Introduction
Hepatocellular carcinoma (HCC) is the third most
common malignancy and second leading cause of cancer-associated
mortality worldwide (1). Notably,
only 10–20% of patients with HCC can be treated surgically, whereas
the majority of patients are treated exclusively with chemotherapy
(2). In addition, the various
genetic backgrounds of individuals lead to unsatisfactory outcomes
following chemotherapy. Cinobufagin, a natural inhibitor of
Na+/K+-ATPase (NKA), is a cardiac glycoside
(CG) that exhibits potential anticancer activity (3,4). A
clinical trial revealed that the interaction between digoxin and
cisplatin had a synergistic effect on cervical cancer cells
(5).
Numerous studies have reported that mutations in
various genes can markedly alter the efficacy of chemotherapy,
including p53, B-Raf proto-oncogene, serine/threonine kinase and
erb-b2 receptor tyrosine kinase 2 (6–9).
Mutations in p53 occur frequently in numerous types of malignant
tumor (10–12). HCC cells have been reported to
express three genotypes of p53: Wild-type p53, mutant p53 and
p53-null (13). Our previous study
revealed that CGs induce cell cycle S phase arrest and lead to
chromosome segregation distortion in HCC HepG2 cells expressing
wild-type p53 by inhibiting the function of aurora kinase A (AURKA)
and activating p53 signaling (14).
As a member of the aurora kinase family, AURKA is a
serine/threonine kinase, which primarily serves roles in regulation
of the cell cycle, separation and maturation of centrosomes, and
establishment of the spindle. Numerous studies have reported the
abnormal amplification and overexpression of AURKA in various types
of malignant tumor, including liver, lung and breast cancer
(15–18). The overexpression of AURKA is
associated with aggressive, poorly differentiated tumors (19,20).
p53 is a tumor suppressor gene (TSG) downstream of AURKA that is
involved in the regulation of cellular activities via
phosphorylation, including cell cycle arrest, DNA repair and
apoptosis (21). Various mutations
in p53 have been reported in malignant tumors; >50% of mutations
are in the form of single nucleotide mutations (22). Mutant p53 loses its TSG function
and acts as an oncogene, promoting the proliferation of tumor
cells, increasing the invasive and metastatic abilities of tumor
cells, and inducing resistance to chemotherapy (23). Notably, p73, a member of the p53
family, can be activated to perform TSG functions in place of the
mutant p53, regulating the transcription of p21, growth arrest and
DNA damage-inducible protein 45 and B-cell lymphoma 2
(Bcl-2)-associated X protein (Bax) (24). The effects of cinobufagin on HCC
cells expressing mutant p53 and the underlying mechanisms remain
unclear. Therefore, the anticancer properties of cinobufagin in
Huh-7 cells with mutant p53 were investigated in this study.
Materials and methods
Chemicals and reagents
Cinobufagin (cat. no. C1272-1MG) and the rabbit
polyclonal anti-phosphorylated (p)-p53 (S315; cat. no. SAB4504500)
antibody were purchased from Sigma-Aldrich (Merck KGaA, Darmstadt,
Germany). Dulbecco's modified Eagle's medium (DMEM) and fetal
bovine serum (FBS) were purchased from Gibco (Thermo Fisher
Scientific, Inc., Waltham, MA, USA). The AURKA inhibitor
PF-03814735 and the following antibodies were purchased from Abcam
(Cambridge, UK): Rabbit monoclonal anti-p21 (cat. no. ab109520),
anti-p73 (cat. no. ab40658), anti-mouse double minute 2 homolog
(MDM2; cat. no. ab178938), anti-p-p53 (S392; ab33889), rabbit
polyclonal anti-p-heterogeneous nuclear ribonucleoprotein K
(hnRNPK; S284; cat. no. ab74066), anti-p53 upregulated modulator of
apoptosis (PUMA; cat. no. ab9643), mouse monoclonal anti-β-actin
(cat. no. ab8226), anti-phorbol-12- myristate-13-acetate-induced
protein 1 (Noxa; cat. no. ab13654) anti-hnRNPK (cat. no. ab39975),
rabbit monoclonal anti-Bcl-2 (cat. no. ab32124), rabbit monoclonal
anti-Bax (cat. no. ab32503), rabbit monoclonal anti-CDK1 (cat. no.
ab18), rabbit monoclonal anti-CylinB1 (cat. no. ab32053) and rabbit
monoclonal anti-PCNA (cat. no. ab92552).
The rabbit monoclonal anti-AURKA (cat. no. 14475),
rabbit polyclonal anti-p-p73 (Y99; cat. no. 4665) and anti-p-MDM2
(S166; cat. no. 3521) antibodies were purchased from Cell Signaling
Technology, Inc. (Danvers, MA, USA). The mouse monoclonal anti-p53
(cat. no. MF267038) antibody was purchased from Mei5 Biotechnology,
Co., Ltd. (Beijing, China). The propidium iodide (PI) kit (cat. no.
Y-6002-T), dimethyl sulfoxide (DMSO; cat. no. D8370) and MTT
reagents (cat. no. M1020) were purchased from Beijing Solarbio
Science & Technology Co., Ltd. (Beijing, China). The
PrimeSTAR® HS DNA Polymerase (cat. no. R010Q) was
purchased from Takara Bio, Inc., Otsu, Japan.
Cell culture
Huh-7 cells (possessing an Tyr220Cys point mutation
in the p53 gene) (25,26) were purchased from the American Type
Culture Collection (Manassas, VA, USA) and were cultured in DMEM
supplemented with 10% FBS, penicillin (100 U/ml) and streptomycin
(100 µg/ml). The cells were cultured at 37°C in a humidified 5%
CO2 atmosphere.
Cell transfection
Full-length AURKA cDNA was obtained from Huh-7 cells
via polymerase chain reaction (PCR) using the following primer
pairs: Forward 5′-CGGGATCCATGGACCGATCTAAG-3′ and reverse,
5′-CCGCTCGAGCTAAGACTGTTTGCTAG-3′. The PCR was performed in a 50 µl
volume with PrimeSTAR® HS DNA Polymerase mix (Takara
Bio, Inc.). The PCR was performed in a GeneAMP@PCR
System 9700 (Applied Biosystems; Thermo Fisher Scientific, Inc.),
according to the following protocol: 5 min at 95°C and 35 cycles of
10 sec at 98°C, 15 sec at 60°C and 2 min at 72°C, followed by 10
min at 72°C. The PCR product, flanked by BamHI and
XhoI sites, was cloned into a pCDNA3.1–3×Flag plasmid
(BioVector NTCC Inc., Beijing, China) to construct the expression
vector pCDNA3.1×3Flag-AURKA. Huh-7 cells were cultured in 6-well
plates overnight and were transfected with 2 µg
pCDNA3.1×3Flag-AURKA or blank vector using 5 µl
Lipofectamine® 3000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) at 37°C for 36 h. Stable Huh-7/AURA cell lines
were established using 1,000 µg/ml G418 for screening. Huh-7 cells
treated with the Huh-7 cells treated with the 50 nmol/l AURKA
inhibitor PF-03814735 at 37°C for 24 h were termed the
Huh7/PF-03814735 group. A series of experiments included six
groups, including Huh-7, Huh-7/AURKA and Huh-7/PF-03814735 cells,
untreated or co-treated with 5 µmol/l cinobufagin at 37°C for 24
h.
Cell viability assay
An MTT assay was performed to determine cell
viability. A total of 5×103 cells suspended in 100 µl
DMEM supplemented with 10% FBS were seeded into 96-well plates and
cultured for 24 h. The medium was replaced with fresh DMEM
containing 10% FBS and various concentrations of cinobufagin (0.75,
1.25, 2.5, 5 and 10 µmol/l). After treatment with cinobufagin or
both 5 µmol/l cinobufagin and 50 nmol/l PF-03814735 at 37°C for 24
h, MTT reagent (20 µl) was added to each well, and the plate was
incubated for a further 3 h. Subsequently, the medium was
discarded, 100 µl DMSO was added, and the optical density (OD) at
490 nm was detected using an immunosorbent assay microplate reader
(SpectraMax M2; Molecular Devices, LLC, Sunnyvale, CA, USA). Cell
viability rates were calculated using the following equation:
Inhibitory rate (IR) (%)=100×
[(ODTreatment-ODBlank)/(ODControl-ODBlank)].
