin's lymphoma (3). The
pathogenesis of CTCL involves the deregulation of signaling
pathways, including signal transducer and activator of
transcription (STAT), Src kinases, c-Myc, cyclooxygenase-2, nuclear
factor-κB, GATA binding protein 3 (GATA-3), thymocyte
selection-associated high mobility group box (TOX), and embryonic
stem cell regulators (4,5). GATA factors can function in
undifferentiated progenitor cells and are involved in their
expansion, or they can direct the coordinated maturation and cell
cycle withdrawal in terminally differentiating cells. Therefore,
alterations of GATA factors contribute to the development of cancer
in humans. GATA3 functions in T lymphocytes, but it is also a
critical regulator of mammary epithelial cells (6). The type 2 T helper cell-specific
transcription factor GATA-3 is overexpressed in patients with CTCL
and peripheral T-cell lymphoma (7,8). The
overexpression of GATA-3 also occurs in other cancer/tumor cells,
including human glioblastoma and other T-cell lymphomas (9,10).
Furthermore, activated GATA-3 can promote T-cell proliferation in
patients with Sézary syndrome (11), and the overexpression of GATA-3 can
develop CD4+/CD8+ double-positive T-cell
lymphoma (10). Therefore, the
overexpression of GATA-3 promotes cancer or tumor cell
proliferation and differentiation (12). However, the gene that regulates
GATA-3 in CTCL and the way in which GATA-3 and its co-activators
and/or co-repressors regulate the expression of disease-associated
genes remain to be fully elucidated.
MicroRNAs (miRNAs), which are a class of small
non-coding RNAs (18–22 nt length), are ubiquitous in eukaryotes and
can regulate protein expression at the mRNA level (13–15).
In CTCL, the expression levels of several miRNAs and proteins are
altered (16,17). miRNA-135a (miR-135a) is involved in
the regulation of several diseases, including colorectal cancer
(18), blood lipid and
inflammatory changes (19),
senescent vascular endothelial cell calcification (20), and lung cancer metastasis and
invasion (21). A low expression
level of miR-135a in classic Hodgkin's lymphoma (cHL) is associated
with a high likelihood of relapse and a short disease-free survival
period (22). Increased expression
of GATA-3 and decreased expression of miR-135a are observed in
T-cell lymphoma. However, the association between miR-135a and
GATA-3 remains to be fully elucidated.
In the present study, reverse
transcription-quantitative polymerase chain reaction (RT-qPCR),
western blot, cell counting, dual luciferase, and pre-miR-135a
assays were used to detect Hut78 cell proliferation and the gene
and protein expression levels in the Hut78 cell line to investigate
the role of miR-135a in regulating GATA-3 mRNA translation. The
results provide evidence that the novel tumor suppressor miR-135a
represses the mechanism underlying the expression of GATA-3 in
CTCL.
Materials and methods
Cell culture
The cultured Hut78 human CTCL cell line (23) was purchased from American Type
Culture Collection (Manassas, VA, USA). The non-malignant T-cell
line (normal T lymphocytes as control cells), were established in
the medical laboratory of The First Hospital of Zibo City (Zibo,
China) from patients with mycosis fungoides and Sézary syndrome
according to Woetmann's methods (24). Fresh blood samples were obtained
from a 46-year-old male patient with mycosis fungoides and Sézary
syndrome at the First Hospital of Zibo City (Zibo, China) in April
2015. The fresh blood was diluted with an equal volume of PBS and
was gently dropped onto the lymphocyte separation medium (Anhui
Haoyang Chemical Group Co., Ltd., Fuyang, China) which was in a
15-ml centrifuge tube. The non-malignant T-cell line was obtained
from the interlayer between the blood and lymphocyte separation
medium following 15-ml tube centrifugation for 15 min at 500 × g
and 25°C. Briefly, in 5% CO2 at 37°C, this non-malignant
T-cell line was cultured in RPMI 1640 medium, containing 10% (v/v)
heat-inactivated fetal bovine serum (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), L-glutamine and antibiotics
(100 IU/ml of penicillin, 100 µg/ml of streptomycin). The present
study was performed with the approval of the First Hospital of Zibo
City. Signed written consent was obtained from the patient prior to
recruitment to the study.
Reagents and instruments
The following reagents and instruments were used in
the present study: miRcute miRNA isolation kit, miRcute miRNA cDNA
first-strand synthesis kit, miRcute miRNA quantitative fluorescence
detection kit, SuperReal PreMix (SYBR Green), and TIANScript II
cDNA first-strand synthesis kit; all were purchased from Tiangen
Biotech Co., Ltd. (Beijing, China). The RT-qPCR instrument (BIOER
FQD-96A), GATA-3 (cat. no. ab106625), TOX (cat. no. ab155768) and
β-actin (cat. no. ab8227) primary antibodies and secondary antibody
(cat. no. ab7090; Abcam, Cambridge, MA, USA), TRIzol reagent
(Yisheng Biology, Shanghai, China), BCA protein assay reagent kit
(Zhongke Ruitai, Beijing, China), and serum RNA extraction miRNeasy
serum/plasma kit (Jianlun Biology, Guangzhou, China) were also
used. All plasmids/agomiR were designed and synthesized by Shanghai
Biological Technology Co., Ltd. (Shanghai, China). Pre-miR-135a,
miR-135a mimic, miR-135a inhibitor, and their negative controls
were purchased from GeneChem Co. (Shanghai, China; Table I). Xfect transfection reagents were
purchased from Takara Biotechnology Co., Ltd. (Dalian, China).
