Introduction
Pneumonia causes ~1.3 million mortalities in
children each year (1,2). Human adenovirus (HAdV) infection is
one of the main causes of community-acquired pneumonia in children
and young adults (3,4). Adenoviruses are DNA viruses that
typically cause infections in the upper or lower respiratory and
gastrointestinal tracts, and the conjunctiva (5–7).
Fatality rates for untreated severe HAdV-associated pneumonia may
be >50% each year (8). The
traditional diagnosis of HAdV infection is based on evidence of
positive, multiplex HAdV polymerase chain reaction (PCR) products
from respiratory tract samples, including sputum or bronchoalveolar
lavage (BAL) fluid (3). However,
this technique has certain limitations, and is unable to provide an
early and effective response to adenovirus infections of the lower
respiratory tract. Therefore, it is necessary to identify novel
methods for the diagnosis of HAdV pneumonia.
MicroRNAs (miRNAs or miRs) are single-stranded,
19–24-nucleotide (nt)-long RNA molecules that regulate gene
expression and function at the post-transcriptional level (9). Serum miRNAs have been reported to be
stable; certain miRNAs have been implicated in cancer pathogenesis,
including breast and colorectal cancer (10–12).
The use of serum/plasma miRNAs as novel, non-invasive biomarkers
for the diagnosis of several diseases has been explored in previous
studies (13,14).
In the present study, to screen potential biomarkers
for the diagnosis of HAdV pneumonia, exosomal miRNA profiles
extracted from the serum of HAdV-infected or healthy children were
compared. Exosomal miRNA expression was investigated as using the
exosomal fraction increases the sensitivity of miRNA detection
(15). Pairs of miRNAs were
selected, which could be considered as candidate diagnostic
biomarkers. These selected miRNA pairs could distinguish
HAdV-infected children from uninfected children, indicating that
such miRNAs may be used for the diagnosis of HAdV pneumonia in
children.
Materials and methods
Preparation of serum samples
The blood samples from 29 healthy children and 30
HAdV patients used in the present study were obtained from
Guangzhou Women and Children's Medical Center (Guangzhou, China)
between January 2015 and January 2019. The ages of all patients and
healthy volunteers, including 33 males and 26 females, ranged from
1 to 13 years. HAdV pneumonia was diagnosed using the following
criteria: i) Lower respiratory and/or systemic symptoms of
infection; ii) computed tomography (CT) scan or lung infiltration
on chest radiography; and iii) positive results for HAdV
immunoglobulin M (IgM) antibody in serum and/or HAdV DNA by PCR in
throat swabs and/or BAL fluid. We also excluded the samples from
mixed infection patients who were infected with HAdV together with
other microorganisms. Serum samples were prepared by centrifugation
at 1,000 × g for 10 min. The supernatants were collected and stored
in aliquots at −80°C. Study approval was granted by the Ethics
Committee at Guangzhou Women and Children's Medical Center
(approval no. 2014121815), and all the parents or legal guardians
of the patients signed written informed consent forms and agreed to
its content.
Extraction of serum exosomes
Serum from all participants was collected for
exosome isolation using the ExoQuick Exosome Precipitation kit
(System Biosciences, LLC, Palo Alto, CA, USA) according to the
manufacturer's protocol (16).
Exosome characterization
Transmission electron microscopy
(TEM)
Exosome suspensions in PBS (100 µl) were added to a
copper mesh placed on a clean wax plate. After 4 min, the copper
mesh was removed and placed in 2% phosphotungstic acid for 5 min,
while the mesh was dried on filter paper. The morphology of the
exosomes was examined using a JEM-1230 transmission electron
microscope (JEOL Ltd., Tokyo, Japan).
Western blot analysis
The exosome pellet was dissolved in
radioimmunoprecipitation assay lysis buffer (Cell Signaling
Technology, Inc., Danvers, MA, USA) for 30 min at 4°C, and the
protein concentration was determined using a Bradford protein assay
kit (Bio-Rad Laboratories, Inc., Hercules, CA, USA) (17,18).
The proteins (20 µg/lane) were separated via 15% SDS-PAGE and then
transferred to polyvinylidene difluoride membranes. Membranes were
blocked in 5% non-fat dried milk for 1 h, followed by incubation
for 1 h with anti-CD9 (1:1,000; cat. no. 13174, Cell Signaling
Technology, Inc.) and anti-heat shock protein 90α (HSP90α; 1:1,000;
cat. no. 8165, Cell Signaling Technology, Inc.) primary antibodies,
and subsequent incubation for 1 h with the secondary antibodies
(horseradish peroxidase-conjugated anti-rabbit immunoglobulin G;
1:1,000; cat. no. 7074, Cell Signaling Technology, Inc.). The bands
were visualized using the SuperSignal chemiluminescence system
(Thermo Fisher Scientific, Inc., Waltham, MA, USA). All steps were
performed at room temperature.
RNA extraction from exosomes
RNA was extracted from the exosome pellets using
TRIzol® (Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol (17).
miRNA sequencing (miRNA-seq) and data
analysis
Small RNA libraries were generated using
NEBNext® Multiplex Small RNA Library Prep Set for
Illumina® (New England BioLabs, Inc., Ipswich, MA, USA);
all subsequent steps were conducted according to the manufacturer's
protocol. Briefly, the multiplex 3′ small RNA (SR) adapters, which
the reverse transcription (RT) primers were hybridized to, were
ligated to the miRNA. The RT step was performed upon ligation of
the multiplex 5′ SR adapter, and 15 cycles of PCR were performed to
enrich those DNA fragments that had adapter molecules on both ends.
PCR was performed as follows: 30 sec at 94°C, then 15 sec at 94°C,
30 sec at 62°C, and 15 sec at 70°C for 15 cycles. The library
fragments were size-selected by 6% PAGE extraction. The purified
DNAs of ~140 nt constituted the miRNA library. The libraries were
quantified with using a Qubit 3.0 fluorometer (Invitrogen; Thermo
Fisher Scientific, Inc.) and validated with an Agilent 2100
Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA),
prior to being sequenced on an Illumina HiSeq 2500 Sequencing
System (Illumina, Inc., San Diego, CA, USA) for 50 cycles.