The half maximal inhibitory concentration (IC50) of
cinobufagin in Huh-7 cells was also calculated. Another MTT assay
was subsequently performed using transfected or 50 nmol/l
PF-03814735-treated cells in the presence or absence of 5 µmol/l
cinobufagin, according to the aforementioned method.
Cell apoptosis assay
Cells were seeded into 6-well plates at a density of
5×104 cells/well and were cultured for 12 h followed by
treatment with 5 µmol/l cinobufagin and/or 50 nmol/l PF-03814735 at
37°C for 24 h. Cells were washed with ice-cold PBS and were
subsequently incubated with 10 µg/ml Hoechst 33342 at 37°C for 10
min. Chromatin condensation was observed under a fluorescence
microscope (BX41; Olympus Corporation, Tokyo, Japan).
Cell cycle analysis
Cells were trypsinized and fixed in a mixture of 75%
ethanol and 25% PBS at 4°C for 10 min. A total of 2×105
cells were incubated for 45 min with a solution containing 25 µg/ml
PI and 40 µg/ml RNase A at 37°C. The cell cycle distribution was
detected using a flow cytometer (FACSCalibur; BD Biosciences,
Franklin Lakes, NJ, USA) and analyzed using FlowJo V10 software
(FlowJo LLC, Ashland, OR, USA).
Western blot analysis
Cells were cultured until they reached 85%
confluence and were then treated with 5 µmol/l cinobufagin at 37°C
for 12 h in the presence or absence of 50 nmol/l PF-03814735.
Subsequently, cells were lysed in radioimmunoprecipitation assay
buffer [50 mmol/l Tris-HCl (pH 7.4), 150 mmol/l NaCl, 1 mmol/l
EDTA, 5% (v/v) β-mercaptoethanol, 1% (v/v) NP-40, 0.25% (v/v)
sodium deoxycholate, complete protease inhibitor cocktail (cat. no.
P8340; Sigma-Aldrich; Merck KGaA], ultrasonicated at 25% maximum
power (20 kHz) (10 sec burst and 20 sec rest, 30 cycles) for 15 min
on ice and centrifuged at 4°C with 13,000 × g for 10 min. The
supernatant was collected and stored at −80°C. Total protein
concentration was determined using the bicinchoninic acid assay
(Pierce; Thermo Fisher Scientific, Inc.). Protein lysates (60
µg/lane) were separated by 10% SDS-PAGE and transferred to
nitrocellulose membranes. The membranes were blocked using 5%
bovine serum albumin (BSA; cat. no. 0218054950; GE Healthcare Life
Sciences, Little Chalfont, UK) for 1 h at 37°C and incubated with
the primary antibodies at a dilution of 1:1,000 overnight at 4°C.
After four washes with PBS/0.1% Tween 20, the membranes were
incubated at room temperature for 1 h with horseradish
peroxidase-conjugated secondary antibodies (1:5,000; cat. no.
01-15-06; KPL, Inc., Gaithersburg, MD, USA). Protein bands were
visualized using an enhanced chemiluminescence immunoblotting
reagent (EMD Millipore, Billerica, MA, USA), and signals were
captured using the Amersham Imager 600 system (GE Healthcare Life
Sciences, Little Chalfont, UK). Protein expression was
semi-quantified using Scion Image version 4.0 (Scion Corporation,
Frederick, MD, USA), with β-actin as the reference control.
Immunofluorescence and confocal
microscopy
A total of 3×103 cells were seeded onto
confocal dishes and treated as aforementioned. Cells were fixed in
4% paraformaldehyde in PBS at 4°C for 30 min and then permeabilized
with 0.5% Triton X-100 in PBS. Cells were blocked with 5% BSA in
PBS at room temperature for 1 h and were then incubated with rabbit
anti-p73 (1:1,000) overnight at 4°C, followed by incubation with a
fluorescein isothiocyanate-conjugated goat anti-rabbit
immunoglobulin G antibody (1:200; cat. no. 5230-0301; KPL, Inc.) at
37°C for 1 h. Cells were washed three times with PBS (10 min/wash),
and the fluorescence was visualized using laser scanning confocal
microscopy (Leica TCS SP8; Leica Microsystems GmbH, Wetzlar,
Germany). The gray intensity of the images was analyzed using
Image-Pro Plus 6.0 software (Media Cybernetics, Inc., Rockville,
MD, USA).
Statistical analysis
Data were analyzed using SPSS version 22.0 software
(IBM Corp., Armonk, NY, USA). Data were analyzed using one-way
analyses of variance followed by a Tukey's test against the control
group to adjust for multiple comparisons. Data were presented as
the means ± standard error of the mean of three independent
experiments. P<0.05 was considered to indicate a statistically
significant difference.
Results
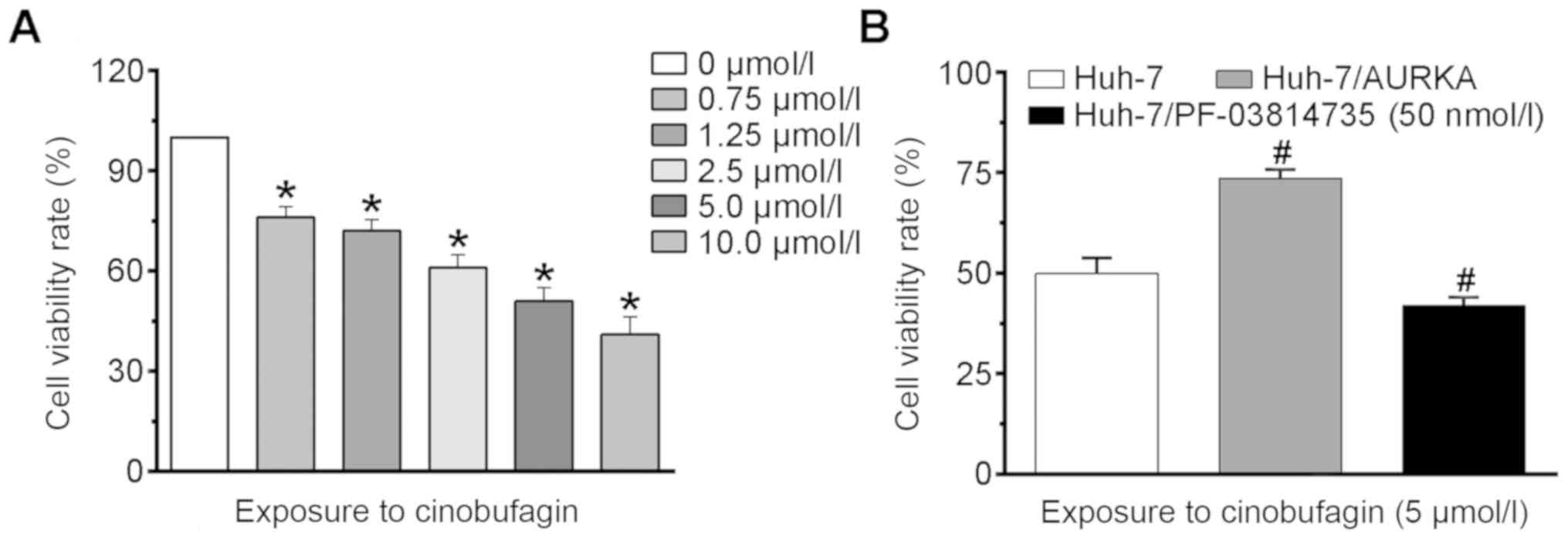
Decreased viability of Huh-7 cells
following cinobufagin treatment is associated with AURKA
activity
An MTT assay was performed to analyze the effects of
cinobufagin on the viability of Huh-7 cells. As presented in
Fig. 1A, treatment with >0.75
µmol/l cinobufagin significantly decreased the viability of cells
compared with the control (P<0.05). The viability rate of the
control group was normalized as 100%, and the viability rates of
Huh-7 cells following treatment with 0.75, 1.25, 2.5, 5 and 10
µmol/l cinobufagin for 24 h were reduced by 24.1±3.2, 28.1±3.5,
39.1±3.8, 49.3±4.1 and 60.3±5.22%, respectively. The
IC50 value of cinobufagin in Huh-7 cells was determined
to be 5.1 µmol/l at 24 h. Following treatment with 5 µmol/l
cinobufagin for 24 h, the viability rate of Huh-7 cells was
determined to be 49.5±3.9% compared with that of the control group;
however, transfection with the AURKA expression vector
significantly increased the viability rate of Huh-7 cells to
73.6±2.2% (Fig. 1B). Conversely,
treatment with 50 nmol/l PF-03814735 significantly decreased the
cell viability rate to 41.9±2.2%. The results of the MTT assay
indicated that cinobufagin inhibited the viability of mutant p53
HCC cells. Additionally, overexpressing and inhibiting AURKA
inhibited and promoted the anti-viability effects of cinobufagin on
Huh-7 cells, respectively.