 | Table I.miR-135a mimics, miR-135a inhibitor
and their NC sequences. |
Table I.
miR-135a mimics, miR-135a inhibitor
and their NC sequences.
| Name | Sequence
(5′-3′) |
|---|
| miR-135a
mimics |
UAUGGCUUUUUAUUCCUAUGUGA |
| Mimics NC |
UUCUCCGAACGUGUCACGUTT |
| miR-135a
inhibitor |
UCACAUAGGAAUAAAAAGCCAUA |
| Inhibitor NC |
CAGUACUUUUGUGUAGUACAA |
RT-qPCR analysis
The Hut78 cells and normal T lymphocytes were
collected, and total RNAs were extracted with TRIzol®
(Invitrogen; Thermo Fisher Scientific, Inc.) using the
phenol-chloroform extraction method. The RNA integrity was examined
by gel electrophoresis, and the RNA purity was assessed according
to the 260/280 ratio by spectrophotometry. Total RNA (2 µg)
underwent RT to synthesize cDNA using Oligo-dT (10 µM) and Super
Pure dNTPs (10 mM), according to the protocol of the TIANScript II
RT kit (cat. no. KR107; Tiangen Biotech Co., Ltd.). The sequences
of the GATA-3, GAPDH, miR-135a, and U6 primers used for RT-qPCR
analysis are shown in Table II.
GAPDH and U6 were used as internal controls for GATA-3 and
miR-135a, respectively. The reaction mixture was prepared as
follows, according to SuperReal PreMix Plus (SYBR Green) (cat. no.
FP205; Tiangen Biotech Co., Ltd.): 10 µl SYBR Ex Taq II, 0.4 µl ROX
Reference Dye, 0.8 µl each primer (final concentration, 250
nmol/l), 7 µl ddH2O and 1.0 µl cDNA. The PCR procedure
for GATA-3 was as follows: Pre-denaturation for 10 min at 95°C,
followed by 40 cycles of denaturation for 30 sec at 95°C, annealing
for 20 sec at 55°C, and extension for 30 sec at 72°C. The reaction
conditions for miR-135a were pre-denaturation at 95°C for 5 min, 40
cycles of denaturation at 95°C for 20 sec, and annealing at 60°C
for 30 sec. The relative levels of GATA-3 and miR-135a were
calculated using the 2−ΔΔCq method (25).
 | Table II.Primer sequences. |
Table II.
Primer sequences.
| Name | Primer
sequence |
|---|
| GATA-3 | Forward:
5′-AAGAGTGCCTCAAGTATCAG-3′ |
|
| Reverse:
5′-GCGGATAGGTGGTAATGG-3′ |
| GAPDH | Forward:
5′-CCCTCAATGACCACTTTGTG-3′ |
|
| Reverse:
5′-GGTTTGAGGGCTCTTACTCCT-3′ |
| MicroRNA-135a | Forward:
5′-GCGCCGTATGGCTTTTTATTCCTA-3′ |
|
| Reverse:
5′-TGCAGAGATGTCCAGTCAGC-3′ |
| U6 | Forward:
5′-AACGCTTCACGAATTTGCGT-3′ |
|
| Reverse:
5′-CTCGCTTCGGCAGCACA-3′ |
Western blot analysis
Total protein was extracted by protein lysis, and
its concentration was measured using the BCA protein assay reagent
kit. Subsequently, the proteins (20 µg) were separated by 10%
SDS-polyacrylamide electrophoresis and transferred onto a PVDF
membrane. The membrane was blocked by 5% skim milk for 2 h.
Following blocking, the primary antibodies (1:1,000) targeting
GATA-3, TOX, and β-actin were added and incubated overnight at 4°C.
Subsequently, the secondary antibody (1:10,000) was added and
incubated at room temperature for 1 h. The membrane was developed
in ECL luminescent liquid, and the developed film was scanned using
a GT 2500 scanner (Epson America, Inc., Long Beach, CA, USA) and
analyzed using ImageJ 1.50i (National Institutes of Health,
Bethesda, MD, USA). The relative expression level of GATA-3 with
respect to β-actin was calculated based on the grey value that was
obtained from Image J software.
Bioinformatics prediction of the
regulatory upstream miRNA for GATA-3
Bioinformatics prediction was used to identify the
upstream miRNA for GATA-3. The following gene prediction software
programs were used for bioinformatics prediction: miRanda
(http://www.microma.org/rnicroma/home.do), TargetScan
(www.targetscan.org), PiTa (http://genie.weizmann.ac.il/pubs/mir07/mir07_data.html),
RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/),
and PicTar (http://pictar.mdc-berlin.de/). miR-135a was identified
as the potential upstream gene of GATA-3.
Dual luciferase assay
The wild and mutant types of the miR-135a binding
sequence in the 3′-untranslated region (3′-UTR) of the
GATA-3 gene were constructed by in vitro chemical
synthesis. The cleavage sites of Spe1 and HindIII
were respectively added on both ends. The two DNA fragments were
cloned into pMIR-REPORT luciferase plasmids. Using the liposome
method, the plasmids with the wild-type 3′-UTR and mutant-type
3′-UTR sequences were transfected into 293T cells, which were
purchased from the Cell Bank of the Institute of Culture Collection
of the Chinese Academy of Sciences (Shanghai, China). Subsequently,
the agomiR-135a (100 nM) was transfected into cells and incubated
for 24 h. The fluorescence values were measured using the GloMax
20/20 luminometer. The Renilla fluorescent activity was used as the
internal control, and all procedures were performed in strict
accordance with the dual luciferase assay kit instructions.
