Introduction
Bladder cancer is one of the most common urological
tumors. The incidence of bladder cancer ranks as the ninth most
common malignant tumor worldwide, and a large number of new cases
re diagnosed every year (1,2). In
China, bladder cancer is the most common malignant tumor of the
urinary system, and >90% bladder cancer is transitional cell
carcinoma with a high recurrence rate and invasive ability
(3). In recent years, the
incidence of bladder cancer in some cities in China has had a
steady upward trend. At present, the predominant treatment methods
for bladder cancer are surgical resection, radiotherapy and
chemotherapy. The higher recurrence rate and tumor progression rate
still greatly impact the prognosis of patients with bladder cancer.
Radical cystectomy can effectively reduce the recurrence rate of
patients with bladder cancer, improve their long-term survival rate
and is the standard treatment method for invasive bladder cancer
(4). However, surgical treatment
can seriously affect the quality of life of patients (5,6).
Therefore, it is necessary to provide a quality transitional care
service (7) to ensure the smooth
transition of patients with bladder cancer from hospital to home,
and to improve the lack of professional guidance available for home
rehabilitation, to meet the needs of patients and improve the
quality of life and prognosis of the patients. Currently, the
transitional care service of China is still in its infancy, a lack
of continuity and coordination of nursing activities for patients
with bladder cancer. Therefore, the search for novel and effective
strategies for the treatment of bladder cancer is urgent and is of
major clinical significance.
MicroRNAs (miRNAs), a family of small non-coding
single-stranded RNAs (20–22 nucleotides in length), have a critical
role in post-transcriptionally regulating gene expression by
binding to the 3′ untranslated region (3′UTR) of target genes
(8–10). Studies demonstrated that abnormal
miRNA expression is associated with the occurrence of numerous
types of tumor, including bladder cancer (11–13).
miRNAs can act as oncogenes or tumor suppressors, and the altering
abnormal expression of an miRNA can inhibit tumorigenesis and
progression (14). With the status
and role of miRNA in tumor research attracting increasing
attention, research on miRNA and bladder cancer has also developed
and deepened.
A previous study reported that miRNA-663b (miR-663b)
was highly expressed in the plasma of patients with bladder cancer,
and may be a promising novel circulating biomarker for the clinical
detection of bladder cancer (15).
However, the expression of miR-663b in the tumor tissues of bladder
cancer patients and its specific role remain unclear.
Numerous previous studies demonstrated that tumor
suppressor 2, mitochondrial calcium regulator (TUSC2) is a tumor
suppressor gene (16,17). TUSC2 was identified to induce G1
phase cell cycle arrest and apoptosis (18), to regulate calcium signaling
(19) and tyrosine kinase activity
(20), and to affect gene
expression (21). TUSC2 is
associated with various signaling pathways, including the p53
pathway (22). However, to the
best of the authors' knowledge, the association between miR-663b
and TUSC2 in bladder cancer cells has not been previously
investigated.
Therefore, the aim of the current study was to
investigate the expression and specific role of miR-663b in bladder
cancer, and further to explore the associated mechanisms. The
findings may help to provide novel and effective targets, and an
additional theoretical basis for the treatment of bladder
cancer.
Materials and methods
Clinical samples
A total of 25 paired bladder cancer tissues and the
adjacent normal tissues were obtained from 25 patients with bladder
cancer (18 male, 7 female; age range, 27–61 years old) at the
Beijing Luhe Hospital (Beijing, China) between May 2015 and May
2017. Informed consent was obtained from every patient, and the
study was approved by The Ethics Committee of Beijing Luhe
Hospital.
Cell culture and cell
transfection
Bladder cancer cell lines T24, 5637, J82, UMUC3, and
a normal uroepithelium cell line (SV-HUC-1) were obtained from the
Shanghai Institute of Cell Biology, Chinese Academy of Sciences
(Shanghai, China). The cell lines were cultured in Dulbecco's
Modified Eagle's Medium (Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) supplement with 10% fetal bovine serum
(Sigma-Aldrich; Merck KGaA, USA) and 1% streptomycin-penicillin mix
solution, and incubated at 37°C with 5% CO2.
T24 cells were seeded in 6-well plates at a density
of 1×106 cells/well and cultured at 37°C for 24 h. T24
cells were transfected with 100 nM miR-663b inhibitor (cat. no.
CIH0743; Guangzhou Weijia Technology Co., Ltd., Guangzhou, China),
100 nM negative control (NC; cat. no. CS8005; Biomics
Biotechnologies Co., Ltd., Nantong, China), 10 µM control-small
interfering RNA (siRNA; cat. no. sc-36869; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA), 10 µM TUSC2-siRNA (cat. no.
sc-77910; Santa Cruz Biotechnology, Inc.), or miR-663b inhibitor +
TUSC2-siRNA at 37°C for 48 h by using Lipofectamine®
2000 (Thermo Fisher Scientific, Inc.) according to the
manufacturer's instruction. The working concentration was
determined following a dose-response analysis (data not shown).
Cells without any treatment were used as the control. Subsequent
analyses were performed at 48 h after cell transfection.
Dual luciferase reporter assay
A bioinformatics software (TargetScan; version 7.2;
http://www.targetscan.org/vert_72/)
was used to predict the potential targets of miR-663b, and an
miR-663b target site was identified in the TUSC2 3′UTR. To confirm
this prediction, a dual luciferase reporter assay was performed.