The adapter sequences were removed from all reads.
Reads with Phred quality scores were <10 were truncated at their
first nucleotides. Reads <17 nt were discarded. The remaining
reads were mapped to a human miRNA reference sequence in the
miRBase database (http://www.mirbase.org/) using the FANSe2 algorithm
with parameters-L60-E2-U1-S10 (FANSe v2.0; http://bioinformatics.jnu.edu.cn/software/fanse2/).
miRNA expression was quantified as reads per million. The
differentially expressed miRNAs were detected using the edgeR
package version 3.8 (R, http://bioconductor.org/packages/release/bioc/html/edgeR.html).
miRNAs with >2 or <0.5-fold change, and P<0.01 (no
correction for Type I error was applied) were considered as
differentially expressed miRNAs (19,20).
RT-quantitative PCR (RT-qPCR)
RNA was extracted from exosome pellets as
aforementioned and the isolated RNAs were reverse transcribed using
a SuperScript™ First-Strand Synthesis System for RT-PCR kit (cat.
no. 11904018, Invitrogen; Thermo Fisher Scientific, Inc.) according
to the manufacturer's protocol. qPCR was performed using the Power
SYBR Green PCR Master Mix (Applied Biosystems; Thermo Fisher
Scientific, Inc.) (17). Each
reaction was performed in a 20-µl volume system including 5 µl
cDNA, 10 µl Power SYBR Green PCR Master Mix, 0.5 µl each primer and
4.0 µl double distilled water. PCR was performed as follows: 5 min
at 94°C, then 20 sec at 94°C and 30 sec at 60°C for 39 cycles. The
relative expression of miRNAs normalized to other miRNAs was
calculated using the 2−∆∆Cq method (15,21).
All RT-qPCR primers used in the study are listed in Tables I–III.
 | Table I.Nucleotide sequence of miRs. |
Table I.
Nucleotide sequence of miRs.
| miR |
Sequencea (5′-3′) | Accession
numberb |
|---|
| hsa-miR-152-3p |
UCAGUGCAUGACAGAACUUGG | MIMAT0000438 |
|
hsa-miR-103a-3p |
AGCAGCAUUGUACAGGGCUAUGA | MIMAT0000101 |
| hsa-miR-185-5p |
UGGAGAGAAAGGCAGUUCCUGA | MIMAT0000455 |
| hsa-miR-98-5p |
UGAGGUAGUAAGUUGUAUUGUU | MIMAT0000096 |
| hsa-let-7f-5p |
UGAGGUAGUAGAUUGUAUAGUU | MIMAT0000067 |
| hsa-miR-206-5p |
UGGAAUGUAAGGAAGUGUGUGG | MI0000490 |
|
hsa-miR-450a-5p |
UUUUGCGAUGUGUUCCUAAUAU | MIMAT0001545 |
| hsa-miR-145-5p |
GUCCAGUUUUCCCAGGAAUCCCU | MIMAT0000437 |
|
hsa-miR-103b-5p |
UCAUAGCCCUGUACAAUGCUGCU | MIMAT0007402 |
 | Table III.Primer sequence of nine miRs for
quantitative polymerase chain reaction. |
Table III.
Primer sequence of nine miRs for
quantitative polymerase chain reaction.
| miRNA | Primer sequence
(5′-3′) |
|---|
| hsa-miR-152-3p | Forward:
TCAGTGCATGACAGAA |
|
| CTTGG |
|
| Reverse:
GTGCAGGGTCCGAGGT |
|
hsa-miR-103a-3p | Forward:
AGCAGCATTGTACAGG |
|
| GCTATGA |
|
| Reverse:
GTGCAGGGTCCGAGGT |
| hsa-miR-185-5p | Forward:
TGGAGAGAAAGGCAG |
|
| TTCCTGA |
|
| Reverse:
GTGCAGGGTCCGAGGT |
| hsa-miR-98-5p | Forward:
TGAGGTAGTAAGTTGT |
|
| ATTGTT |
|
| Reverse:
GTGCAGGGTCCGAGGT |
| hsa-let-7f-5p | Forward:
TGAGGTAGTAGATTGT |
|
| ATAGTT |
|
| Reverse:
GTGCAGGGTCCGAGGT |
| hsa-miR-206-5p | Forward:
TGGAATGTAAGGAAGT |
|
| GTGTGG |
|
| Reverse:
GTGCAGGGTCCGAGGT |
|
hsa-miR-450a-5p | Forward:
TTTTGCGATGTGTTCCT |
|
| AATAT |
|
| Reverse:
GTGCAGGGTCCGAGGT |
| hsa-miR-145-5p | Forward:
GTCCAGTTTTCCCAGG |
|
| AATCCCT |
|
| Reverse:
GTGCAGGGTCCGAGGT |
|
hsa-miR-103b-5p | Forward:
TCATAGCCCTGTACAAT |
|
| GCTGCT |
|
| Reverse:
GTGCAGGGTCCGAGGT |
Statistical analysis
Data was analyzed using SPSS software version 18.0
(SPSS, Inc., Chicago, IL, USA). Paired t-tests and two-way analyses
of variance were performed to analyze the relative expression of
exosomal miRNAs. P<0.01 was considered to indicate a
statistically significant difference.