Cell transfection
The plasmids pre-miR-135a, miR-135a mimic, miR-135a
inhibitor, and their negative controls were transfected into cells
using Xfect transfection reagents, according to the manufacturer's
protocol.
Statistical analysis
Processed by SPSS18.0 (SPSS, Inc., Chicago, IL,
USA), all data are presented as the mean ± standard deviation and a
normality test performed. Multiple groups of measurement data were
analyzed using one-way analysis of variance (26). P<0.05 was considered to indicate
a statistically significant difference.
Results
mRNA and protein expression of
GATA-3
To detect the mRNA and protein expression levels of
GATA-3 in the Hut78 CTCL cells and normal T lymphocytes, RT-qPCR
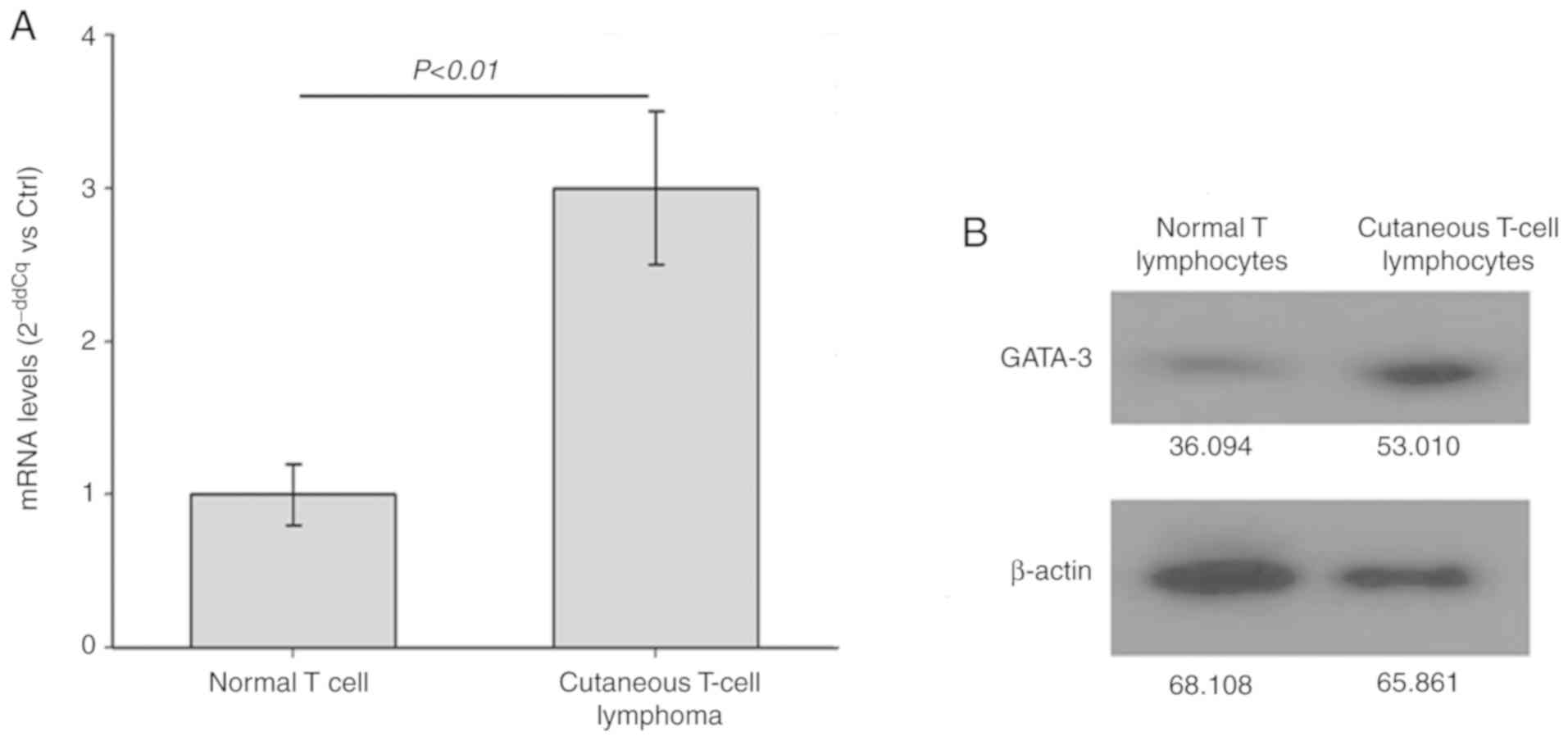
and western blot analyses were performed, respectively. As shown in
Fig. 1, the mRNA (Fig. 1A) and protein (Fig. 1B) levels of GATA-3 in the CTCL
Hut78 cells were significantly increased compared with those in the
normal T lymphocytes (P<0.01). Therefore, the expression level
was GATA-3 is increased in the CTCL Hut78 cells.