Wild-type (WT; 5′-AGGAGCCCAGGCUGGGGCCACA-3′) and mutant (MUT;
5′-AGGAGCCCAGGCUGAUAUAGAA-3′) TUSC2 3′UTRs were synthesized using
the PrimeScript reverse transcription (RT) reagent kit (Takara Bio,
Inc., Otsu, Japan) according to the manufacturer's protocol, using
RNA extracted from T24 cells. The reverse transcription was
performed by incubating the reaction at 50°C for 15 min and at 85°C
for 2 min. The cDNA obtained was subsequently used as template to
clone the 3′UTRs into the psiCHECK-2 reporter. Point mutations in
the binding site for miR-663b in the 3′UTR of TUSC2 were generated
using the QuikChange Site-Directed Mutagenesis kit (Stratagene;
Agilent Technologies, Inc., Santa Clara, CA, USA) according to the
manufacturer's protocol. miR-663b mimic (cat. no. HmiR-SN0775-SN;
GeneCopoeia, Inc., Rockville, MD, USA) at a concentration of 50 nM
or mimic control (cat. no. CmiR-SN0001-SN; GeneCopoeia, Inc.) at a
concentration at 50 nM and 10 µl miR-663b-TUSC2-WT 3′UTR or 10 µl
miR-663b-TUSC2-MUT 3′UTR vector were co-transfected into T24 cells
(5×104 cells/well) using Lipofectamine® 2000
(Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. At 48 h after the transfection, the luciferase activity
was analyzed by performing the dual-luciferase reporter assay
system (Promega Corporation, Madison, WI, USA) according to the
manufacturer's instruction, and normalized to Renilla
luciferase activity.
Cell viability assay
At 48 h following transfection, a Cell Counting
Kit-8 (CCK-8) (cat. no. C0038; Beyotime Institute of Biotechnology,
Haimen, China) assay was performed to detect cell viability
ability. In brief, T24 cells were collected, re-suspended and then
re-plated into 96-well culture plates (5×103 cells/well)
(Corning Incorporated, Corning, NY, USA). The cells were incubated
for 24 h at 37°C with 5% CO2. Subsequently, CCK-8
solution (10 µg/ml) was added to each well, and then incubated at
37°C for 2 h. Finally, the optical density value at 450 nm was
determined using a microplate reader (Thermo Fisher Scientific,
Inc.). Each experiment was repeated at least three times.
Apoptosis analysis assay
At 48 h after cell transfection, cell apoptosis
analysis was performed. The transfected T24 cells were washed with
PBS, labeled with 5 µl Annexin V-fluorescein isothiocyanate and 5
µl propidium iodide with the Annexin V-FITC Apoptosis Detection kit
(cat. no. C1062; Beyotime Institute of Biotechnology), and then
incubated at room temperature for 30 min in dark. A flow cytometer
(BD Biosciences, Franklin Lakes, NJ, USA) was used to analyze cell
apoptosis, and the proportion of apoptotic cells was calculated.
Experiments were repeated at least three times.
Western blot analysis
At 48 h after cell transfection, western blotting
was used to detect protein expression. Radioimmunoprecipitation
assay buffer (Auragene Bioscience, Changsha, China) was used to
extract the total cellular proteins. A bicinchoninic acid protein
quantitative kit (Thermo Fisher Scientific, Inc.) was performed to
detect protein concentration following the manufacturer's
instructions. Protein samples (25 µg/lane) were separated by using
SDS-PAGE on 11% gels and then transferred onto polyvinylidene
fluoride membranes. Following blocking with 5% skim milk at room
temperature for 1 h, the membranes were then incubated with a
primary antibody: Anti-TUSC2 (cat. no. sc-517369; 1:1,000; Santa
Cruz Biotechnology, Inc.), anti-p53 (cat. no. 2527; 1:1,000; Cell
Signaling Technology, Inc., Danvers, MA, USA), anti-p21 (cat. no.
2947; 1:1,000; Cell Signaling Technology, Inc.), anti-β-actin(cat.
no. 4970; 1:2,000; Cell Signaling Technology, Inc.) at 4°C
overnight. Subsequently, the membranes were washed with TBS-Tween
solution three times, then the membranes were incubated with an
anti-rabbit immunoglobulin G horseradish peroxidase-conjugated
secondary antibody (1:5,000; cat no. 7074; Cell Signaling
Technology, Inc.) at room temperature for 4 h. Finally, the protein
bands were visualized using an enhanced chemiluminescence kit
(Applygen Technologies, Inc., Beijing, China) according to the
manufacturer's protocol.
RT-quantitative polymerase chain
reaction (qPCR)
Total RNA from tissues and cells was extracted by
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) following the manufacturer's instructions.
Absorbance 260/280 ratio was calculated to assess the integrity and
quality of the RNA. U6 (for miRNA) and GAPDH (for mRNA) were used
as the internal controls. Total RNA was reverse transcribed into
cDNA using the TaqMan microRNA RT kit (Thermo Fisher Scientific,
Inc.) according to the manufacturer's instructions. mRNA was
reverse transcribed into cDNAs using the PrimeScript RT reagent kit
(Takara Bio, Inc.) according to the manufacturer's protocol. The
reverse transcription was performed by incubating the reaction at
50°C for 15 min and at 85°C for 2 min. RT-qPCR was performed using
SYBR Premix Ex Taq (Takara Bio, Inc.) according to the
manufacturer's instructions. The thermocycling conditions were as
follows: Initial denaturation at 95°C for 5 min, followed by 40
cycles of denaturation at 95°C for 15 sec and annealing/elongation
at 60°C for 30 sec. The primer sequences used for PCR were as
follows: GAPDH, forward 5′CTTTGGTATCGTGGAAGGACTC3′, reverse
5′GTAGAGGCAGGGATGATGTTCT3′; U6, forward
5′GCTTCGGCAGCACATATACTAAAAT3′, reverse 5′CGCTTCACGAATTTGCGTGTCAT3′;
TUSC2, forward 5′GGAGACAATCGTCACCAAGAAC3′, reverse
5′TCACACCTCATAGAGGATCACAG3′; p53, forward
5′CTGCCCTCAACAAGATGTTTTG3′, reverse 5′CTATCTGAGCAGCGCTCATGG3′; p21,
forward 5′ATGAAATTCACCCCCTTTCC3′, reverse 5′CCCTAGGCTGTGCTCACTTC3′;
miR-663b, forward 5′CATAATAAATAGGCGGGGCG3′, reverse
5′CAGAGCAGGGTCCGAGGTA3′. The relative gene expression was
determined using the 2−ΔΔCq method (23).