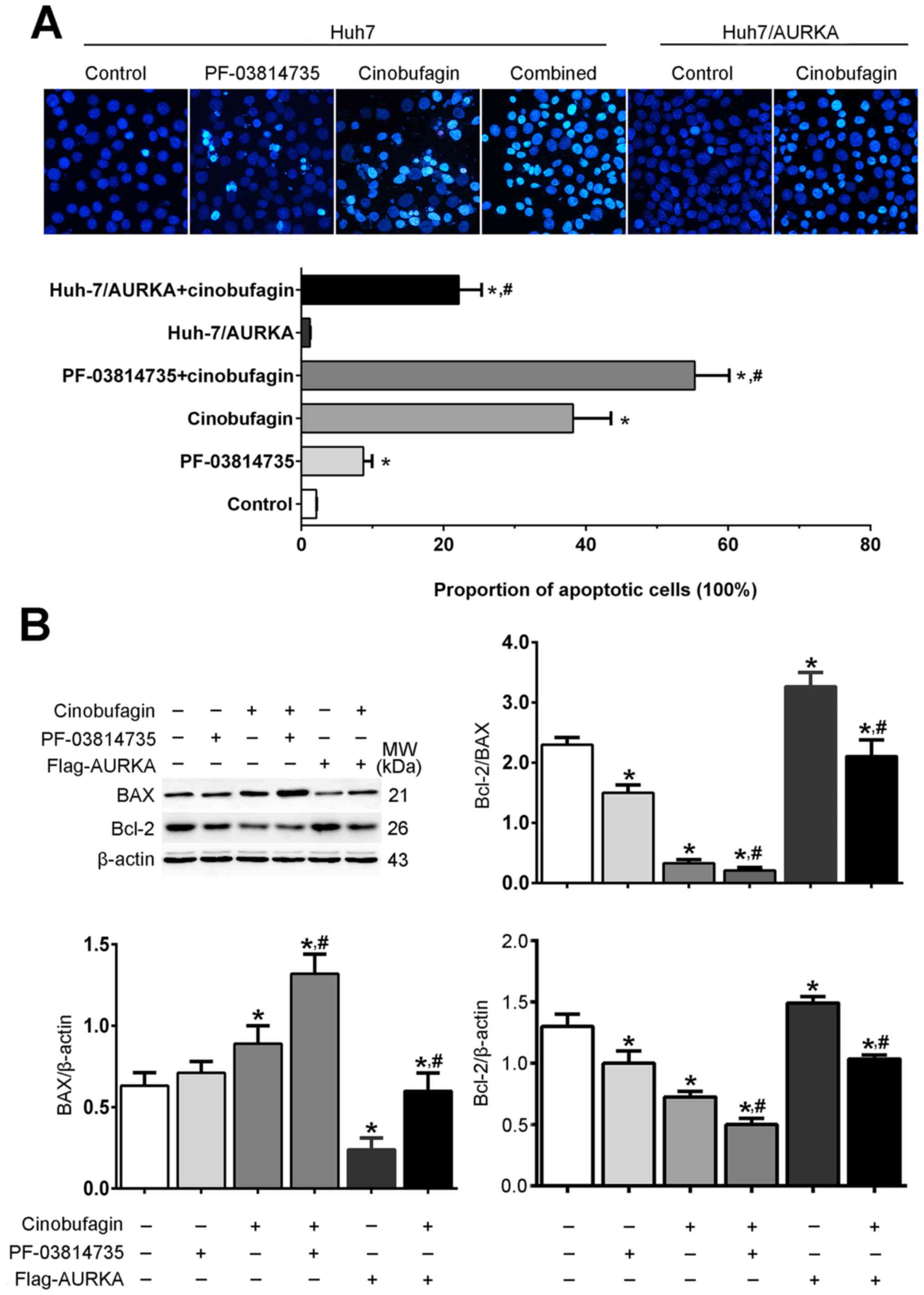
Cinobufagin-induced apoptosis of Huh-7
cells is associated with AURKA activity
Hoechst 33342 staining revealed that the nuclear
morphologies of Huh-7 and Huh-7/AURKA cells presented uniform blue
fluorescence, dispersed chromatin and oval nuclei, whereas cells
treated with cinobufagin and/or PF-03814735 detached from the
culture plate, shrunk and formed small apoptotic bodies (Fig. 2A). Other characteristics of
apoptosis were also observed following cinobufagin treatment,
including nuclear condensation and fragmentation. The proportion of
apoptotic cells in the PF-03814735, cinobufagin and PF-03814735 +
cinobufagin groups were significantly increased compared with in
the control group (P<0.05); however, there was no significant
difference between the control and Huh-7/AURKA groups. Furthermore,
the proportion of apoptotic cells in the PF-03814735 + cinobufagin
group was significantly increased compared with in the cinobufagin
group (P<0.05), whereas that in the Huh-7/AURKA + cinobufagin
group was significantly decreased (P<0.05). Western blot
analysis revealed that cinobufagin treatment significantly
downregulated the expression levels of the antiapoptotic protein
Bcl-2 in Huh-7 cells compared with the control, and upregulated
those of the proapoptotic protein Bax (P<0.05; Fig. 2B). In addition, the Bcl-2/Bax ratio
was significantly decreased following cinobufagin treatment
compared with the control (P<0.05). Furthermore, the
overexpression and inhibition of AURKA in cinobufagin-treated Huh-7
cells significantly increased and decreased the Bcl-2/Bax ratio
compared with cinobufagin treatment alone, respectively
(P<0.05). These results indicated that cinobufagin induced the
apoptosis of Huh-7 cells in an AURKA-dependent manner.