Bioinformatics and dual luciferase
assay
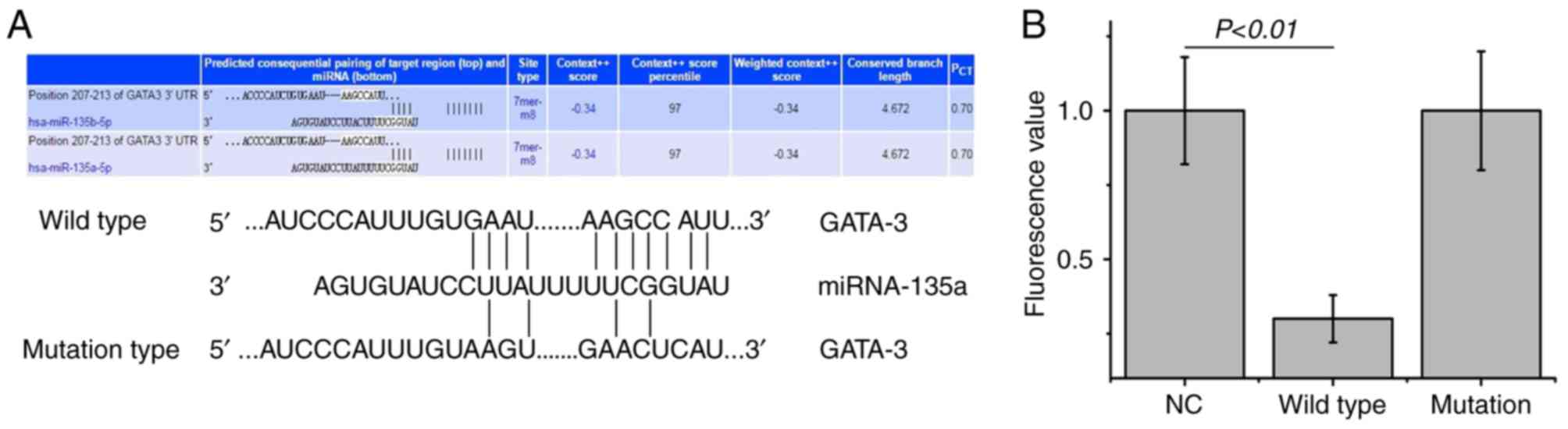
To identify the upstream regulatory miRNA of GATA-3,
bioinformatics prediction was performed. miR-135a was found to be
the potential upstream gene of GATA-3. The wild and mutation types
of the binding sequence are shown in Fig. 2A. To determine whether miRNA-135
directly targets GATA-3, a dual luciferase assay was performed. The
fluorescence values were significantly decreased following
co-transfection with agomiR-135a and the pMIR-REPORT plasmid
(P<0.01; Fig. 2B). No
statistically significant differences in fluorescence values were
observed compared with the mutation group (P>0.05); however,
miR-135a was shown to bind directly with GATA-3 at the 3′-UTR to
regulate its expression.
Expression of miR-135a in Hut78
cells
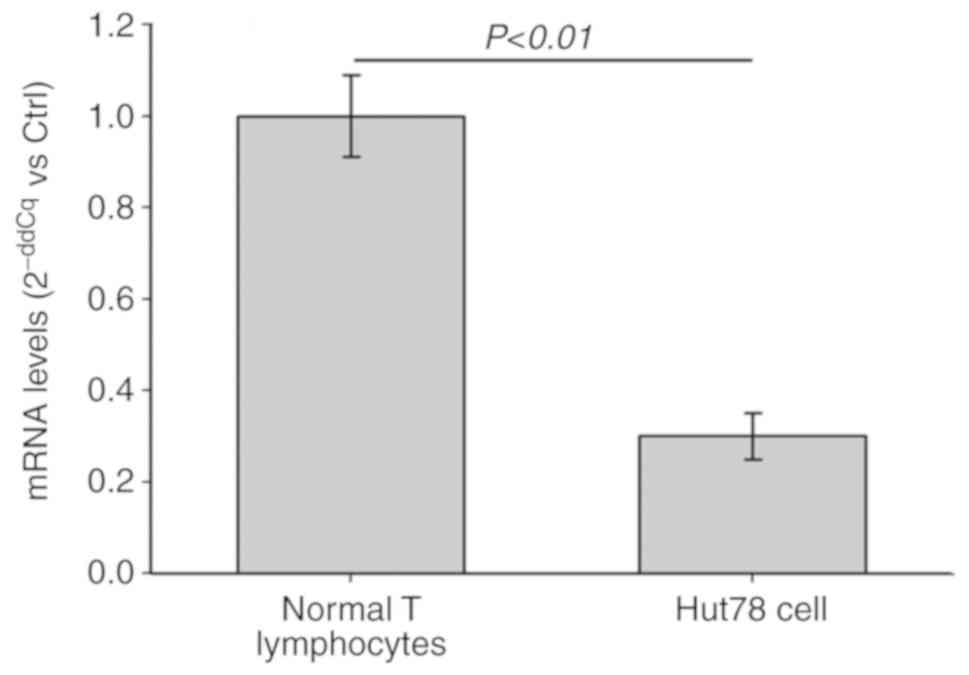
To detect changes in the expression of miR-135a in
the CTCL Hut78 cells, RT-qPCR analysis was performed. The
expression level of miR-135a in the CTCL Hut78 cells was
significantly decreased compared with that in the normal T
lymphocytes (P<0.01; Fig. 3).
Therefore, miR-135a may have a regulatory role in the pathological
process of CTCL through GATA-3.