Results
Characteristics of patients with HAdV
pneumonia
All patients enrolled in the present study were
diagnosed with pneumonia based on clinical symptoms and lobar
infiltrates on chest radiography. The main clinical symptoms were
fever and cough, while two patients exhibited wheezing and
tachypnea. All patients were HAdV-positive, with results from the
HAdV IgM antibody test using serum and/or HAdV DNA PCR analysis
using throat swabs and/or BAL fluid. The clinical characteristics
of the cases are summarized in Table
IV. Abnormal chest radiographs were also observed in all cases.
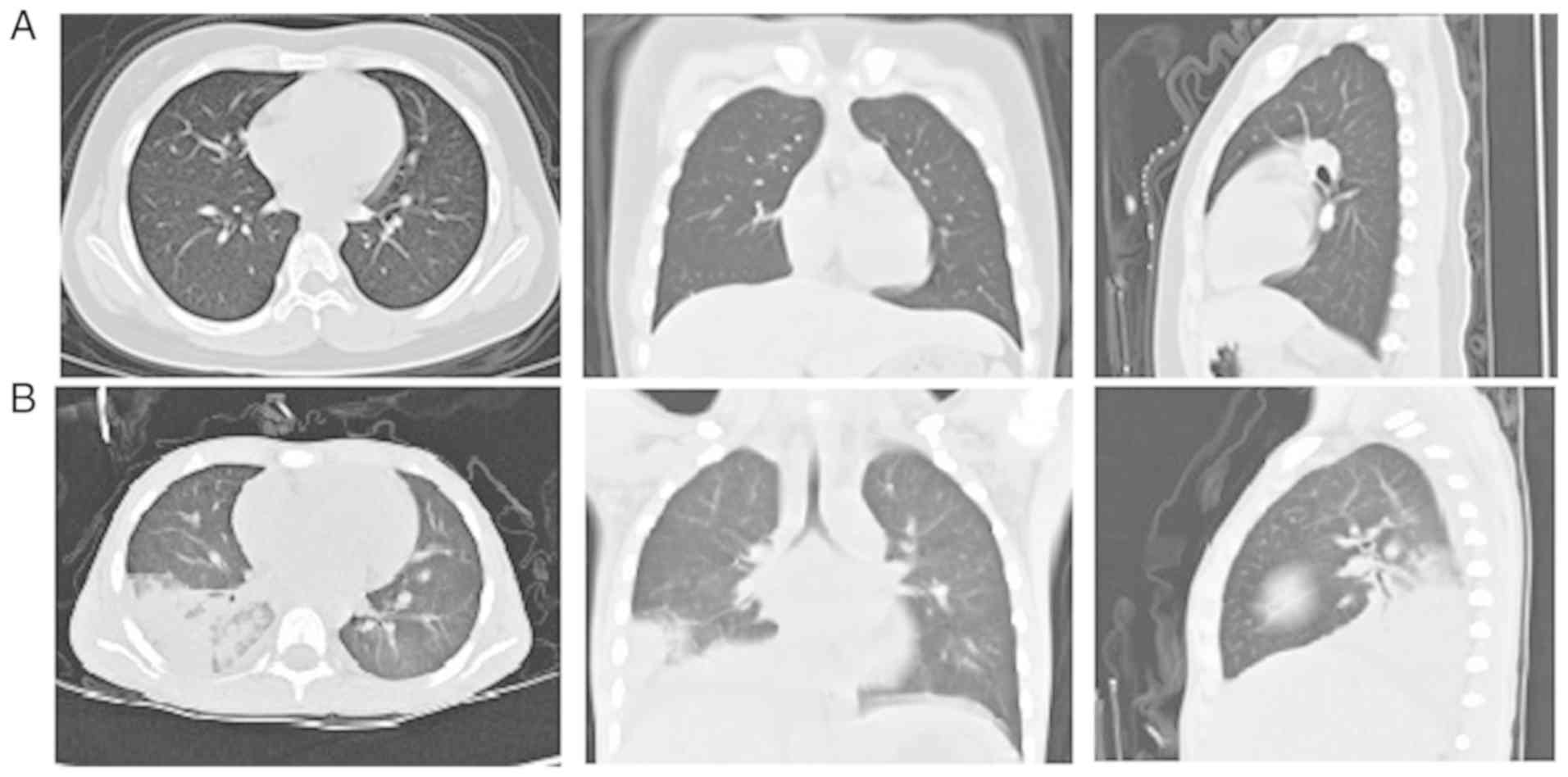
Chest scans revealed diffuse infiltration of the lungs.
Representative high-resolution CT scans of the chest of healthy
children and patients are shown in Fig. 1.
 | Table IV.Clinical features of patients. |
Table IV.
Clinical features of patients.
|
|
|
|
| Laboratory
characteristicsb |
Radiologyg |
|---|
|
|
|
|
|
|
|
|---|
| Patient | Age | Gender | Fever
durationa (days) | WBC
(×109/l) | HsCRPc (mg/l) | Specific
IgMd (IU/ml) | HAdV
DNAe (throat
swabs/BALF) | LDH
(U/l)f | Consolidation | Hydrothorax |
|---|
| 1 | 1 year 3
months | Male | 3 | 5.5 | 0.54 | − | +/+ | 278 | + | − |
| 2 | 1 year 2
months | Female | 5 | 11.7 | 1.7 | + | −/+ | 305 | + | + |
| 3 | 4 years | Male | 3 | 8.3 | 2.8 | + | +/+ | 211 | + | − |
| 4 | 3 years 1
month | Male | 6 | 4.5 | 1.9 | − | +/+ | 107 | + | + |
| 5 | 2 years 5
months | Female | 5 | 8.2 | 11.8 | + | +/+ | 308 | + | − |
Isolation and validation of serum
exosomes