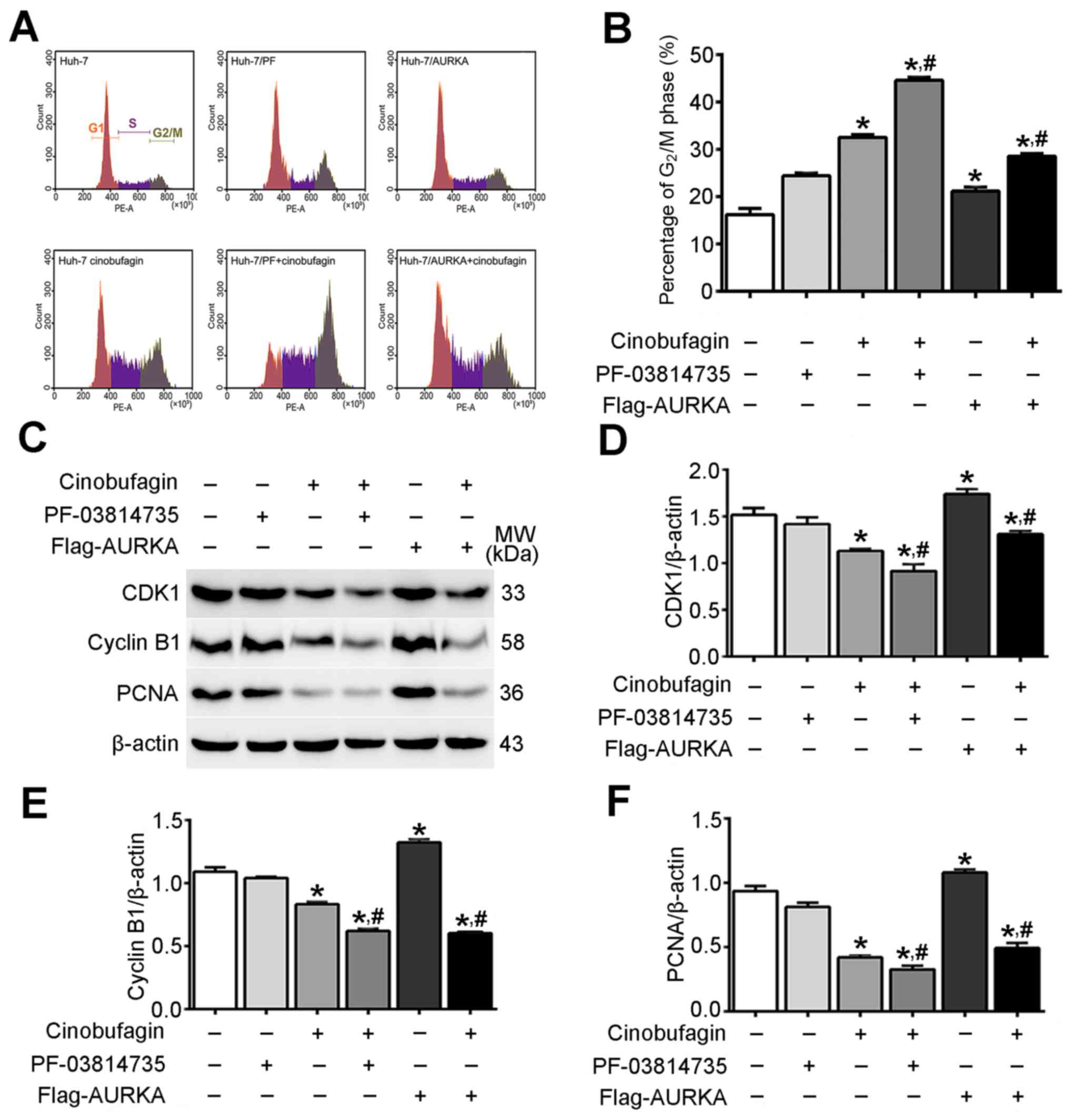
Cinobufagin induces cell cycle arrest
in Huh-7 cells by inhibition of AURKA signaling
As presented in Fig.
3A, cinobufagin induced the arrest of Huh-7 cells in
G2/M phase. The proportion of cells in the
G2/M phase was 16.17±0.77, 24.39±0.35 and 21.14±0.48 in
the control, PF-03814735-treated and Flag-AURKA-transfected groups,
respectively; treatment with cinobufagin increased the proportions
in the respective groups to 32.52±0.34, 44.58±0.41 and 28.53±0.31%.
The proportion of cells in G2/M phase in the cinobufagin
group was significantly increased compared with in the control
group (P<0.05; Fig. 3B).
Additionally, overexpressing or inhibiting AURKA in
cinobufagin-treated Huh-7 cells significantly decreased or
increased the proportion of G2/M phase cells,
respectively, compared with cinobufagin treatment alone
(P<0.05). Western blot analysis revealed that cinobufagin
decreased the expression levels of cyclin-dependent kinase 1
(CDK1), cyclin B1 and proliferating cell nuclear antigen (PCNA) in
Huh-7 cells compared with the control (P<0.05; Fig. 3C-F). Additionally, overexpression
or inhibition of AURKA in cinobufagin-treated Huh-7 cells
significantly promoted or weakened the changes in the expression of
these changed proteins between Huh-7 cells treated with cinobufagin
and the control group, respectively (P<0.05). The results
indicated that cinobufagin induced cell cycle G2/M phase
arrest in Huh-7 cells via downregulation of CDK1, cyclin B1 and
PCNA in an AURKA-dependent manner.
Anticancer effects of cinobufagin on
Huh-7 cells are not dependent on the activation of p53
signaling
The present study revealed that the expression
levels of total p53 and hnRNPK were not affected following
cinobufagin treatment, or overexpression or inhibition of AURKA
(Fig. 4A). Conversely, the
expression levels of AURKA, p-p53 (S315), p-p53 (S392) and p-hnRNPK
were significantly downregulated in cinobufagin-treated Huh-7 cells
compared with the control (P<0.05; Fig. 4B). The expression of AURKA was
significantly upregulated following transfection with the
Flag-AURKA vector. Additionally, it was demonstrated that p-p53
(S315)/p53, p-p53 (S392)/p53 and p-hnRNPK (S284)/hnRNPK ratios were
significantly decreased following cinobufagin treatment compared
with the control. Overexpression and inhibition of AURKA in
cinobufagin-treated cells significantly increased and decreased the
p-p53 (S315)/p53 ratio compared with cinobufagin treatment alone,
respectively; however, the p-p53 (S392)/p53 ratio was significantly
increased in cinobufagin-treated cells following transfection with
Flag-AURKA or treatment with PF-03814735 compared with cinobufagin
treatment alone, whereas the p-hnRNPK (S284)/hnRNPK ratio was not
significantly different between the cinobufagin and cinobufagin +
PF-03814735 groups. The results suggested that cinobufagin
downregulated the expression of AURKA, and inhibited the
phosphorylation of p53 (S315 and S392) and hnRNPK (S284).
Furthermore, overexpression of AURKA upregulated phosphorylation of
p53 (S315), p53 (S392) and hnRNPK (S284) compared with cinobufagin
treatment; however, PF-03814735 + cinobufagin treatment further
decreased the phosphorylation of p53 (S315) only. Notably, the
expression of total p53 was not altered, and the levels of p-p53
(S315 and S392) were significantly decreased in the cinobufagin
group. The ratios of p-P53(S315)/p53 and p-P53(S392)/p53 in Huh-7
treated with cinobufagin were significantly decreased compared with
the control group (P<0.05). Compared with Huh-7 treated with
cinobufagin, the ratio of p-P53(S315)/p53 was significantly
decreased, and the ratio of p-P53(S392)/p53 was significantly
increased in Huh-7 cells treated with PF-03814735 + cinobufagin
treatments (P<0.05). However, compared with Huh-7 cells treated
with cinobufagin, the ratios of p-P53(S315)/p53 and p-P53(S392)/p53
in Flag-AURKA-transfected Huh-7 cells treated with cinobufagin were
significantly increased (P<0.05), compared with the classic
anticancer signaling of p53 (27,28).