Statistical analysis
SPSS 16.0 statistical software (SPSS, Inc., Chicago,
IL, USA) was used for statistical analyses. Data are presented as
the mean ± standard deviation. Student's t-test or one-way analysis
of variance followed by Tukey's test was performed to compare
differences between groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
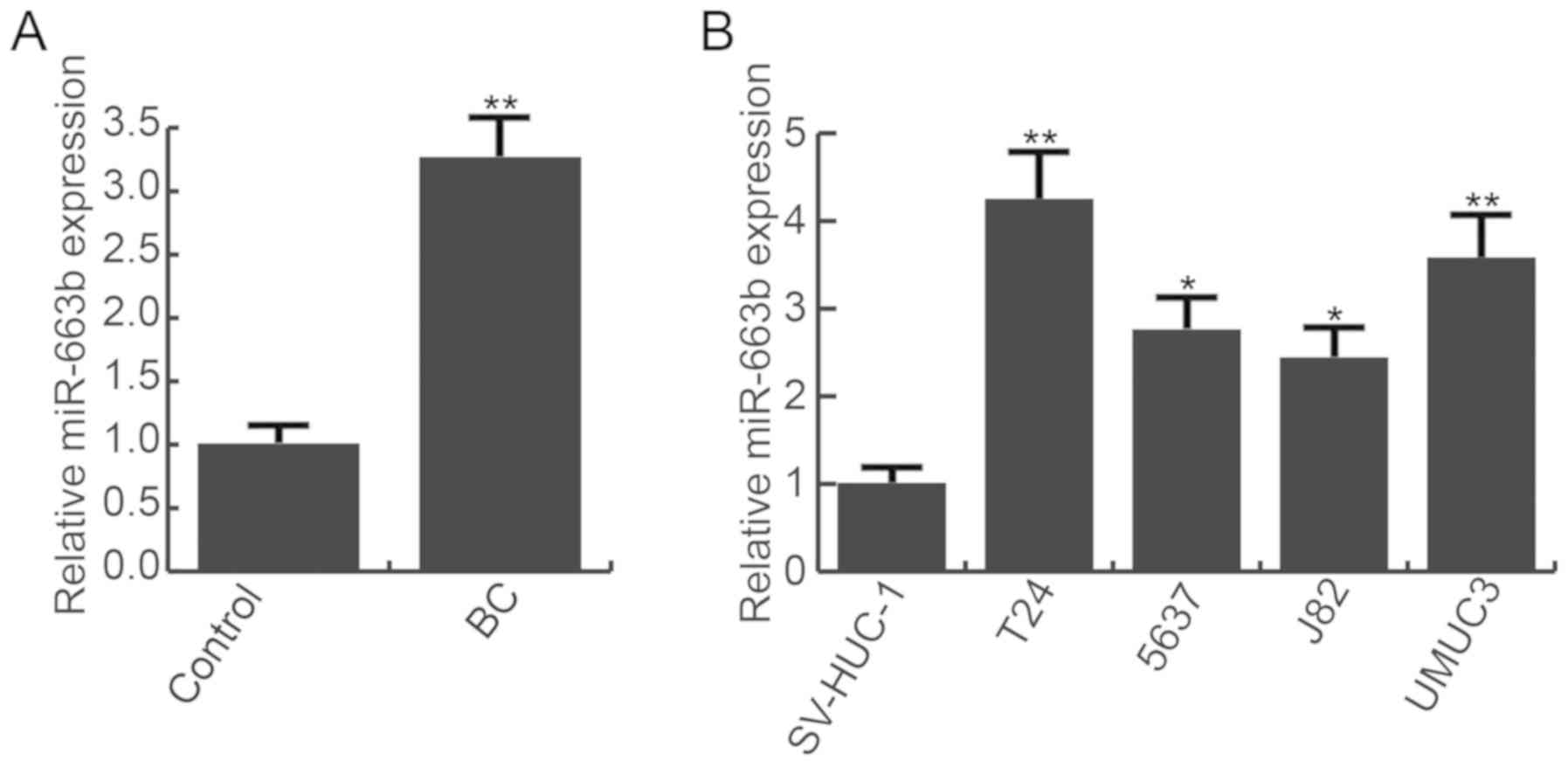
miR-663b is upregulated in bladder
cancer
RT-qPCR was performed to detect the level of
miR-663b in bladder cancer tissues and cell lines (T24, 5637, J82,
UMUC3) and the normal bladder cell line (SV-HUC-1). Compared with
adjacent normal tissues, the level of miR-663b was significantly
increased in bladder cancer tissues (Fig. 1A). Additionally, bladder cancer
cell lines (T24, 5637, J82, UMUC3) exhibited higher expression of
miR-663b than the normal bladder cell line (SV-HUC-1). miR-663b was
highest in T24 cells (Fig. 1B).
The data indicated that miR-663b was upregulated in bladder cancer.
T24 cells were used for subsequent analysis.