Exosomes were isolated from the serum of four
healthy children and five adenovirus-infected patients. The
patients were included in the study according to the criteria in
Table IV. The isolated exosomes
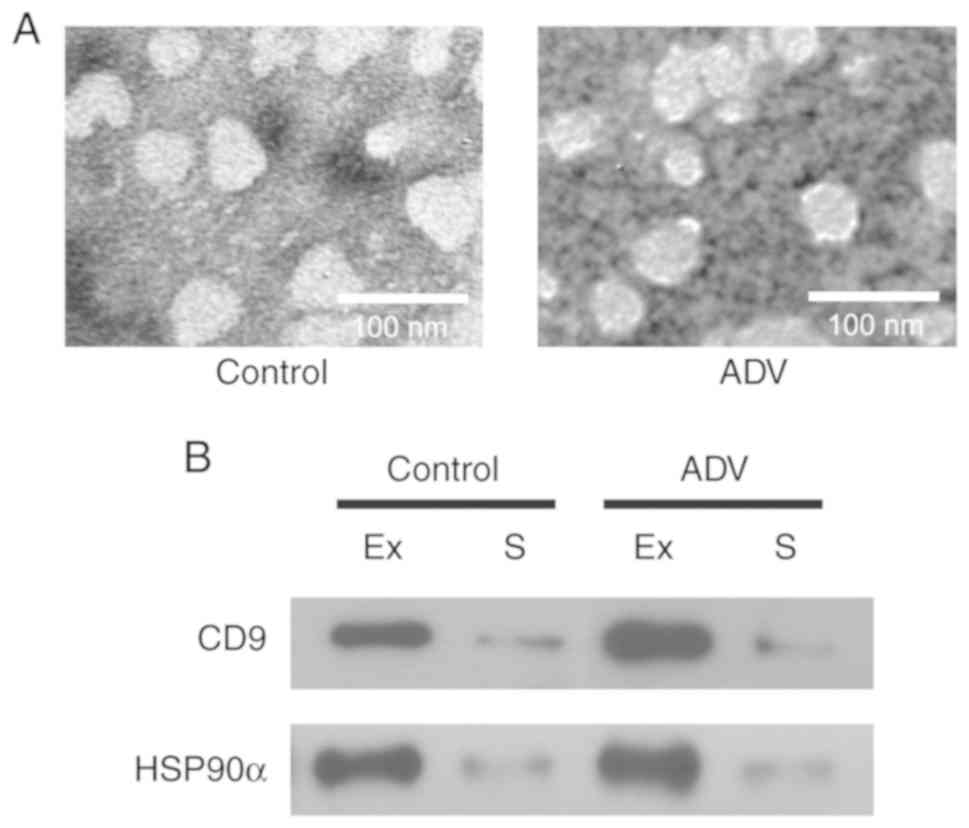
were characterized by TEM, and appeared as spherical vesicles of
30–100 nm in diameter in all samples (Fig. 2A), which is consistent with
previously reported characteristics of exosomes (20,22).
In addition, exosome protein markers, namely CD9 and HSP90α
(22,23) were also detected in our exosome
samples (Fig. 2B). CD9 and HSP90α
were highly expressed in the isolated exosomes compared with their
levels in serum. These results suggest that exosomes were
successfully isolated in the present study.
miRNA signatures of different
exosomes
To compare the different miRNA signatures of
patients with pneumonia caused by adenovirus infection with those
of healthy volunteers, a total of 300 ng RNA isolated from 400 µl
serum of each of the samples was used for miRNA-seq to compare the
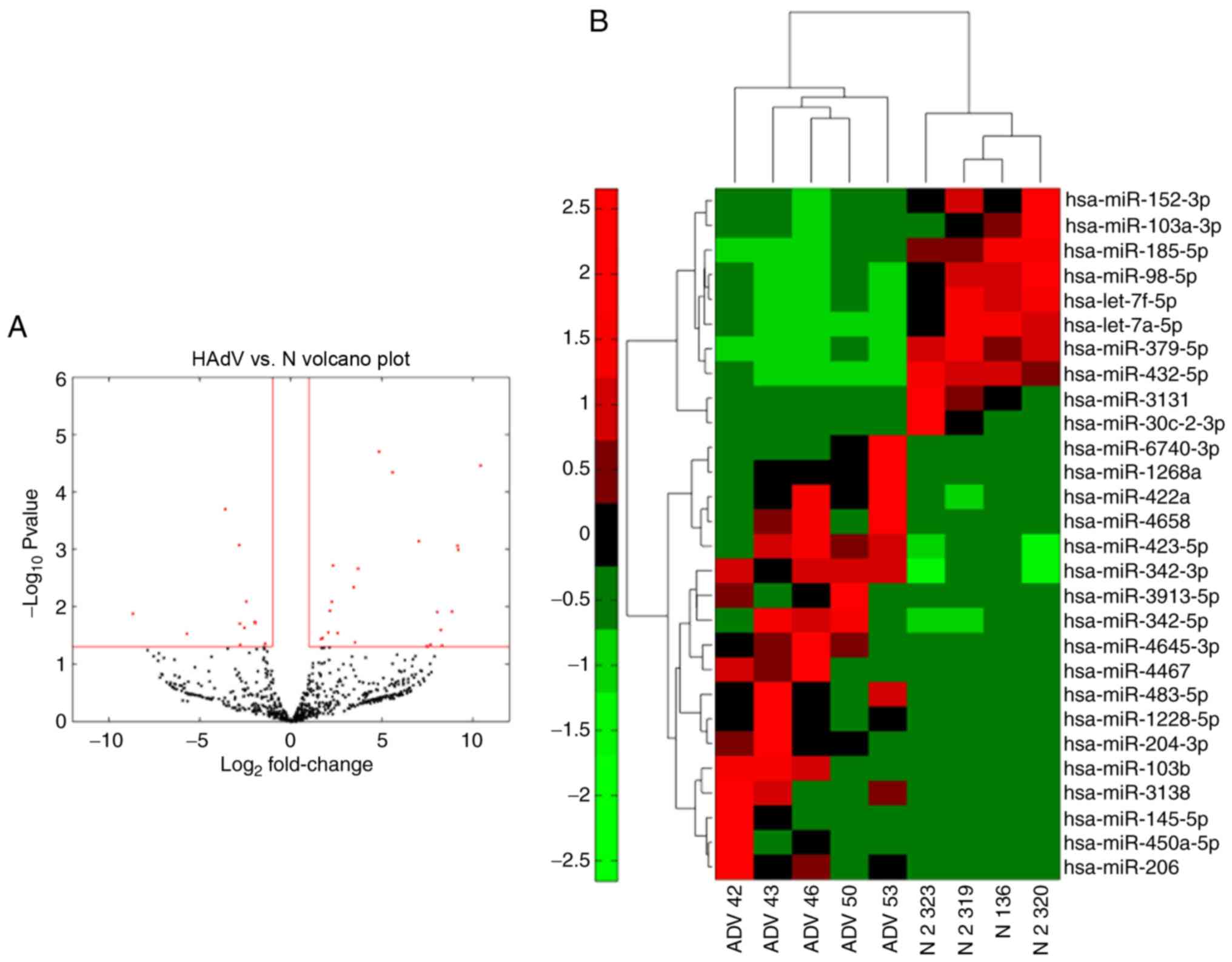
differentially expressed miRNAs in HAdV-infected patients. The
expression of the miRNAs analyzed by miRNA-seq was compared between
samples from four healthy controls and samples from five patients
with HAdV infection (Fig. 3A). The
majority of miRNAs exhibited significantly altered expression in
HAdV-infected patients, providing a clear distinction of HAdV
pneumonia. Among the miRNAs evaluated, 18 were significantly
upregulated while 10 were downregulated in patients compared with
the normal controls (Fig. 3B).