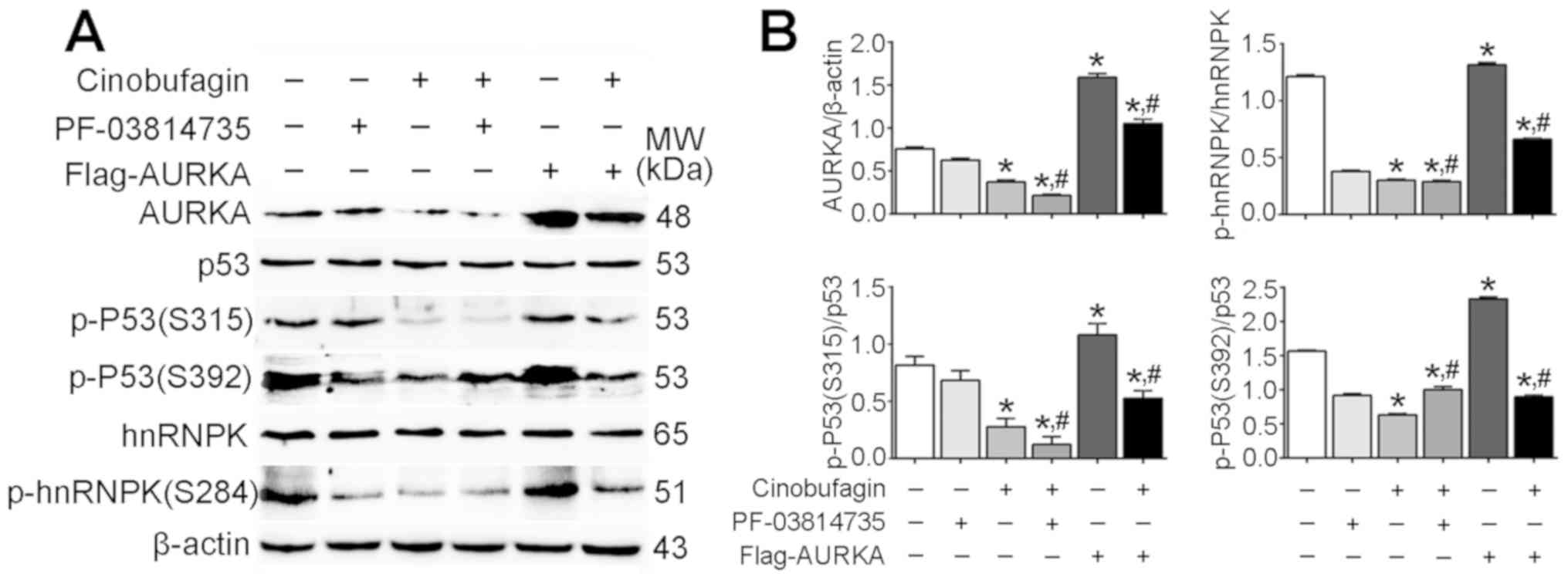 | Figure 4.Anticancer effects of cinobufagin on
Huh-7 cells are not dependent on the activation of p53 signaling.
(A) Protein expression levels of AURKA, p53, p-p53 (S315), p-p53
(S392), hnRNPK and p-hnRNPK (S284) in cells following treatment
with 5 µmol/l cinobufagin, as determined using western blotting.
(B) Densitometric analysis of AURKA, p-p53 and p-hnRNPK. Data are
presented as the means ± standard error of the mean of three
independent experiments. *P<0.05 vs. control,
#P<0.05 vs. cinobufagin. AURKA, aurora kinase A;
hnRNPK, heterogeneous nuclear ribonucleoprotein K; p,
phosphorylated. |
Anticancer properties of cinobufagin
in Huh-7 cells are associated with the activation of p73
signaling
Dabiri et al (29) demonstrated that p73 served as a
substitute for p53 in bortezomib-induced apoptosis in p53-deficient
or mutated cells, implicating that p73 could be a potential
therapeutic target for treatment of colorectal cancer, in
particular those lacking functional p53. The somatic mutation
frequency of p53 is 11.2% in Huh-7 cells (30). It was hypothesized that the p53
mutation may result in a loss of function, leading to p53 losing
its tumor-suppressive properties and acting as an oncogene. p73 is
a proapoptotic protein that serves an important role during
tumorigenesis, mimicking the tumor suppressor activities of p53 due
to its structural similarity (31). The p-p73 (Y99)/p73 ratio was
significantly increased in Huh-7 cells following cinobufagin
treatment compared with the control, whereas that of p-MDM2
(S166)/MDM2 was significantly decreased (Fig. 5). Additionally, cinobufagin
upregulated the expression of p21, Puma and Noxa compared with the
control (P<0.05). Furthermore, the overexpression or inhibition
of AURKA reduced or promoted the effects of cinobufagin,
respectively (P<0.05).
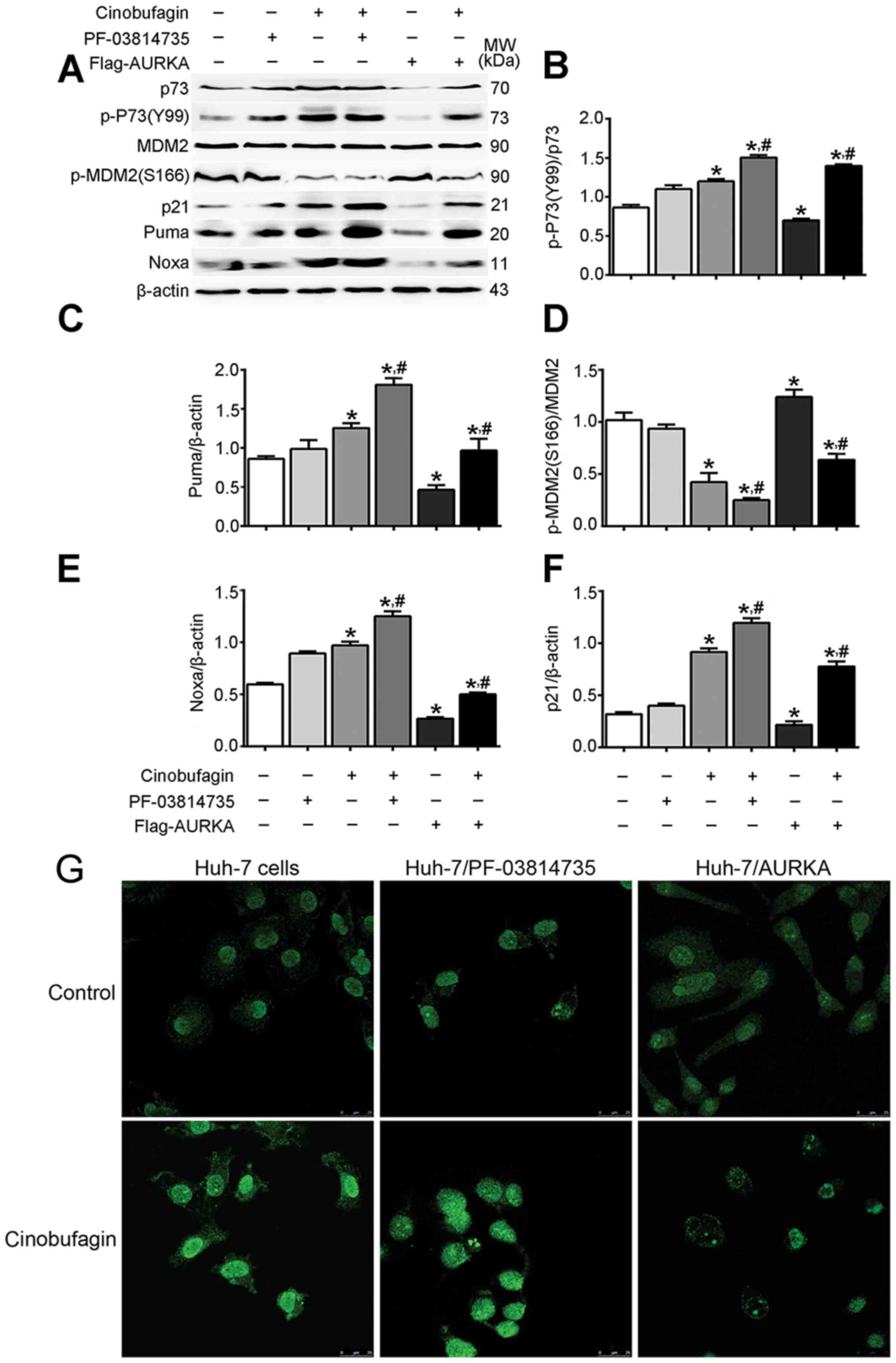 | Figure 5.Cinobufagin may induce anticancer
effects on Huh-7 cells via activation of p73 signaling. (A) Protein
expression levels of p73, p-p73 (Y99), MDM2, p-MDM2 (S166), p21,
Puma and Noxa in cells in cells following treatment with 5 µmol/l
cinobufagin for 24 h, as determined by western blotting.