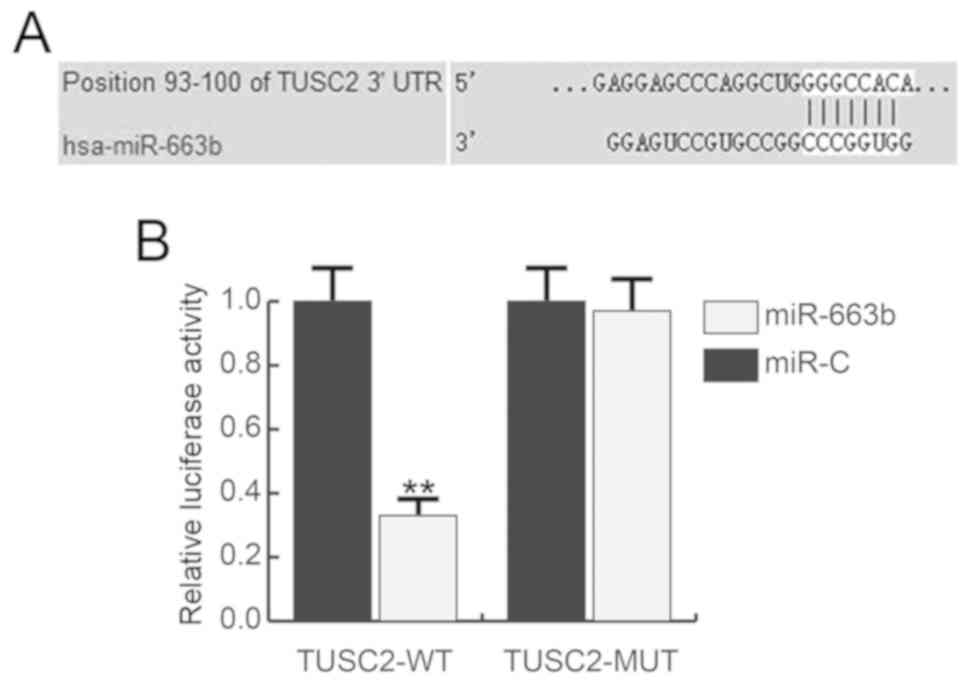
TUSC2 is a target of miR-663b
To study the precise role of miR-663b in bladder
cancer, bioinformatics software (TargetScan) we used to predict the
potential targets of miR-663b, and TUSC2 was revealed to have an
miR-663b binding site in the 3′UTR (Fig. 2A). The luciferase reporter gene
assay confirmed that miR-663b binds the TUSC2 3′UTR to reduce TUSC2
expression (Fig. 2B).
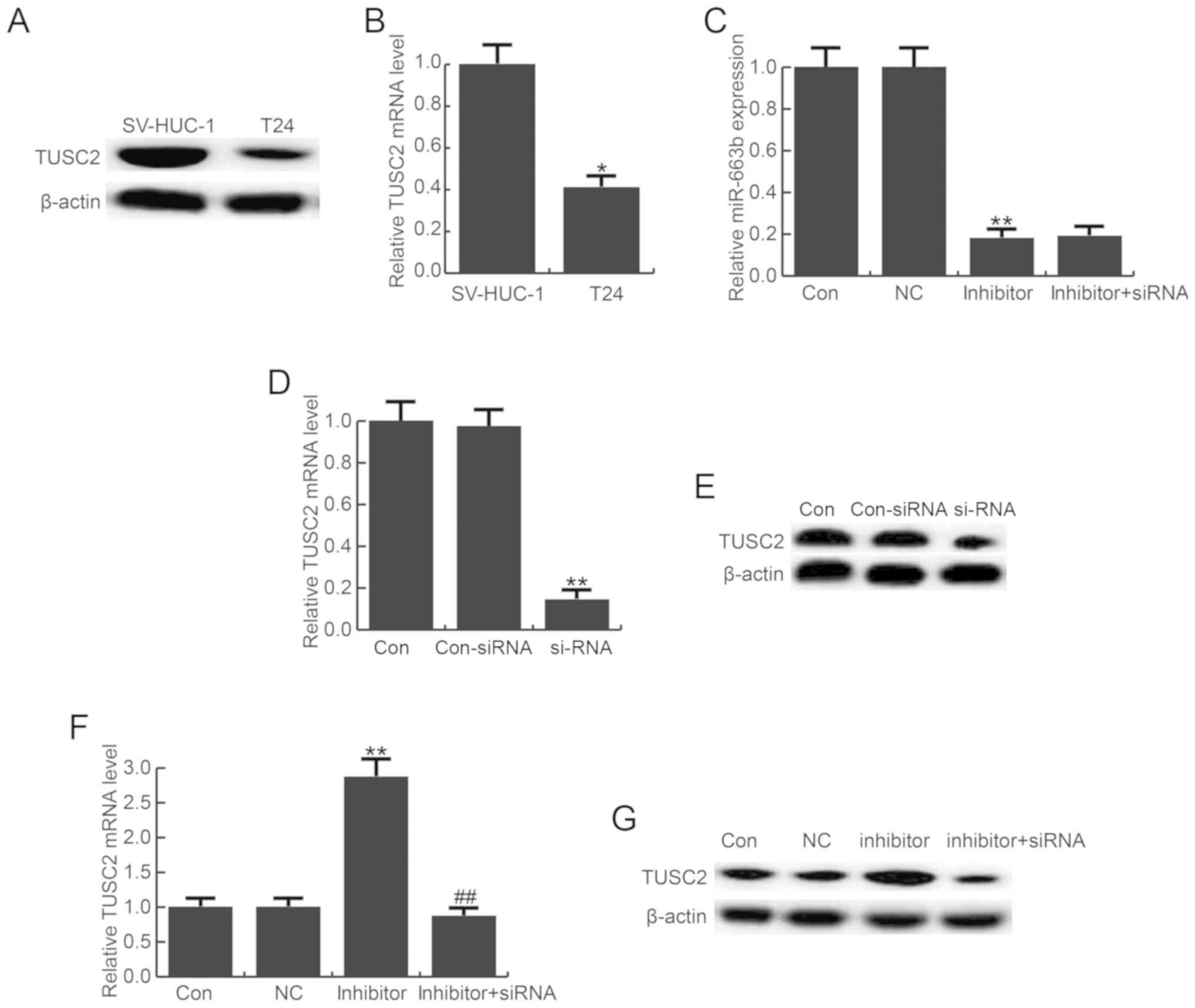
Furthermore, the level of TUSC2 was detected in
bladder cancer cell line T24 and the normal bladder cell line
SV-HUC-1, and the findings suggested TUSC2 was significantly
down-regulated in T24 cells at the protein and mRNA level compared
with the SV-HUC-1 cells (Fig. 3A and
B). To analyze the role of miR-663b, T24 cells were transfected
with miR-663b inhibitor, a negative control, control-siRNA,
TUSC2-siRNA, or miR-663b inhibitor + TUSC2-siRNA for 48 h. The
transfection efficiency of miR-663b inhibitor was evaluated using
RT-qPCR, and the results indicated that miR-663b inhibitor
significantly decreased the level of miR-663b in T24 cells
(Fig. 3C). TUSC2-siRNA
significantly reduced the mRNA and protein level of TUSC2 in T24
cells (Fig. 3D and E). miR-663b
inhibitor markedly enhanced the expression level of TUSC2, and this
increase was eliminated by TUSC2-siRNA (Fig. 3F and G).