These results indicated that these miRNAs may serve as biomarkers
for HAdV-infected patients.
Pairwise RT-qPCR analysis of potential
miRNA biomarkers for HAdV pneumonia
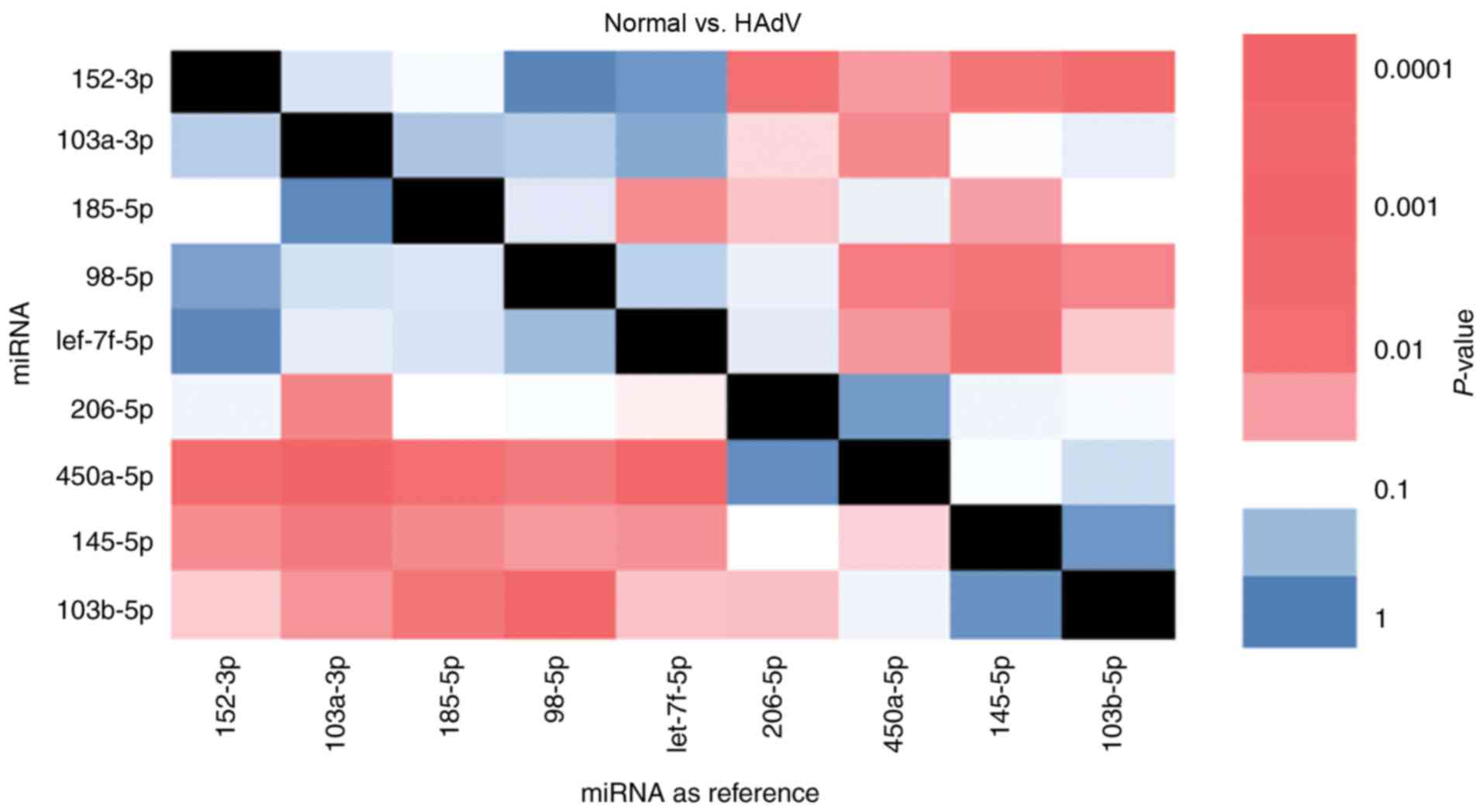
The present study further analyzed the miRNAs with
significantly different expression in HAdV-infected patients
compared with healthy children. In total, the top nine
differentially expressed miRNAs in order to analyze their
association with pneumonia caused by HAdV infection. The expression
of these nine miRNAs was evaluated in samples from five healthy
children and five HAdV-infected patients. Importantly, an internal
reference is essential in RT-qPCR experiments. However, in the case
of miRNAs isolated from exosomes, an internal reference is not
available. To solve this issue, pairwise analysis of each miRNA was
used. It was observed that miR-103b-5p/miR-98-5p and
miR-450a-5p/miR-103a-3p exhibited opposite expression trends.
miR-103b-5p and miR-450a-5p were upregulated in HAdV samples while
miR-98-5p and miR-103a-3p were downregulated in HAdV samples,
indicating that this ratio may serve as a biomarker for HAdV
pneumonia (Fig. 4). Notably,
miR-103b-5p is a serum biomarker for colorectal adenocarcinoma
(24). Thus, the present results
suggested that the selected miRNAs may reflect HAdV infection.