Densitometric analysis of (B) p-p73, (C) Puma, (D) p-MDM2, (E) Noxa
and (F) p21. (G) Expression of p73 in Huh-7 cells, as determined by
immunocytochemistry. Representative images are shown at ×200
magnification. Data are presented as the means ± standard error of
the mean of three independent experiments. *P<0.05 vs. control,
#P<0.05 vs. cinobufagin. AURKA, aurora kinase A;
MDM2, mouse double minute 2 homolog; Noxa,
phorbol-12-myristate-13-acetate-induced protein 1; p,
phosphorylated; Puma, p53 upregulated modulator of apoptosis. |
Immunocytochemistry demonstrated that cinobufagin
treatment markedly upregulated p73 expression compared with the
control, whereas overexpression or inhibition of AURKA eliminated
or promoted these cinobufagin-induced effects (Fig. 5G). These results indicated that the
anticancer effects of cinobufagin in p53-mutant HCC cells were
associated with the activation of p73, but not p53 signaling.
Discussion
At present, only 10-20% of patients with HCC can be
treated surgically, whereas the majority of patients are treated
exclusively with chemotherapy (2);
However, the treatment of HCC with anticancer agents, including
sorafenib, capecitabine and oxaliplatin, is limited by multidrug
resistance and individual heterogeneity (32). Notably, numerous studies have
reported that CGs, as NKA inhibitors, exert anticancer properties
against various types of cancer that are not susceptible to
chemotherapy (33,34). CGs are synthetic or naturally
occurring steroid hormones observed in plant or animal species,
including ouabain, bufalin and cinobufagin (35). A number of studies reported that
the survival rate of patients undergoing chemotherapy against HCC
with mutant p53 is decreased compared with patients with wild-type
p53 (26,36). Our previous study (14) revealed that CGs reduce the
viability and induce the apoptosis of HCC cells with wild-type p53
by inhibiting AURKA signaling. In the present study, the anticancer
effects of CGs were investigated in HCC Huh-7 cells with mutant
p53.
Previous studies have reported that the
overexpression or abnormal amplification of AURKA may serve an
important role in the pathogenesis of various types of cancer
(20,37). AURKA is a serine/threonine kinase
that phosphorylates numerous target proteins involved in the
establishment of the mitotic spindle, centrosome duplication,
centrosome separation and cytokinesis, including BRCA1 DNA repair
associated, cell division cycle 25B, kinesin family member 2A,
large tumor suppressor kinase 2, p53 and TPX2 microtubule
nucleation factor (38). In the
present study, it was demonstrated that cinobufagin reduced the
viability, arrested the cell cycle and induced the apoptosis of
Huh-7 HCC cells possessing mutant p53. Furthermore, the
overexpression or inhibition of AURKA suppressed or promoted the
anticancer effects of cinobufagin on Huh-7 cells. The cyclin
B1/CDK1 complex is required for regulation of the G2/M
transition phase of the cell cycle (39). The present study demonstrated that
cinobufagin induced cell cycle G2/M arrest by
downregulating the expression of cyclin B1, CDK1 and PCNA, and
upregulating the expression of p21. Additionally, the expression
levels of p53 in Huh-7 cells were markedly unaltered following
cinobufagin treatment, whereas the expression of p-p53 (S315) was
significantly decreased. Therefore, it was suggested that the
anticancer effects of cinobufagin in Huh-7 cells with mutant p53
did not depend on activation of the p53 signaling pathway.
Our previous study reported that the expression
levels of p53 are significantly increased in HepG2 cells with
wild-type p53 following CG treatment (14); however, the present study revealed
that the expression of p53 was not significantly altered in
CG-treated Huh-7 cells, and that the phosphorylation levels of p53
(S315 and S392) were decreased. Li et al (26) observed that the wild type p53 could
inhibit cell proliferation and colony formation, but mutant p53
served a pro-oncogene function in several kinds of cancer cells. A
separate study demonstrated that mutant p53 (R175H) exhibits
pro-oncogenic properties, increasing the sensitivity of malignant
ovarian cancer cells to growth factors and promoting their growth
(40). Numerous studies revealed
that p53, but not p73, is frequently mutated during tumorigenesis
(41,42).
As a member of the p53 family, p73, a proapoptotic
protein, serves an important role during tumorigenesis, mimicking
the biological activities of p53 as a result of their structural
similarity (43). Previous studies
have reported that the knockdown of AURKA can induce increased p73
expression and increase the expression levels of its downstream
molecules, including p21/WAF1, PUMA and Noxa (44,45).
Meanwhile, Y99 phosphorylation of p-73 could bind to the PUMA
promoter to induce PUMA expression, leading to apoptosis induction
in cancer cells (46). In the
present study, it was revealed that cinobufagin downregulated AURKA
expression, and increased the expression and phosphorylation (Y99)
levels of p73. MDM2 is an E3 ubiquitin ligase localized to the
nucleus that inhibits p73-mediated cell cycle arrest and apoptosis
by binding its transcriptional activation domain (47). The present study revealed that
cinobufagin inhibited the phosphorylation of MDM2. The upregulation
of p-p73 increased the expression levels of downstream molecules,
including Puma, Noxa and p21. Puma binds antiapoptotic Bcl-2 family
members to induce mitochondrial dysfunction and caspase activation
(48). Noxa promotes alterations
in the mitochondrial membrane and the release of apoptogenic
proteins from the mitochondria (49). p21 binds and inhibits the activity
of CDK2 or CDK4, thereby regulating cell cycle progression
(50). Additionally, it was
demonstrated that the overexpression or inhibition of AURKA
significantly opposed or promoted the anticancer effects of
cinobufagin in Huh-7 cells, respectively.
Based on these findings, a schematic diagram was
produced to present the hypothesized mechanisms underlying the
anticancer effects of cinobufagin in Huh-7 cells, including
inhibition of AURKA and p53 signaling, and activation of p73
signaling, which is associated with the activity of AURKA (Fig. 6). These results indicated that
cinobufagin has potential as a novel anticancer agent for the
treatment of patients with HCC possessing mutant p53. Future
studies aim to establish animal models to further investigate the
anticancer mechanisms of CGs.