Overexpression of miR-135a in Hut78
cells
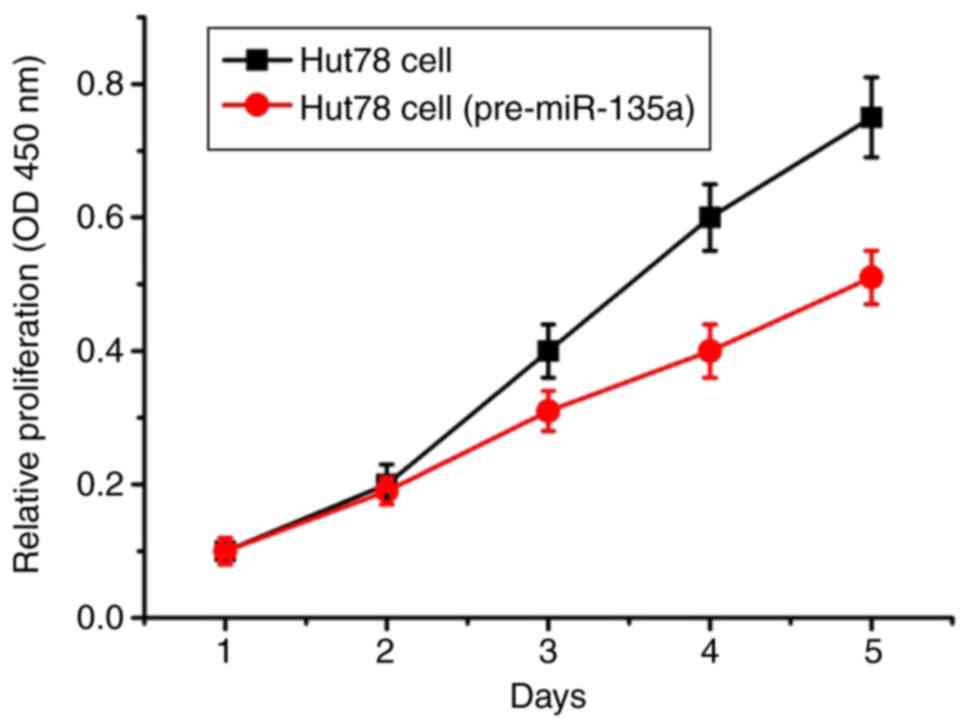
The proliferation curves of Hut78 CTCL cells were
drawn according to the instructions of the Cell Counting Kit-8. The
proliferation of the Hut78 CTCL cells transfected with the
pre-miR-135a plasmid was increasingly inhibited on the third day
(Fig. 4). On the fourth and fifth
days, the proliferation of cells was further increased.
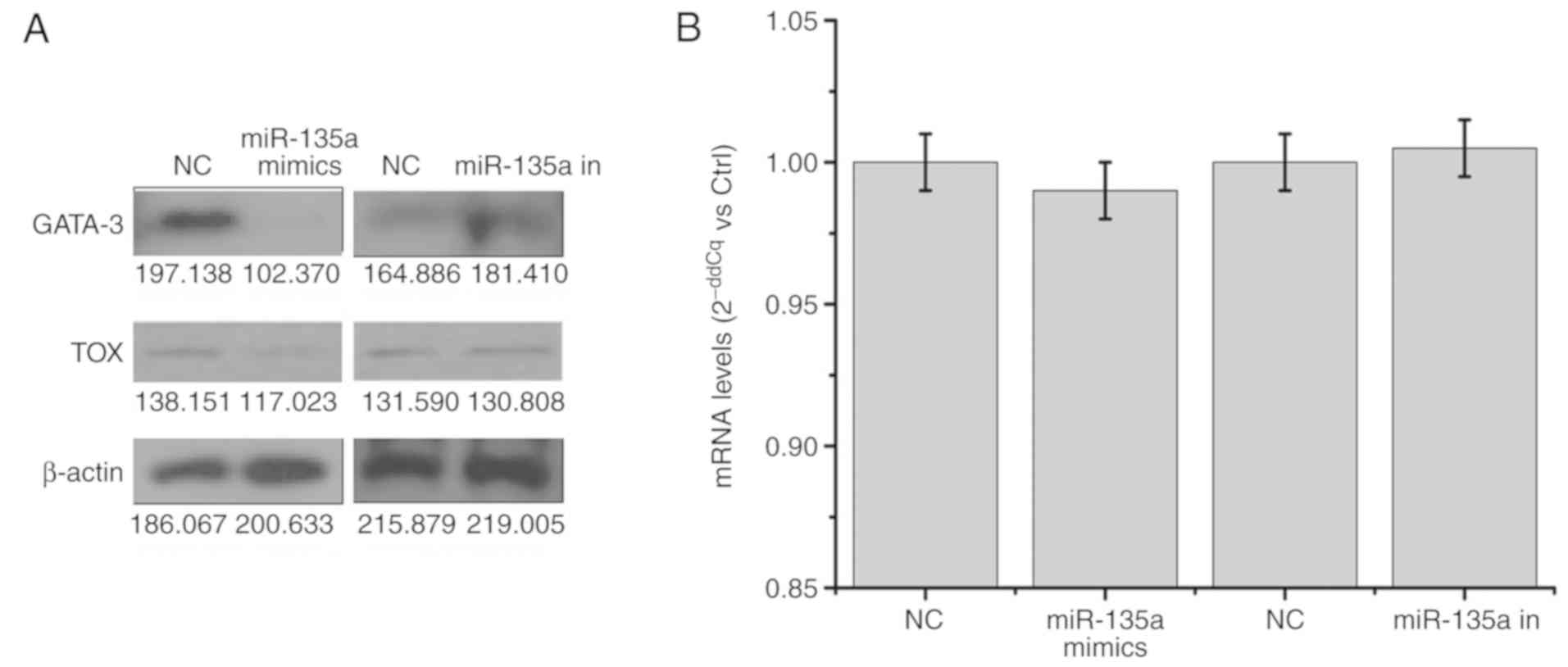
miR-135a downregulates the levels of
GATA-3 and TOX
According to the bioinformatics prediction, one of
the target genes of miR-135a was identified as GATA-3. The
potential binding sequence for miR-135a was identified in the
3′-UTR of GATA-3 (Fig. 2A). As
predicted, the results of the western blot analysis showed that the
overexpression of miR-135a mimics decreased the expression of
GATA-3, whereas miR-135a increased its expression upon transfection
(Fig. 5A). Simultaneously, the
expression level of TOX, which is a downstream gene of
GATA-3, was also decreased in the miR-135a-overexpressing cells
(Fig. 5A). Additionally the
results of the RT-qPCR assay showed that the mRNA level of GATA-3
in the miR-135a mimics group was marginally decreased compared with
its level in the other groups (Fig.