Validation of potential biomarkers for
HAdV pneumonia diagnosis
To verify the diagnostic capability of serum miRNAs
as potential biomarkers in the present study, blood samples from 20
healthy children and 20 HAdV-infected patients were collected. The
RT-qPCR results of the two pairs of miRNAs identified above were
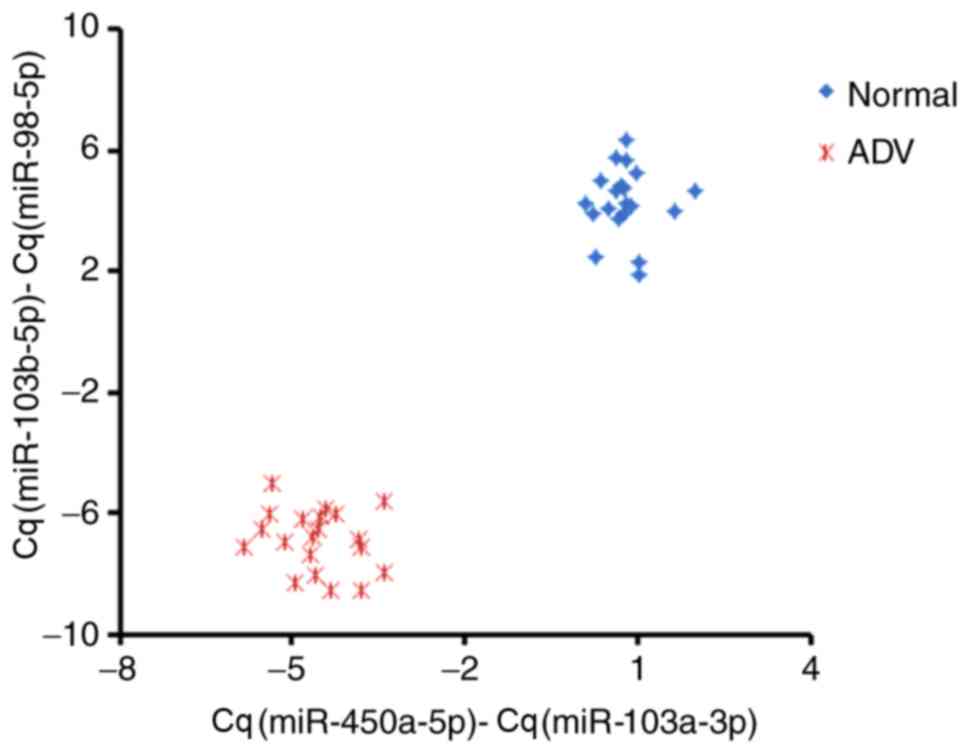
analyzed for all the samples in the validation cohort (Fig. 5). Relative Cq values for the miRNAs
in the two pairs were subtracted from one another
[Cq(miR-450a-5p)-Cq(miR-103a-3p) and
Cq(miR-103b-5p)-Cq(miR-98-5p)]; subtracted values were used instead
of ratios to simplify clinical analysis. The two pairs of miRNAs
analyzed between healthy controls and HAdV patients were clearly
separated as two clusters, suggesting that the miRNAs selected in
the present study could be considered as good diagnostic biomarkers
for HAdV pneumonia, at least in the cohort experiment. Importantly,
neither pair of miRNAs could independently distinguish
HAdV-infected patients from healthy children, highlighting the
requirement for combining the two miRNA pairs identified in the
present study.
Discussion
Serum miRNAs have been used as biomarkers for the
diagnosis of various types of cancer (13). The associations between serum
miRNAs and viruses have also been discussed in previous studies
(25–27); however, there is no report thus far
regarding serum miRNAs and pneumonia caused by HAdV infection.
HAdVs are common pathogens that cause acute respiratory infections
(1). The treatment of HAdV
infections is controversial, as prospective, randomized therapeutic
trials have not been conducted. Furthermore, severe HAdV pneumonia
exhibits rapid development and a prolonged course in a proportion
of patients with long-term respiratory problems, including
bronchiectasis, bronchiolitis obliterans and hyperlucent lung
(28,29). In addition, an early diagnosis of
pneumonia associated with HAdV infection is crucial but difficult.
Therefore, it is important to identify biomarkers for HAdV
pneumonia in children.
Recently, investigations of the function of miRNAs
in lung diseases or those caused by viral infections have increased
gradually (30–33). Furthermore, the roles of exosomal
miRNAs have received increasing attention (34). Numerous studies have reported that
changes in serum exosomes reflect different diseases and disease
status (35,36). Based on the available reports,
miRNAs appear to be differentially enriched in serum exosomes.
Additionally, the expression patterns of serum miRNAs change among
different diseases. Therefore, exosomal miRNAs are considered
potential diagnostic biomarkers for several diseases (30,32).
The use of exosomal miRNAs as biomarkers may be useful for the
early diagnosis of diseases.
Our study screened HAdV-specific serum miRNAs.
Although exosomal miRNAs were proposed as potential biomarkers for
diseases several years ago, limited information has been identified
and applied in clinical practice for the management of pneumonia in
adenovirus-infected children to date. To the best of our knowledge,
this is the first study to identify two pairs of exosomal miRNAs
isolated from serum that can be considered as biomarkers for
pneumonia in adenovirus-infected children. Using this approach, the
present study identified that these two pairs of miRNAs were
differentially expressed in the serum of HAdV-infected children
compared with normal controls. However, additional samples should
be evaluated in the future to identify other potential biomarkers.
Our study has provided promising, non-invasive biomarkers for HAdV
pneumonia.
The present study identified a unique expression
profile for HAdV infection-associated serum miRNAs, namely
miR-450a-5p/miR-103a-3p and miR-103b-5p/miR-98-5p. The results of
our study agree with previously published reports to some extent.