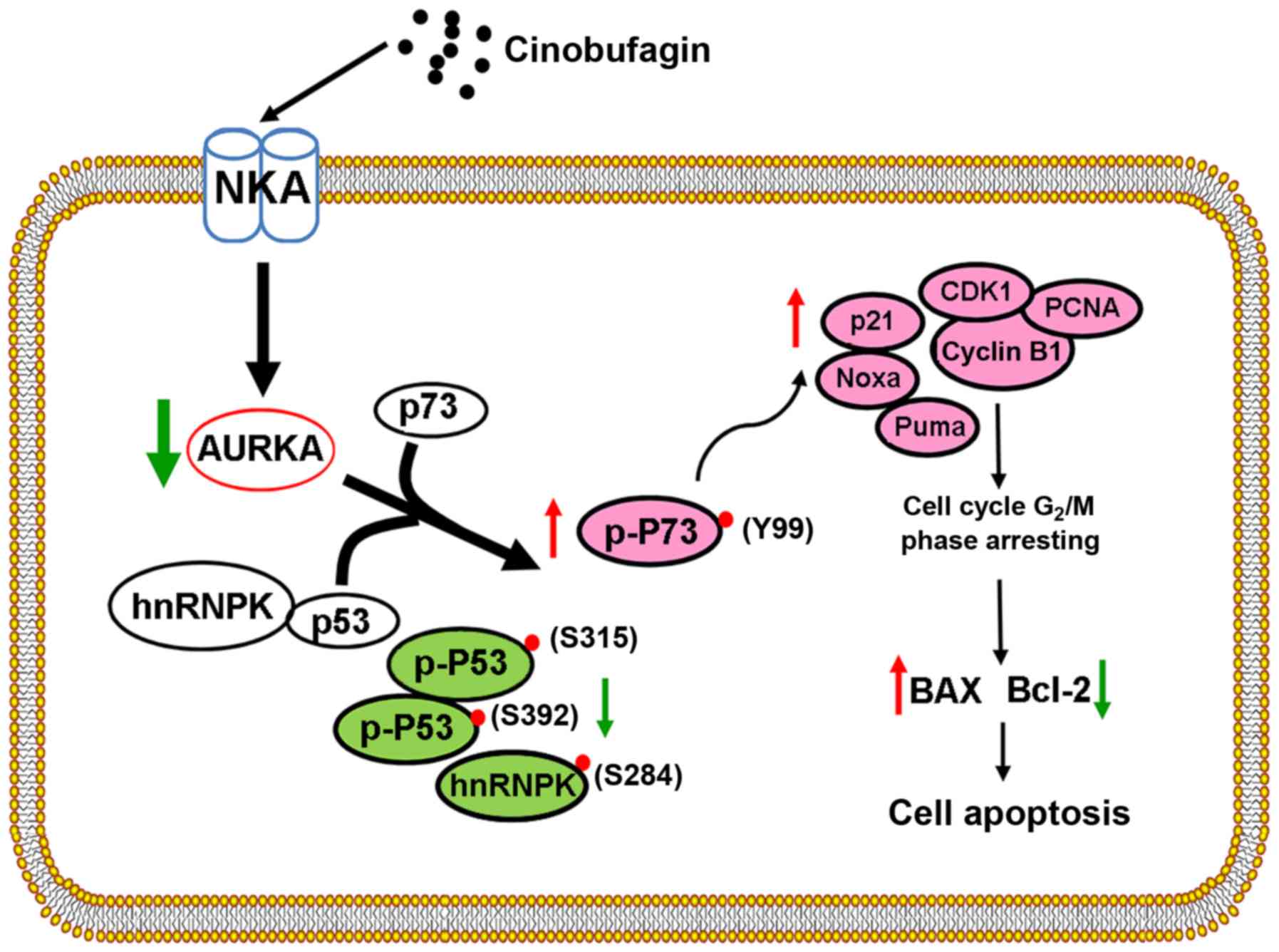 | Figure 6.Schematic diagram of
cinobufagin-induced effects on hepatocellular carcinoma Huh-7 cells
with mutant p53. Cinobufagin inhibits the viability, arrests the
cell cycle and induces the apoptosis of Huh-7 cells by inhibiting
AURKA and p53 signaling, and activating p73 signaling. AURKA,
aurora kinase A; Bax, Bcl-2-associated X protein; Bcl-2, B-cell
lymphoma 2; CDK1, cyclin-dependent kinase 1; hnRNPK, heterogeneous
nuclear ribonucleoprotein K; NKA,
Na+/K+-ATPase; Noxa,
phorbol-12-myristate-13-acetate-induced protein 1; p,
phosphorylated; PCNA, proliferating cell nuclear antigen; Puma, p53
upregulated modulator of apoptosis. |
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81273552, 81673651
and 81273745) and the Natural Science Foundation of Tianjin City
(grant nos. 18JCZDJC36500 and 11JCYBJC10900).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
FW and ED were responsible for study design. LZ, LF
and ZX performed the experiments and completed the manuscript
draft. RFa, RX, RFu, SZ, CW, YaZ, JW, JB, ZW, XH and YuZ conducted
data interpretation and analysis. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
AURKA
|
aurora kinase A
|
|
CG
|
cardiac glycoside
|
|
HCC
|
hepatocellular carcinoma
|
|
NKA
|
Na+/K+-ATPase
|
|
TSG
|
tumor suppressor gene
|
References
|
1
|
Clark T, Maximin S, Meier J, Pokharel S
and Bhargava P: Hepatocellular carcinoma: Review of epidemiology,
screening, imaging diagnosis, response assessment, and treatment.
Curr Probl Diagn Radiol. 44:479–486. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Roxburgh P and Evans TR: Systemic therapy
of hepatocellular carcinoma: Are we making progress? Adv Ther.
25:1089–1104. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Baek SH, Kim C, Lee JH, Nam D, Lee J, Lee
SG, Chung WS, Jang HJ, Kim SH and Ahn KS: Cinobufagin exerts
anti-proliferative and pro-apoptotic effects through the modulation
ros-mediated mapks signaling pathway. Immunopharmacol
Immunotoxicoz. 37:265–273. 2015. View Article : Google Scholar
|
|
4
|
Yu CH, Kan SF, Pu HF, Jea Chien E and Wang
PS: Apoptotic signaling in bufalin- and cinobufagin-treated
androgen- dependent and -independent human prostate cancer cells.
Cancer Sci. 99:2467–2476. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pereira DG, Salgado MAR, Rocha SC, Santos
HL, Villar JAFP, Contreras RG, Fontes CFL, Barbosa LA and Cortes
VF: Involvement of Src signaling in the synergistic effect between
cisplatin and digoxin on cancer cell viability. J Cell Biochem.
119:3352–3362. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gan PP, Zhou YY, Zhong MZ, Peng Y, Li L
and Li JH: Endoplasmic reticulum stress promotes autophagy and
apoptosis and reduces chemotherapy resistance in mutant p53 lung
cancer cells. Cell Physiol Biochem. 44:133–151. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bahrami A, Hesari A, Khazaei M, Hassanian
SM, Ferns GA and Avan A: The therapeutic potential of targeting the
braf mutation in patients with colorectal cancer. J Cell Physiol.
233:2162–2169. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li BT, Ross DS, Aisner DL, Chaft JE, Hsu
M, Kako SL, Kris MG, Varella-Garcia M and Arcila ME: Her2
amplification and her2 mutation are distinct molecular targets in
lung cancers. J Thorac Oncol. 11:414–419. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhu VW, Cui JJ, Fernandez-Rocha M, Schrock
AB, Ali SM and Ou SI: Identification of a novel t1151k alk mutation
in a patient with alk-rearranged nsclc with prior exposure to
crizotinib and ceritinib. Lung Cancer. 110:32–34. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kang N, Wang Y, Guo S, Ou Y, Wang G, Chen
J, Li D and Zhan Q: Mutant TP53 G245C and R273H promote cellular
malignancy in esophageal squamous cell carcinoma. BMC Cell Biol.
19:162018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tuna M, Amos CI and Mills GB: Genome-wide
analysis of head and neck squamous cell carcinomas reveals HPV,
TP53, smoking and alcohol-related allele-based acquired uniparental
disomy genomic alterations. Neoplasia. 21:197–205. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Amelio I, Mancini M, Petrova V, Cairns RA,
Vikhreva P, Nicolai S, Marini A, Antonov AA, Le Quesne J, Baena
Acevedo JD, et al: p53 mutants cooperate with HIF-1 in
transcriptional regulation of extracellular matrix components to
promote tumor progression. Proc Natl Acad Sci USA.
115:E10869–E10878. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Na K, Sung JY and Kim HS: Tp53 mutation
status of tubo-ovarian and peritoneal high-grade serous carcinoma
with a wild-type p53 immunostaining pattern. Anticancer Res.
37:6697–6703. 2017.PubMed/NCBI
|
|
14
|
Xu Z, Wang F, Fan F, Gu Y, Shan N, Meng X,
Cheng S, Liu Y, Wang C, Song Y and Xu R: Quantitative proteomics
reveals that the inhibition of na(+)/k(+)-atpase activity affects
s-phase progression leading to a chromosome segregation disorder by
attenuating the aurora a function in hepatocellular carcinoma
cells. J Proteome Res. 14:4594–4602. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Goos JA, Coupe VM, Diosdado B, Delis-Van
Diemen PM, Karga C, Beliën JA, Carvalho B, van den Tol MP, Verheul
HM, Geldof AA, et al: Aurora kinase a (AURKA) expression in
colorectal cancer liver metastasis is associated with poor
prognosis. Br J Cancer. 109:2445–2452. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Woo JK, Kang JH, Shin D, Park SH, Kang K,
Nho CW, Seong JK, Lee SJ and Oh SH: Daurinol enhances the efficacy
of radiotherapy in lung cancer via suppression of aurora kinase A/B
expression. Mol Cancer Ther. 14:1693–1704. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Treekitkarnmongkol W, Katayama H, Kai K,
Sasai K, Jones JC, Wang J, Shen L, Sahin AA, Gagea M, Ueno NT, et
al: Aurora kinase-a overexpression in mouse mammary epithelium
induces mammary adenocarcinomas harboring genetic alterations
shared with human breast cancer. Carcinogenesis. 37:1180–1189.