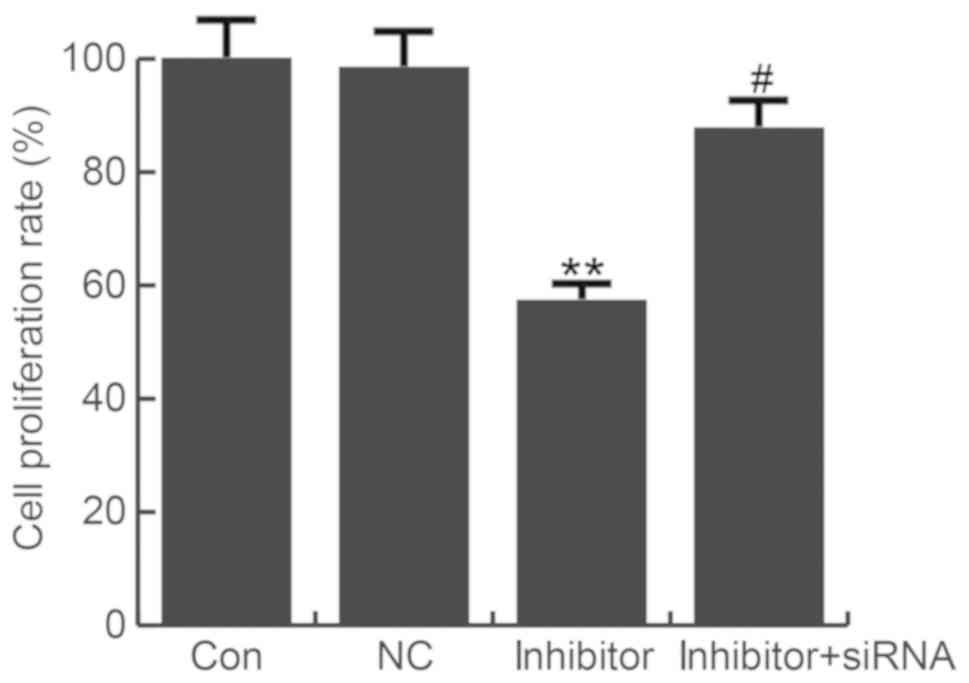
miR-663b inhibitor reduced T24 cell
viability
A CCK-8 assay was used to evaluate the effect of
miR-663b on T24 cell proliferation. miR-663b inhibitor
significantly reduced T24 cell viability, which was eliminated by
TUSC2-siRNA (Fig. 4).
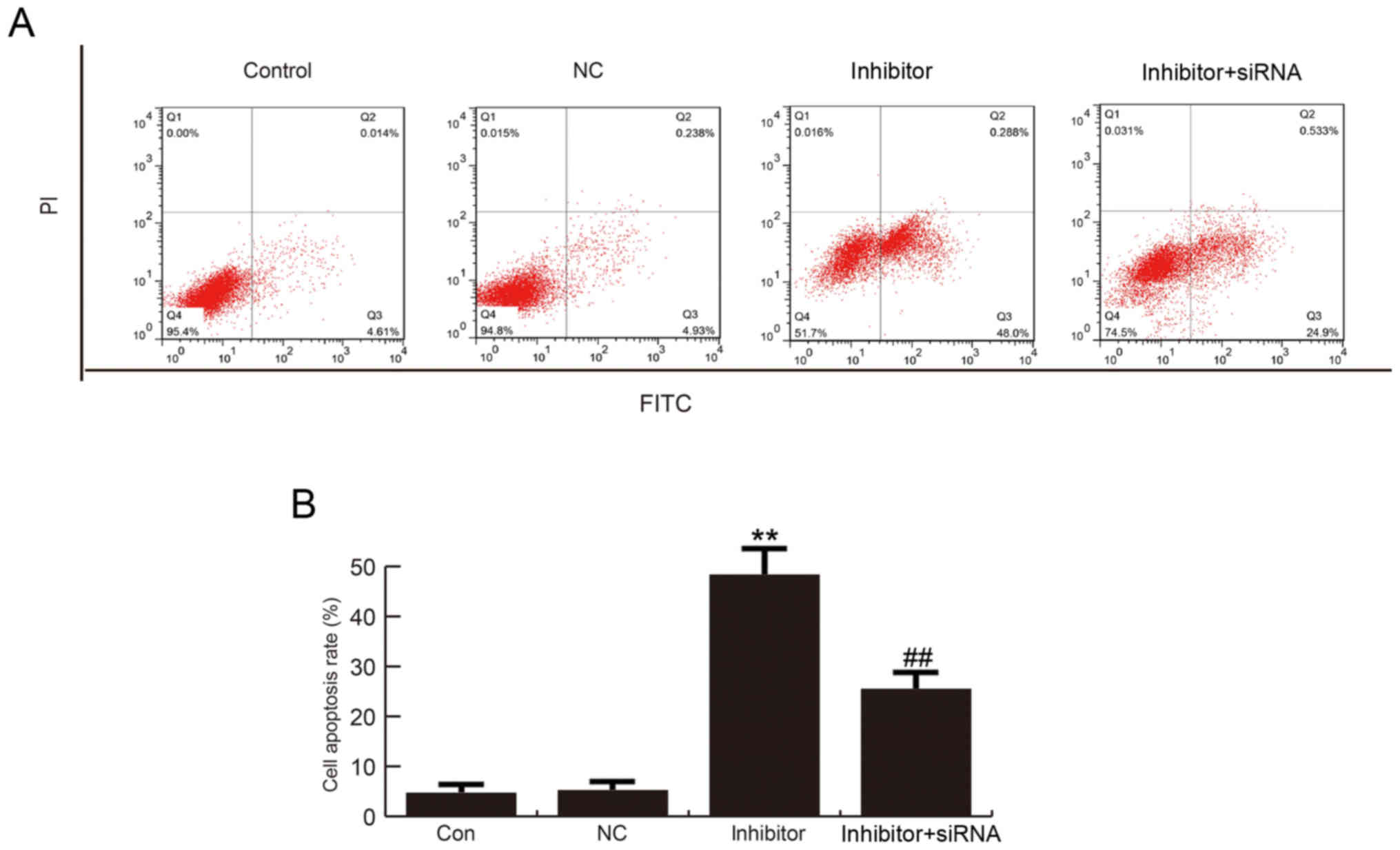
miR-663b inhibitor induced T24 cell
apoptosis
The effect of miR-663b on T24 cell apoptosis was
analyzed by using flow cytometry. The findings suggested that
compared with the control group, miR-663b inhibition significantly
induced T24 cell apoptosis, and this increase was reversed by
TUSC2-siRNA (Fig. 5A and B).