In particular, miR-103b-5p has been reported as a serum biomarker
for colorectal adenocarcinoma (24). miR-103a-3p was identified in serum,
and was considered to serve an important role in lung
adenocarcinoma (37). Serum
miR-450a-5p has been reported to be a biomarker for several types
of cancer (38); however, the
function of serum miR-98-5p has not been discussed thus far. In
addition to their potential role as biomarkers, miR-450a-5p was
reported to control the mRNA expression of signal transducer and
activator of transcription 1 (39)
and miR-98-5p inhibited the activation of interferon (IFN)-α
signaling (40), indicating that
miR-450a-5p and miR-98-5p may serve roles in the IFN signaling
pathway and may influence the progression of HAdV. Furthermore,
serum miR-103a-3p is reported to be involved in the differentiation
of Th2/Th17 cells in severe equine asthma (41), indicating that miR-103a-3p may
affect the immune reaction in patients with HAdV infection.
However, as a limited number of samples were included in the
present study, additional samples should be evaluated to confirm
the diagnostic capabilities of the aforementioned miRNAs for
adenovirus infection-associated pneumonia, and further experiments
should be performed to clarify the underlying mechanism in the
future.
In conclusion, the expression profile of serum
miRNAs may provide novel biomarkers for the diagnosis of pneumonia
in adenovirus-infected children. miR-450a-5p/miR-103a-3p and
miR-103b/miR-98-5p could be considered as potential diagnostic
biomarkers for adenovirus infection-associated pneumonia.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Science and Technology Major Project (grant no.
2018ZX10101004003001), the National Natural Science Foundation of
China (grant nos. 81601759 and 81701990), the Science and
Technology Project of Guangdong Province, China (grant no.
2014A020212020) and the General Projects of Guangzhou Scientific
Research (grant no. 201707010183).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
GL made significant contributions towards the design
of the present study. DY and HF participated in sample diagnosis
and collection. FH, JZ, JB and TS performed the experiments. LH and
HF were responsible for the statistical analysis. FH, JZ and GL
drafted, revised and edited the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee at
Guangzhou Women and Children's Medical Center (Guangzhou, China).
All the parents or legal guardians of the patients signed written
informed consent forms and agreed to its content.
Patient consent for publication
All the parents or legal guardians of the patients
signed written informed consent forms and agreed to its
content.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Agweyu A, Kibore M, Digolo L, Kosgei C,
Maina V, Mugane S, Muma S, Wachira J, Waiyego M and Maleche-Obimbo
E: Prevalence and correlates of treatment failure among Kenyan
children hospitalised with severe community-acquired pneumonia: A
prospective study of the clinical effectiveness of WHO pneumonia
case management guidelines. Trop Med Int Health. 19:1310–1320.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bhutta ZA, Das JK, Walker N, Rizvi A,
Campbell H, Rudan I and Black RE; Lancet Diarrhoea and Pneumonia
Interventions Study Group, : Interventions to address deaths from
childhood pneumonia and diarrhoea equitably: What works and at what
cost? Lancet. 381:1417–1429. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kim SJ, Kim K, Park SB, Hong DJ and Jhun
BW: Outcomes of early administration of cidofovir in
non-immunocompromised patients with severe adenovirus pneumonia.
PLoS One. 10:e01226422015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nascimento-Carvalho CM: Etiology of
childhood community acquired pneumonia and its implications for
vaccination. Braz J Infect Dis. 5:87–97. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Elenius V, Peltola V, Ruuskanen O,
Ylihärsilä M and Waris M: Plasma procalcitonin levels in children
with adenovirus infection. Arch Dis Child. 97:582–583. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Girouard G, Garceau R, Thibault L, Bourque
C, Bastien N and Li Y: Province-wide adenovirus type 3 outbreak
with severe cases in New Brunswick. Can J Infect Dis Med Microbiol.
22:e4–e6. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lynch JP III, Fishbein M and Echavarria M:
Adenovirus. Semin Respir Crit Care Med. 32:494–511. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lynch JP III and Kajon AE: Adenovirus:
Epidemiology, global spread of novel serotypes, and advances in
treatment and prevention. Semin Respir Crit Care Med. 37:586–602.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yan S, Han B, Gao S, Wang X, Wang Z, Wang
F, Zhang J, Xu D and Sun B: Exosome-encapsulated microRNAs as
circulating biomarkers for colorectal cancer. Oncotarget.
8:60149–60158. 2017.PubMed/NCBI
|
|
11
|
Tanaka T, Tanaka M, Tanaka T and
Ishigamori R: Biomarkers for Colorectal Cancer. Int J Mol Sci.
11:3209–3225. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Clancy C, Joyce MR and Kerin MJ: The use
of circulating microRNAs as diagnostic biomarkers in colorectal
cancer. Cancer Biomark. 15:103–113. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Keller A, Leidinger P, Gislefoss R, Haugen
A, Langseth H, Staehler P, Lenhof HP and Meese E: Stable serum
miRNA profiles as potential tool for non-invasive lung cancer
diagnosis. RNA Biol. 8:506–516. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Geekiyanage H, Jicha GA, Nelson PT and
Chan C: Blood serum miRNA: Non-invasive biomarkers for Alzheimer's
disease. Exp Neurol. 235:491–496. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gallo A, Tandon M, Alevizos I and Illei
GG: The majority of microRNAs detectable in serum and saliva is
concentrated in exosomes. PLoS One. 7:e306792012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rekker K, Saare M, Roost AM, Kubo AL,
Zarovni N, Chiesi A, Salumets A and Peters M: Comparison of serum
exosome isolation methods for microRNA profiling. Clin Biochem.