2016.PubMed/NCBI
|
|
18
|
Zhang J, Li B, Yang Q, Zhang P and Wang H:
Prognostic value of aurora kinase a (AURKA) expression among solid
tumor patients. A systematic review and meta-analysis. Jpn J Clin
Oncol. 45:629–636. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lo Iacono M, Monica V, Saviozzi S, Ceppi
P, Bracco E, Papotti M and Scagliotti GV: Aurora kinase a
expression is associated with lung cancer histological-subtypes and
with tumor de-differentiation. J Transl Med. 9:1002011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen C, Song G, Xiang J, Zhang H, Zhao S
and Zhan Y: AURKA promotes cancer metastasis by regulating
epithelial-mesenchymal transition and cancer stem cell properties
in hepatocellular carcinoma. Biochem Biophys Res Commun.
486:514–520. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fischer M: Census and evaluation of p53
target genes. Oncogene. 36:3943–3956. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yue X, Zhao Y, Xu Y, Zheng M, Feng Z and
Hu W: Mutant p53 in cancer: Accumulation, gain-of-function, and
therapy. J Mol Biol. 429:1595–1606. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Singh S, Vaughan CA, Frum RA, Grossman SR,
Deb S and Palit Deb S: Mutant p53 establishes targetable tumor
dependency by promoting unscheduled replication. J Clin Invest.
127:1839–1855. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Botchkarev VA and Flores ER: P53/p63/p73
in the epidermis in health and disease. Cold Spring Harb Perspect
Med. 4(pii): a0152482014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kasai F, Hirayama N, Ozawa M, Satoh M and
Kohara A: HuH-7 reference genome profile: Complex karyotype
composed of massive loss of heterozygosity. Hum Cell. 31:261–267.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li Q, Liu X, Jin K, Lu M, Zhang C, Du X
and Xing B: Nat10 is upregulated in hepatocellular carcinoma and
enhances mutant p53 activity. BMC Cancer. 17:6052017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cavic M, Spasic J, Krivokuca A, Boljevic
I, Kuburovic M, Radosavljevic D and Jankovic R: TP53 and DNA-repair
gene polymorphisms genotyping as a low-cost lung adenocarcinoma
screening tool. J Clin Pathol. 72:75–80. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hsieh Li SM, Liu ST, Chang YL, Ho CL and
Huang SM: Metformin causes cancer cell death through downregulation
of p53-dependent differentiated embryo chondrocyte 1. J Biomed Sci.
25:812018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dabiri Y, Kalman S, Gurth CM, Kim JY,
Mayer V and Cheng X: The essential role of TAp73 in
bortezomib-induced apoptosis in p53-deficient colorectal cancer
cells. Sci Rep. 7:54232017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kandoth C, McLellan MD, Vandin F, Ye K,
Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, et al:
Mutational landscape and significance across 12 major cancer types.
Nature. 502:333–339. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zawacka-Pankau J, Kostecka A, Sznarkowska
A, Hedström E and Kawiak A: P73 tumor suppressor protein: A close
relative of p53 not only in structure but also in anti-cancer
approach? Cell Cycle. 9:720–728. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Babula P, Masarik M, Adam V, Provaznik I
and Kizek R: From na+/k+-atpase and cardiac glycosides to
cytotoxicity and cancer treatment. Anticancer Agents Med Chem.
13:1069–1087. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kaushik V, Yakisich JS, Azad N, Kulkarni
Y, Venkatadri R, Wright C, Rojanasakul Y and Iyer AKV: Anti-tumor
effects of cardiac glycosides on human lung cancer cells and lung
tumorspheres. J Cell Physiol. 232:2497–2507. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kaushik V, Azad N, Yakisich JS and Iyer
AK: Antitumor effects of naturally occurring cardiac glycosides
convallatoxin and peruvoside on human er+ and triple-negative
breast cancers. Cell Death Discov. 3:170092017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Calderon-Montano JM, Burgos-Moron E, Orta
ML, Maldonado- Navas D, Garcia-Dominguez I and Lopez-Lazaro M:
Evaluating the cancer therapeutic potential of cardiac glycosides.
Biomed Res Int. 2014:7949302014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang ZY, Hong D, Nam SH, Kim JM, Paik YH,
Joh JW, Kwon CH, Park JB, Choi GS, Jang KY, et al: Sirt1 regulates
oncogenesis via a mutant p53-dependent pathway in hepatocellular
carcinoma. J Hepatol. 62:121–130. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li T, Chen Y, Zhang J and Liu S: LncRNA
TUG1 promotes cells proliferation and inhibits cells apoptosis
through regulating AURKA in epithelial ovarian cancer cells.
Medicine (Baltimore). 97:E121312018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zheng F, Yue C, Li G, He B, Cheng W, Wang
X, Yan M, Long Z, Qiu W, Yuan Z, et al: Nuclear aurka acquires
kinase-independent transactivating function to enhance breast
cancer stem cell phenotype. Nat Commun. 7:101802016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lee MH, Cho Y, Kim DH, Woo HJ, Yang JY,
Kwon HJ, Yeon MJ, Park M, Kim SH, Moon C, et al: Menadione induces
G2/M arrest in gastric cancer cells by down-regulation of CDC25C
and proteasome mediated degradation of CDK1 and cyclin B1. Am J
Transl Res. 8:5246–5255. 2016.PubMed/NCBI
|
|
40
|
Padmanabhan A, Candelaria N, Wong KK,
Nikolai BC, Lonard DM, O'Malley BW and Richards JS: USP15-dependent
lysosomal pathway controls p53-R175H turnover in ovarian cancer
cells. Nat Commun. 9:12702018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Melino G, Bernassola F, Ranalli M, Yee K,
Zong WX, Corazzari M, Knight RA, Green DR, Thompson C and Vousden
KH: P73 induces apoptosis via puma transactivation and bax
mitochondrial translocation. J Biol Chem. 279:8076–8083. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yoon MK, Ha JH, Lee MS and Chi SW:
Structure and apoptotic function of p73. BMB Rep. 48:81–90. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Huang L, Li A, Liao G, Yang F, Yang J,
Chen X and Jiang X: Curcumol triggers apoptosis of p53 mutant
triple-negative human breast cancer MDA-MB 231 cells via activation
of p73 and PUMA. Oncol Lett. 14:1080–1088. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Katayama H, Wang J, Treekitkarnmongkol W,
Kawai H, Sasai K, Zhang H, Wang H, Adams HP, Jiang S, Chakraborty
SN, et al: Aurora kinase-A inactivates DNA damage-induced apoptosis
and spindle assembly checkpoint response functions of p73. Cancer
Cell. 21:196–211. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Dar AA, Belkhiri A, Ecsedy J, Zaika A and
El-Rifai W: Aurora kinase a inhibition leads to p73-dependent
apoptosis in p53-deficient cancer cells. Cancer Res. 68:8998–9004.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Riley MF, You MJ, Multani AS and Lozano G:
Mdm2 overexpression and p73 loss exacerbate genomic instability and
dampen apoptosis, resulting in B-cell lymphoma. Oncogene.
35:358–365. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Knickelbein K, Tong J, Chen D, Wang YJ,
Misale S, Bardelli A, Yu J and Zhang L: Restoring PUMA induction
overcomes KRAS-mediated resistance to anti-EGFR antibodies in
colorectal cancer. Oncogene. 37:4599–4610. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Bean GR, Ganesan YT, Dong Y, Takeda S, Liu
H, Chan PM, Huang Y, Chodosh LA, Zambetti GP, Hsieh JJ and Cheng
EH: PUMA and BIM are required for oncogene inactivation-induced
apoptosis. Sci Signal. 6:ra202013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Guikema JE, Amiot M and Eldering E:
Exploiting the pro-apoptotic function of NOXA as a therapeutic
modality in cancer. Expert Opin Ther Targets. 21:767–779. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Cazzalini O, Scovassi AI, Savio M, Stivala
LA and Prosperi E: Multiple roles of the cell cycle inhibitor
p21(CDKN1A) in the DNA damage response. Mutat Res. 704:12–20. 2010.
View Article : Google Scholar : PubMed/NCBI
|