5B), and the protein expression of GATA-3 was substantially
downregulated in the Hut78 cells. The primers for RT-qPCR analysis
were located at the middle of GATA-3 mRNA, and the PCR product
belonged to one of several exons in the GATA-3 pre-mRNA. The
existence of GATA-3 pre-mRNA may have resulted in the marginal
decrease in the mRNA expression of GATA-3 in the RT-qPCR assay.
Furthermore, GATA-3 mRNA was destroyed by miR-135a following
pre-mRNA maturation. Therefore, miR-135a led to the destruction of
GATA-3 mRNA and inhibited the translation of GATA-3 mRNA.
Discussion
GATA-3, which can be upregulated in lymphoma or
CTCL, has binding sites in the TOX promoter (11,27,28).
In CTCL, the expression level of GATA-3 in Hut78 cells was higher
than that in normal T lymphocytes. To understand the mechanism
underlying the changes in the expression of GATA-3, the present
study performed a search for the miRNA that targets GATA-3 mRNA via
bioinformatics analysis; this was found to be miR-135a.
Increasing levels of mature miR-135a can cause cHL
cell apoptosis and growth reduction through miR-135a-regulating
Janus kinase 2 (JAK2) (22). As
confirmed by a dual luciferase assay, an increased level of
miR-135a inhibited Hut78 cell proliferation via the role of
miR-135a in targeting GATA-3 mRNA. However, the level of miR-135a
was lower in CTCL Hut78 cells than in normal T lymphocytes,
suggesting that miR-135a may be involved in the pathogenesis of
CTCL.
The transcription factor of TOX is GATA-3,
which is involved in the signal cascades governing T-cell
development (11,29). In addition, the transcript levels
of TOX are markedly increased in CTCL, compared with those in
normal skin or benign inflammatory dermatoses (4). Therefore, the enhanced transcription
of TOX may be induced by the overexpression of GATA-3 due to the
decreased expression of regulatory miRNA-135a in CTCL.
In the present study, the miRNA functioning as the
upstream regulator of GATA-3 was investigated via bioinformatics
analysis. miRNAs are small, endogenous, non-coding RNAs, which can
dissect and inhibit target mRNA translation for its deregulation
(30,31). miRNAs are vital in regulating
disease development, physiology and pathogenesis (32). According to the bioinformatics
prediction, miR-135a was identified as one of the potential
upstream genes that may regulate the expression GATA-3. The dual
luciferase assay further confirmed that miR-135a can directly bind
with the 3′UTR of GATA-3, suggesting that miR-135a can directly
regulate the translation of GATA-3. Activated GATA-3 can promote
T-cell proliferation in patients with Sézary syndrome (11), and the overexpression of GATA-3
promotes cancer/tumor cell proliferation and differentiation
(10,12). In the present study, the expression
of GATA-3 was upregulated due to the downregulated expression of
miR-135a in the Hut78 CTCL cell line and exhibited enhanced cell
proliferation.
The assays involving miR-135a mimics showed that the
mRNA level of GATA-3 in the miR-135a mimics group was marginally
decreased compared with those in other groups, and the protein
level of GATA-3 was substantially downregulated in the Hut78 cells.
Unfortunately, whether the miR-135a mimics altered the level of
miR-135a in the Hut78 cells was not examined. Previous reports have
confirmed that miRNA mimics can induce the upregulation of target
gene mRNA and decrease the protein level of the target gene
(33,34). The results of the present study
suggest that the enhanced gene transcription of GATA-3 was
activated in the miR-135a mimics assay owing to the decreased
protein expression of GATA-3, which is a key molecule in the
signaling pathway.
Cell apoptosis is a complex, multistage process that
involves numerous genes. Apoptosis can be induced by endoplasmic
reticulum stress, the mitochondrial pathway, and the death receptor
pathway. The mitochondrial pathway has been relatively well
researched and is controlled predominantly by members of caspase-3,
cleaved caspase-3, and B-cell lymphoma 2 (Bcl-2) and
Bcl-2-associated X protein. Certain natural products, including
Euphorbia factor L2 and bruceine D, can induce cell apoptosis
through the mitochondrial pathway (35,36).