47:135–138. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang F, Zhang J, Zhang Y, Geng G, Liang
J, Li Y, Chen J, Liu C and Zhang H: RNA helicase MOV10 functions as
a co-factor of HIV-1 Rev to facilitate Rev/RRE-dependent nuclear
export of viral mRNAs. Virology. 486:15–26. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang J, Huang F, Tan L, Bai C, Chen B,
Liu J, Liang J, Liu C, Zhang S, Lu G, et al: Host protein moloney
leukemia virus 10 (MOV10) acts as a restriction factor of influenza
a virus by inhibiting the nuclear import of the viral
nucleoprotein. J Virol. 90:3966–3980. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chaudhary K, Poirion OB, Lu L and Garmire
LX: Deep learning-based multi-omics integration robustly predicts
survival in liver cancer. Clin Cancer Res. 24:1248–1259. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jia HL, Zeng XQ, Huang F, Liu YM, Gong BS,
Zhang KZ, Zeng JH, Guo DG, Wang ZY and Li YG: Integrated microRNA
and mRNA sequencing analysis of age-related changes to mouse thymic
epithelial cells. IUBMB Life. 70:678–690. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xie XF, Chu HJ, Xu YF, Hua L, Wang ZP,
Huang P, Jia HL and Zhang L: Proteomics study of serum exosomes in
Kawasaki disease patients with coronary artery aneurysms. Cardiol
J. Apr;3.2018.(Epub ahead of print).
|
|
23
|
Stamer WD, Hoffman EA, Luther JM, Hachey
DL and Schey KL: Protein profile of exosomes from trabecular
meshwork cells. J Proteomics. 74:796–804. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zheng G, Wang H, Zhang X, Yang Y, Wang L,
Du L, Li W, Li J, Qu A, Liu Y and Wang C: Identification and
validation of reference genes for qPCR detection of serum microRNAs
in colorectal adenocarcinoma patients. PLoS One. 8:e830252013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhu Z, Qi Y, Ge A, Zhu Y, Xu K, Ji H, Shi
Z, Cui L and Zhou M: Comprehensive characterization of serum
microRNA profile in response to the emerging avian influenza A
(H7N9) virus infection in humans. Viruses. 6:1525–1539. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zekri AN, Youssef AS, El-Desouky ED, Ahmed
OS, Lotfy MM, Nassar AA and Bahnassey AA: Serum microRNA panels as
potential biomarkers for early detection of hepatocellular
carcinoma on top of HCV infection. Tumour Biol. 37:12273–12286.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li LM, Hu ZB, Zhou ZX, Chen X, Liu FY,
Zhang JF, Shen HB, Zhang CY and Zen K: Serum microRNA profiles
serve as novel biomarkers for HBV infection and diagnosis of
HBV-positive hepatocarcinoma. Cancer Res. 70:9798–9807. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rudan I, Chan KY, Zhang JSF, Theodoratou
E, Feng XL, Salomon JA, Lawn JE, Cousens S, Black RE, Guo Y, et al:
Causes of deaths in children younger than 5 years in China in 2008.
Lancet. 375:1083–1089. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lee J, Choi EH and Lee HJ: Clinical
severity of respiratory adenoviral infection by serotypes in Korean
children over 17 consecutive years (1991–2007). J Clin Virol.
49:115–120. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kishore A, Borucka J, Petrkova J and
Petrek M: Novel insights into miRNA in lung and heart inflammatory
diseases. Mediators Inflamm. 2014:2591312014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hassan T, McKiernan PJ, McElvaney NG,
Cryan SA and Greene CM: Therapeutic modulation of miRNA for the
treatment of proinflammatory lung diseases. Expert Rev Anti Infect
Ther. 10:359–368. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shwetha S, Gouthamchandra K, Chandra M,
Ravishankar B, Khaja MN and Das S: Circulating miRNA profile in HCV
infected serum: Novel insight into pathogenesis. Sci Rep.
3:15552013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bargalló ME, Guardo AC, Maleno MJ,
Miralles L, Egaña-Gorroño L, Escribà T, García F, Gatell JM, Arnedo
M and Plana M: Utility of systematic isolation of immune cell
subsets from HIV-infected individuals for miRNA profiling. J
Immunol Methods. 442:12–19. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liao J, Liu R, Yin LH and Pu YP:
Expression profiling of exosomal miRNAs derived from human
esophageal cancer cells by Solexa high-throughput sequencing. Int J
Mol Sci. 15:15530–15551. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Beltrami C, Clayton A, Phillips AO, Fraser
DJ and Bowen T: Analysis of urinary microRNAs in chronic kidney
disease. Biochem Soc Trans. 40:875–879. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Alipoor SD, Mortaz E, Garssen J,
Movassaghi M, Mirsaeidi M and Adcock IM: Exosomes and exosomal
miRNA in respiratory diseases. Mediators Inflamm. 2016:56284042016.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ito S, Kamoto Y, Sakai A, Sasai K, Hayashi
T, Toyooka S and Katayama H: Unique circulating microRNAs in
relation to EGFR mutation status in Japanese smoker male with lung
adenocarcinoma. Oncotarget. 8:114685–114697. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang Y, Yu M, Dai M, Chen C, Tang Q, Jing
W, Wang H and Tian W: miR-450a-5p within rat adipose tissue
exosome-like vesicles promotes adipogenic differentiation by
targeting WISP2. J Cell Sci. 130:1158–1168. 2017.PubMed/NCBI
|
|
39
|
Dernowsek JA, Pereira MC, Fornari TA,
Macedo C, Assis AF, Donate PB, Bombonato-Prado KF, Passos-Bueno MR
and Passos GA: Posttranscriptional interaction between miR-450a-5p
and miR-28-5p and STAT1 mRNA triggers osteoblastic differentiation
of human mesenchymal stem cells. J Cell Biochem. 118:4045–4062.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Dong G, Fan H, Yang Y, Zhao G, You M, Wang
T and Hou Y: 17β-Estradiol enhances the activation of IFN-α
signaling in B cells by down-regulating the expression of
let-7e-5p, miR-98-5p and miR-145a-5p that target IKKε. Biochim
Biophys Acta. 1852:1585–1598. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Pacholewska A, Kraft M, Gerber V and
Jagannathan V: Differential expression of serum MicroRNAs supports
CD4+ T cell differentiation into Th2/Th17 cells in
severe equine asthma. Genes (Basel). 8(pii): E3832017. View Article : Google Scholar : PubMed/NCBI
|