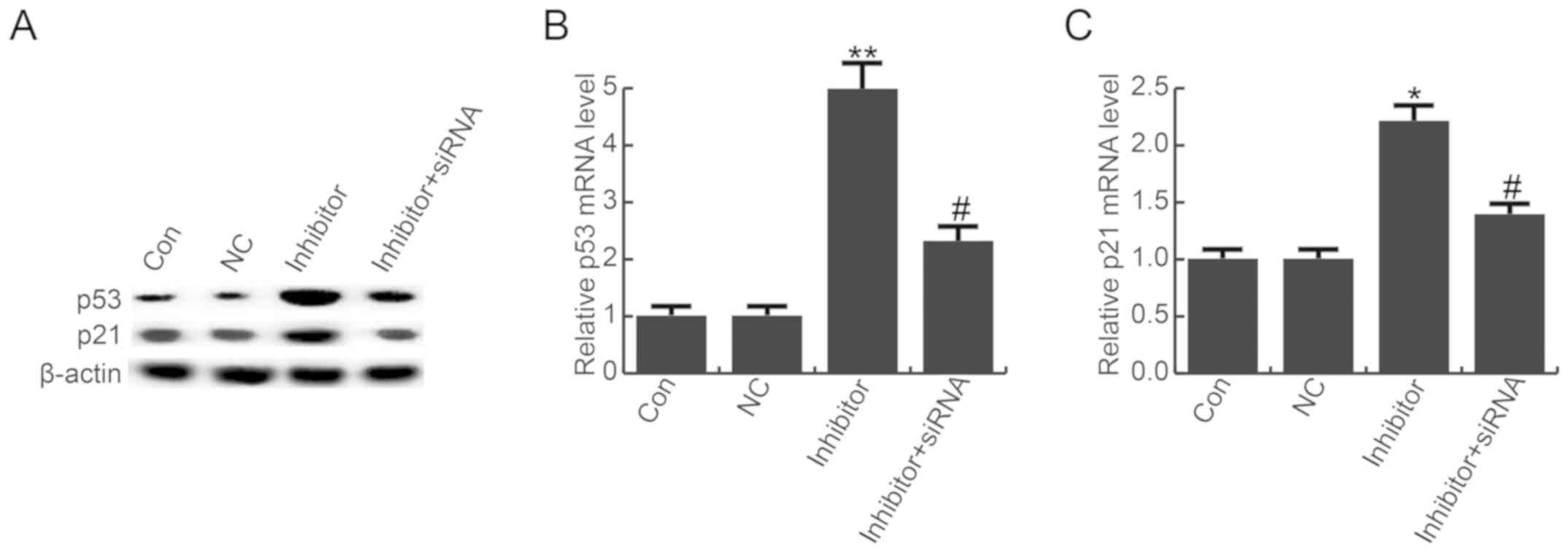
miR-663b down-regulation enhanced the
expression of p53 and p21
A previous study suggested that miR-663b
down-regulation promoted p53 and p21 expression in osteosarcoma
cells (24). Additionally, TUSC2
has been revealed to be associated with the p53 pathway (22). Therefore, to explore the underlying
molecular mechanism of the role of miR-663b in T24 cells,
expression of p53 and p21 was detected by western blot analysis and
RT-qPCR. Compared with the control group, the protein expression
levels of p53 and p21 were notably enhanced by miR-663b inhibitor,
and these effects were eliminated by TUSC2-siRNA (Fig. 6A). The same trend was detected by
RT-qPCR (Fig. 6B and C).
Discussion
Bladder cancer is one of the most common malignant
tumors in humans, with a complex pathogenesis involving numerous
genes, aberrant protein functions and changes in multiple signaling
pathways. Currently, the molecular genetics of the development and
progression of bladder cancer remain unclear. In recent years, the
pathological relevance and importance of miRNAs in bladder cancer
have attracted increasing attention. In the present study, the
expression, biological function and molecular mechanisms of
miR-663b and its target gene in the pathogenesis of bladder cancer
were investigated.
miR-663b inhibition reduced bladder cancer cell
viability and induced cell apoptosis. miR-663b was demonstrated to
directly target TUSC2 and negatively regulated TUSC2 expression.
miR-663b inhibition indirectly promoted the expression of p53 and
p21, which may contribute to the prevention of T24 cell
proliferation and the induction of T24 cell apoptosis. miR-663b in
bladder cancer tissues and cell lines, compared with normal tissue
and a normal bladder cell line, respectively. T24 cells had the
highest miR-663b level of the cell lines. Therefore, in the current
study, T24 cells were used for the investigation of bladder cancer
in vitro. A novel functional link between miR-663b and TUSC2
in the tumorigenesis of bladder cancer was also revealed. miR-663b
inhibitor significantly reduced cell viability and increased
apoptosis of T24 cells in vitro. miR-663b inhibition
significantly enhanced the expression levels of p53 and p21.