Cell apoptosis can be induced by miR-133b and miR-135a in
vitro via a signaling cascade, involving JAK2, STAT3 and Bcl-2
(37). In the present study,
miR-135a also induced cell apoptosis by targeting GATA-3 and
regulating GATA-3/TOX signaling.
In conclusion, the mRNA and protein expression
levels of GATA-3 were markedly increased in CTCL. This finding may
be associated with the downregulated expression of miR-135a,
leading to T-cell deregulation and proliferation through GATA-3/TOX
regulation and subsequently causing CTCL. However,
immunohistochemical analysis is required to further examine the
expression of GATA-3 in more tissue samples of CTCL.
Acknowledgements
Not applicable.
Funding
This study was supported by the Technology
Development Project Plan of Shandong Education Department (grant
nos. J15LM63 and J14LM54).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HW and JW designed the study. Western blot analysis
was conducted by HW and RL. RT-qPCR analysis was performed by HW,
XG and YZ. Other experiments were conducted by HW, BS and JW. HW
and JW analyzed and interpreted the data, and drafted the
manuscript. All authors critically revised the manuscript, and read
and approved the final version of the manuscript.
Ethics approval and consent to
participate
Ethical approval was provided by the Medical Ethics
Committee of The First Hospital of Zibo City (reference no.
201503045).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kotz EA, Anderson D and Thiers BH:
Cutaneous T-cell lymphoma. J Eur Acad Dermatol Venereol.
17:131–137. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Neelis KJ, Schimmel EC, Vermeer MH, Senff
NJ, Willemze R and Noordijk EM: Low-dose palliative radiotherapy
for cutaneous B- and T-cell lymphomas. Int J Radiat Oncol Biol
Phys. 74:154–158. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Willemze R, Jaffe ES, Burg G, Cerroni L,
Berti E, Swerdlow SH, Ralfkiaer E, Chimenti S, Diaz-Perez JL,
Duncan LM, et al: WHO-EORTC classification for cutaneous lymphomas.
Blood. 105:3768–3785. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Huang Y, Su MW, Jiang X and Zhou Y:
Evidence of an oncogenic role of aberrant TOX activation in
cutaneous T-cell lymphoma. Blood. 125:1435–1443. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sibbesen NA, Kopp KL, Litvinov IV, Jønson
L, Willerslev-Olsen A, Fredholm S, Petersen DL, Nastasi C,
Krejsgaard T, Lindahl LM, et al: Jak3, STAT3, and STAT5 inhibit
expression of miR-22, a novel tumor suppressor microRNA, in
cutaneous T-Cell lymphoma. Oncotarget. 6:20555–20569. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chou J, Provot S and Werb Z: GATA3 in
development and cancer differentiation: Cells GATA have it! J Cell
Physiol. 222:42–49. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang W, Wang Z, Luo Y, Zhong D, Luo Y and
Zhou D: GATA3 expression correlates with poor prognosis and
tumor-associated macrophage infiltration in peripheral T cell
lymphoma. Oncotarget. 7:65284–65294. 2016.PubMed/NCBI
|
|
8
|
Kari L, Loboda A, Nebozhyn M, Rook AH,
Vonderheid EC, Nichols C, Virok D, Chang C, Horng WH, Johnston J,
et al: Classification and prediction of survival in patients with
the leukemic phase of cutaneous T cell lymphoma. J Exp Med.
197:1477–1488. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Majewska E, Rola R, Barczewska M, Marquez
J, Albrecht J and Szeliga M: Transcription factor GATA3 expression
is induced by GLS2 overexpression in a glioblastoma cell line but
is GLS2-independent in patient-derived glioblastoma. J Physiol
Pharmacol. 68:209–214. 2017.PubMed/NCBI
|
|
10
|
Nawijn MC, Ferreira R, Dingjan GM, Kahre
O, Drabek D, Karis A, Grosveld F and Hendriks RW: Enforced
expression of GATA-3 during T cell development inhibits maturation
of CD8 single-positive cells and induces thymic lymphoma in
transgenic mice. J Immunol. 167:715–723. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gibson HM, Mishra A, Chan DV, Hake TS,
Porcu P and Wong HK: Impaired proteasome function activates GATA3
in T cells and upregulates CTLA-4: Relevance for Sézary syndrome. J
Invest Dermatol. 133:249–257. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zheng R and Blobel GA: GATA transcription
factors and cancer. Genes Cancer. 1:1178–1188. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jiang XI, Luo Y, Zhao S, Chen Q, Jiang C,
Dai Y, Chen Y and Cao Z: Clinical significance and expression of
microRNA in diabetic patients with erectile dysfunction. Exp Ther
Med. 10:213–218. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jia W, Wu Y, Zhang Q, Gao GE, Zhang C and
Xiang Y: Expression profile of circulating microRNAs as a promising
fingerprint for cervical cancer diagnosis and monitoring. Mol Clin
Oncol. 3:851–858. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Graziano A, Lo Monte G, Piva I, Caserta D,
Karner M, Engl B and Marci R: Diagnostic findings in adenomyosis: A
pictorial review on the major concerns. Eur Rev Med Pharmacol Sci.