Furthermore, the effects of miR-663b inhibitor in T24 cells were
eliminated by TUSC2-siRNA. These results indicated that miR-663b
acts as an oncogene in the development of bladder cancer, and
miR-663b inhibitor had anti-tumor activity; thus, miR-663b may be
as a promising therapeutic target for bladder cancer treatment.
Various studies suggested that miRNAs have critical
roles in the diagnosis, therapy and prognosis of a variety of
cancers and can be involved in tumorigenesis, tumor growth and
tumor metastasis (25,26). Therefore, miRNAs may be promising
treatment targets for cancer. Increasing evidence has indicated
that numerous miRNAs are involved in bladder cancer cell
proliferation, migration and invasion (27–31).
High expression of miR-663b has been reported in the plasma of
patients with bladder cancer (15); however, to the best of our
knowledge, the expression of miR-663b in bladder tumors and its
specific role remain unclear. He findings of the current
demonstrated that miR-663b was significantly upregulated in bladder
cancer tissues and cell lines compared with adjacent tissues and
normal bladder cells, and miR-663b inhibitor markedly repressed the
viability of T24 cells and induced apoptosis.
TUSC2 is a recognized tumor suppressor gene
(16,17). A variety of allele losses and
genetic alterations are observed in various cancers, including
breast cancer, lung cancer and others (17,32,33).
Studies have reported that TUSC2 can induce G1 phase cell cycle
arrest and apoptosis (18),
regulate calcium signaling (19)
and tyrosine kinase activity (20), and affect gene expression (21). TUSC2 is associated with several
pathways, including the p53 pathway (22). However, to the best of our
knowledge, no interaction between miR-663b and TUSC2 in bladder
cancer cells has been reported previously. In the present study,
TUSC2 was identified as a direct target of miR-663b, and it was
demonstrated that miR-663b affects bladder cancer cell viability
and apoptosis by directly targeting TUSC2. Additionally, as TUSC2
is associated with p53 pathway (22), and it was speculated as to whether
the effect of miR-663b on bladder cancer cells was associated with
p53 pathway, thus p53 and p21 expression was analyzed in the
current study. The results indicated that miR-663b inhibition
increased the expression of p53 and p21 in bladder cancer
cells.
In summary, the current study demonstrated that
miR-663b was upregulated in bladder cancer tissues and cell lines,
and inhibition of miR-663b reduced the viability of bladder cancer
cells, induced cell apoptosis and enhanced the expression of p53
pathway proteins by directly targeting TUSC2. Therefore, miR-663b
may serve as a promising therapeutic target for the treatment of
bladder cancer. However, this is a preliminary study of the role of
miR-663b in bladder cancer, and further research is required in
order to make the role of miR-663b in bladder cancer more
convincing. For instance, the association of miR-663b expression
with the clinical characteristics and prognosis of patients with
bladder cancer required evaluation. Additionally, whether miR-663b
has a similar effect on bladder cancer in vivo requires
further investigation. Only one bladder cancer line was used in the
present study, the effect of miR-663b needs to be established in
more bladder cancer cell lines. Furthermore, the effects of TUSC2
and miR-663b overexpression in bladder cancer were not determined.
Furthermore, the in vitro analysis of bladder cancer is
considerably different from the bladder cancer in patients, thus
further in vivo experiments and clinical studies are
required to validate the role of miR-663b in bladder cancer.
In-depth research on these issues will be performed in the
future.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JC contributed to the design of the study, data
collection, statistical analysis and data interpretation. BS
contributed to data collection, manuscript preparation and
literature search. GK contributed to the design of the study and to
the preparation of the manuscript.
Ethics approval and consent to
participate
The study was approved by The Ethics Committee of
Beijing Luhe Hospital (Beijing, China).
Patient consent for publication
All patients provided their informed consent.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Swarup V and Rajeswari MR: Circulating
(cell-free) nucleic acids-a promising, non-invasive tool for early
detection of several human diseases. FEBS Lett. 581:795–799. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Antoni S, Ferlay J, Soerjomataram I, Znaor
A, Jemal A and Bray F: Bladder cancer incidence and mortality: A
global overview and recent trends. Eur Urol. 71:96–108. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Moyer VA and U.S. Preventive Services Task
Force: Screening for bladder cancer: U.S. Preventive Services Task
Force recommendation statement. Ann Intern Med. 155:246–251. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Griffiths TR and Action on Bladder Cancer:
Current perspectives in bladder cancer management. Int J Clin
Pract. 67:435–448. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Smith AB, Crowell K, Woods ME, Wallen EM,
Pruthi RS, Nielsen ME and Lee CT: Functional outcomes following
radical cystectomy in women with bladder cancer: A systematic
review. Eur Urol Focus. 3:136–143. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Schulz GB, Grimm T, Buchner A, Jokisch F,
Grabbert M, Schneevoigt BS, Kretschmer A, Stief CG and Karl A:
Prognostic value of the preoperative platelet-to-leukocyte ratio
for oncologic outcomes in patients undergoing radical cystectomy
for bladder cancer. Clin Genitourin Cancer. 15:e915–e921. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Coleman EA and Boult C; American
Geriatrics Society Health Care Systems Committee, : Improving the
quality of transitional care for persons with complex care need. J
Am Geriatr Soc. 51:556–557. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ambros V: microRNAs: Tiny regulators with
great potential. Cell. 107:823–826. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Valencia-Sanchez MA, Liu J, Hannon GJ and
Parker R: Control of translation and mRNA degradation by miRNAs and
siRNAs. Genes Dev. 20:515–524. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Catto JW, Alcaraz A, Bjartell AS, De Vere
White R, Evans CP, Fussel S, Hamdy FC, Kallioniemi O, Mengual L,
Schlomm T and Visakorpi T: MicroRNA in prostate, bladder, and
kidney cancer: A systematic review. Eur Urol. 59:671–681. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shi Z, Wei Q, Zhang M and She J: MicroRNAs
in bladder cancer: Expression profiles, biological functions,
regulation, and clinical implications. Crit Rev Eukaryot Gene Expr.