19:1146–1154. 2015.PubMed/NCBI
|
|
16
|
Abe F, Kitadate A, Ikeda S, Yamashita J,
Nakanishi H, Takahashi N, Asaka C, Teshima K, Miyagaki T, Sugaya M
and Tagawa H: Histone deacetylase inhibitors inhibit metastasis by
restoring a tumor suppressive microRNA-150 in advanced cutaneous
T-cell lymphoma. Oncotarget. 8:7572–7585. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
McGirt LY, Baerenwald DA, Vonderheid EC
and Eischen CM: Early changes in miRNA expression are predictive of
response to extracorporeal photopheresis in cutaneous T-cell
lymphoma. J Eur Acad Dermatol Venereol. 29:2269–2271. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang Q, Zhang H, Shen X and Ju S: Serum
microRNA-135a-5p as an auxiliary diagnostic biomarker for
colorectal cancer. Ann Clin Biochem. 54:76–85. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Desgagné V, Guay SP, Guérin R, Corbin F,
Couture P, Lamarche B and Bouchard L: Variations in HDL-carried
miR-223 and miR-135a concentrations after consumption of dietary
trans fat are associated with changes in blood lipid and
inflammatory markers in healthy men-an exploratory study.
Epigenetics. 11:438–448. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lin L, He Y, Xi BL, Zheng HC, Chen Q, Li
J, Hu Y, Ye MH, Chen P and Qu Y: MiR-135a suppresses calcification
in senescent VSMCs by regulating KLF4/STAT3 pathway. Curr Vasc
Pharmacol. 14:211–218. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shi H, Ji Y, Zhang D, Liu Y and Fang P:
MiR-135a inhibits migration and invasion and regulates EMT-related
marker genes by targeting KLF8 in lung cancer cells. Biochem
Biophys Res Commun. 465:125–130. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Navarro A, Diaz T, Martinez A, Gaya A,
Pons A, Gel B, Codony C, Ferrer G, Martinez C, Montserrat E and
Monzo M: Regulation of JAK2 by miR-135a: Prognostic impact in
classic Hodgkin lymphoma. Blood. 114:2945–2951. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nasser MI, Masood M, Wei W and Li X, Zhou
Y, Liu B, Li J and Li X: Cordycepin induces apoptosis in SGC-7901
cells through mitochondrial extrinsic phosphorylation of PI3K/Akt
by generating ROS. Int J Oncol. 50:911–919. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Woetmann A, Lovato P, Eriksen KW,
Krejsgaard T, Labuda T, Zhang Q, Mathiesen AM, Geisler C, Svejgaard
A, Wasik MA and Ødum N: Nonmalignant T cells stimulate growth of
T-cell lymphoma cells in the presence of bacterial toxins. Blood.
109:3325–3332. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang Q, Xu J, Chai B, Liang A and Wang W:
Functional comparison of metallothioneins MTT1 and MTT2 from
Tetrahymena thermophila. Arch Biochem Biophys. 509:170–176. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Stamm CL and Safrit MJ: Comparison of
significance tests for repeated measures ANOVA design. Res Q.
46:403–409. 1975.PubMed/NCBI
|
|
27
|
McGirt LY, Degesys CA, Johnson VE, Zic JA,
Zwerner JP and Eischen CM: TOX expression and role in CTCL. J Eur
Acad Dermatol Venereol. 30:1497–1502. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dulmage BO, Akilov O, Vu JR, Falo LD and
Geskin LJ: Dysregulation of the TOX-RUNX3 pathway in cutaneous
T-cell lymphoma. Oncotarget. 2015.
|
|
29
|
Ho IC, Tai TS and Pai SY: GATA3 and the
T-cell lineage: Essential functions before and after
T-helper-2-cell differentiation. Nat Rev Immunol. 9:125–135. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Williams AE, Moschos SA, Perry MM, Barnes
PJ and Lindsay MA: Maternally imprinted microRNAs are
differentially expressed during mouse and human lung development.
Dev Dyn. 236:572–580. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wienholds E and Plasterk RH: MicroRNA
function in animal development. FEBS Lett. 579:5911–5922. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li X, Yu Z, Li Y, Liu S, Gao C, Hou X, Yao
R and Cui L: The tumor suppressor miR-124 inhibits cell
proliferation by targeting STAT3 and functions as a prognostic
marker for postoperative NSCLC patients. Int J Oncol. 46:798–808.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Vasudevan S, Tong Y and Steitz JA:
Switching from repression to activation: MicroRNAs can up-regulate
translation. Science. 318:1931–1934. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Khan AA, Betel D, Miller ML, Sander C,
Leslie CS and Marks DS: Transfection of small RNAs globally
perturbs gene regulation by endogenous microRNAs. Nat Biotechnol.
27:549–555. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lin M, Tang S, Zhang C, Chen H, Huang W,
Liu Y and Zhang J: Euphorbia factor L2 induces apoptosis in A549
cells through the mitochondrial pathway. Acta Pharm Sin B. 7:59–64.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang JY, Lin MT, Tung HY, Tang SL, Yi T,
Zhang YZ, Tang YN, Zhao ZZ and Chen HB: Bruceine D induces
apoptosis in human chronic myeloid leukemia K562 cells via
mitochondrial pathway. Am J Cancer Res. 6:819–826. 2016.PubMed/NCBI
|
|
37
|
Zhou W, Bi X, Gao G and Sun L: miRNA-133b
and miRNA-135a induce apoptosis via the JAK2/STAT3 signaling
pathway in human renal carcinoma cells. Biomed Pharmacother.
84:722–729. 2016. View Article : Google Scholar : PubMed/NCBI
|