24:55–75. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gottardo F, Liu CG, Ferracin M, Calin GA,
Fassan M, Bassi P, Sevignani C, Byrne D, Negrini M, Pagano F, et
al: Micro-RNA profiling in kidney and bladder cancers. Urol Oncol.
25:387–392. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Du M, Shi D, Yuan L, Li P, Chu H, Qin C,
Yin C, Zhang Z and Wang M: Circulating miR-497 and miR-663b in
plasma are potential novel biomarkers for bladder cancer. Sci Rep.
5:104372015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xin J, Zhang XK, Xin DY, Li XF, Sun DK, Ma
YY and Tian LQ: FUS1 acts as a tumor-suppressor gene by
upregulating miR-197 in human glioblastoma. Oncol Rep. 34:868–876.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Uzhachenko R, Shanker A, Yarbrough WG and
Ivanova AV: Mitochondria, calcium, and tumor suppressor Fus1: At
the crossroad of cancer, inflammation, and autoimmunity.
Oncotarget. 6:20754–20772. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li L, Yu C, Ren J, Ye S, Ou W, Wang Y,
Yang W, Zhong G, Chen X, Shi H, et al: Synergistic effects of
eukaryotic coexpression plasmid carrying LKB1 and FUS1 genes on
lung cancer in vitro and in vivo. J Cancer Res Clin Oncol.
140:895–907. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Uzhachenko R, Ivanov SV, Yarbrough WG,
Shanker A, Medzhitov R and Ivanova AV: Fus1/Tusc2 is a novel
regulator of mitochondrial calcium handling, Ca2+-coupled
mitochondrial processes, and Ca2+-dependent NFAT and NF-κB pathways
in CD4+ T cells. Antioxid Redox Signal. 20:1533–1547. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lin J, Sun T, Ji L, Deng W, Roth J, Minna
J and Arlinghaus R: Oncogenic activation of c-Abl in non-small cell
lung cancer cells lacking FUS1 expression: Inhibition of c-Abl by
the tumor suppressor gene product Fus1. Oncogene. 26:6989–6996.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ivanova AV, Ivanov SV, Prudkin L, Nonaka
D, Liu Z, Tsao A, Wistuba I, Roth J and Pass HI: Mechanisms of
FUS1/TUSC2 deficiency in mesothelioma and its tumorigenic
transcriptional effects. Mol Cancer. 8:912009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rimkus T, Sirkisoon S, Harrison A and Lo
HW: Tumor suppressor candidate 2 (TUSC2, FUS-1) and human cancers.
Discov Med. 23:325–330. 2017.PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shu Y, Ye W, Gu YL and Sun P: Blockade of
miR-663b inhibits cell proliferation and induces apoptosis in
osteosarcoma via regulating TP73 expression. Bratisl Lek Listy.
119:41–46. 2018.PubMed/NCBI
|
|
25
|
Rogers K and Chen X: Biogenesis, turnover,
and mode of action of plant microRNAs. Plant Cell. 25:2383–2399.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cho WC: MicroRNAs: Potential biomarkers
for cancer diagnosis, prognosis and targets for therapy. Int J
Biochem Cell Biol. 42:1273–1281. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang X, Shi L, Yi C, Yang Y, Chang L and
Song D: MiR-210-3p inhibits the tumor growth and metastasis of
bladder cancer via targeting fibroblast growth factor receptor-like
1. Am J Cancer Res. 7:1738–1753. 2017.PubMed/NCBI
|
|
28
|
Luo H, Yang R, Li C, Tong Y, Fan L, Liu X
and Xu C: MicroRNA-139-5p inhibits bladder cancer proliferation and
self-renewal by targeting the Bmi1 oncogene. Tumour Biol.
39:10104283177184142017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang W, Shen F and Wang C, Lu W, Wei J,
Shang A and Wang C: MiR-1-3p inhibits the proliferation and
invasion of bladder cancer cells by suppressing CCL2 expression.
Tumour Biol. 39:10104283176983832017.PubMed/NCBI
|
|
30
|
Wu D, Niu X, Tao J, Li P, Lu Q, Xu A, Chen
W and Wang Z: MicroRNA-379-5p plays a tumor-suppressive role in
human bladder cancer growth and metastasis by directly targeting
MDM2. Oncol Rep. 37:3502–3508. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Takai T, Yoshikawa Y, Inamoto T, Minami K,
Taniguchi K, Sugito N, Kuranaga Y, Shinohara H, Kumazaki M, Tsujino
T, et al: A novel combination rnai toward warburg effect by
replacement with miR-145 and silencing of PTBP1 induces apoptotic
cell death in bladder cancer cells. Int J Mol Sci. 18:E1792017.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Meng J, Majidi M, Fang B, Ji L, Bekele BN,
Minna JD and Roth JA: The tumor suppressor gene TUSC2 (FUS1)
sensitizes NSCLC to the AKT inhibitor MK2206 in LKB1-dependent
manner. PLoS One. 8:e770672013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
da Costa Prando E, Cavalli LR and Rainho
CA: Evidence of epigenetic regulation of the tumor suppressor gene
cluster flanking RASSF1 in breast cancer cell lines. Epigenetics.
6:1413–1424. 2011. View Article : Google Scholar : PubMed/NCBI
|