Introduction
Renal cell carcinoma (RCC) is the one of the 10 most
common cancers, accounting for 2–3% of all adult malignancies
(1). Clear cell renal cell
carcinoma (ccRCC) is the most common subtype of RCC, and
approximately 75–80% of patients with RCC are diagnosed with ccRCC
(2). Current therapies for renal
cancer include conventional chemotherapy and radiation, however,
the prognosis is poor (3).
Therefore, to aid in the development of effective treatments for
ccRCC, it is necessary to investigate the underlying molecular
mechanism involved in the pathogenesis of ccRCC.
Only 2% of RNAs encode proteins in human cells. Many
transcripts that do not encode proteins have recently been
identified in large-scale genomic studies (4,5).
Although these RNAs are not translated, they play critical roles in
regulating transcriptional and non-transcriptional processes
(6). These non-coding RNAs
(ncRNAs) can be broadly classified as small ncRNAs (<200
nucleotides, including microRNAs (miRNAs), siRNAs, and piRNAs) or
long non-coding (lncRNAs) (>200 nucleotides). miRNAs are the
most widely studied subclass of small ncRNAs, and they can
post-transcriptionally regulate the expression of multiple genes
via the imperfect complementarity to their target mRNA transcripts
(7). miRNAs have been found to be
involved in tumorigenesis, and evaluating the changes in miRNA
expression could provide useful information to better understand
tumor formation and progression (8). lncRNAs are eukaryotic RNAs with no
coding capacity. Alteration of lncRNA expression has been reported
to be associated with tumor development (9), and certain lncRNAs have been used as
cancer biomarkers and potential targets in several types of tumors
(10).
lncRNAs have been revealed to act as competing
endogenous RNAs (ceRNAs), where the lncRNAs bind to and keep miRNAs
from their cognate mRNA targets (11,12).
Hence, the mRNAs can be prevented from mediating miRNA-mediated
repression (11,12). A previous study indicated that
small concentration changes in miRNA-mRNA or miRNA-ceRNA pairs can
substantially affect the regulation of the gene expression network
(13). For example, the lncRNA
HULC has been previously shown to act as an endogenous sponge and
inhibit miR-372, which plays an important role in tumorigenesis in
liver cancer (14). The
muscle-specific lnRNA, linc-MDI, regulates muscle differentiation
by sequestering miR-133 and miR-135 and thus modulating the
expression of MAML1 and MEF2C (15). Recent advances in the study of the
functions and mechanisms of lncRNAs in physiological and
pathological processes have led to expanding the possible manner in
which lncRNAs function within the already complex system of
miRNA-mediated gene regulation.
Although the expression profile of mRNA and lncRNA
transcripts in ccRCC has been explored and several lncRNAs, such as
HIF-1alpha-AS1, H19, KCQN1OT1, and MALAT1, have been reported to be
involved in renal cancer (16–18),
the regulation mechanism of lncRNA-miRNA-mRNA in ccRCC has not been
elucidated. In this study, we downloaded the data for an important
cohort encompassing more than 300 samples from the Cancer Genomes
Atlas (TCGA) and used a bioinformatics method to predict the
regulation mechanism of lncRNAs-miRNAs-mRNAs in ccRCC.
Materials and methods
Data
RNA-seq data collection and
processing
The TCGA database is a comprehensive, publicly
available, and vast source of information on cancer genetic and
epigenetic profiles. The TCGA RNA-seq expression datasets (level 3)
for ccRCC were downloaded from the TCGA Data Portal (https://tcga-data.nci.nih.gov/tcga/) (June 3,
2016), which contained 606 mRNA expression profiles and 326 miRNA
expression profiles. Since TCGA level 3 data typically represents
aggregated, normalized, and/or segmented data, the level 3 data
downloaded was analyzed without normalization. Based on the
barcodes, the RNA-seq samples were matched to miRNAs, and a total
of 326 samples, including 254 ccRCC samples and 71 normal controls,
were obtained.
lncRNA and mRNA sequencing was performed on the
Illumina Hiseq 2000 platform, and miRNAs were sequenced on the
RNASeqV2 platform (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga/using-tcga/technology/Illumina-HiSeq2000-Data-Sheet).
RNA-seq reads were processed using RNA-Seq by
Expectation-Maximization (RSEM). In addition, lncRNAs and mRNAs
were annotated and identified on the website of the HUGO gene
nomenclature committee (HGNC, http://www.genenames.org/), which contains annotation
information on 2,775 lncRNAs and 19,004 protein-coding genes.
Analysis of differentially expressed
mRNAs (DEGs), lncRNAs (DElncRs), and miRNAs (DEmiRs)
The differential expression of mRNAs, lncRNAs, and
miRNAs was analyzed using the edgeR package (19), which is an R Bioconductor package
that provides statistical routines for examining the differential
expression of replicated count data in RNA-seq data. This package
is based on an overdispersed Poisson model (20) to account for the variability at
both the biological and technical levels. In addition, the
Empirical Bayes methods (21) in
this package were used to moderate the overdispersion across
transcripts, improving the reliability of inferences. The nominal
P-values were estimated based on the method of Benjamini and
Hochberg (22), using the multtest
package (http://www.bioconductor.org/packages/release/bioc/html/multtest.html)
in R. Only RNAs with a false-discovery rate (FDR) <0.05 and
|logFC (fold change)>1| were selected as DEGs, DelncRs, or
DEmiRs.
Correlation analysis between the
clinical characteristics and expression of DemiRs and DElncRs
The clinical characteristics of patients with ccRCC
were extracted from the downloaded data, including sex (female vs.
male), age at diagnosis (≥61 years vs. <61 years), tumor grade
(G3/G4 vs. G1/G2), tumor status (tumor-burden state vs. tumor-free
state), American Joint Committee on Cancer pathologic stage (III/IV
vs. I/II), tumor metastasis (yes vs. no), and overall survival
time. Then, the DElncRs/DemiRs that were significantly correlated
with these clinical characteristics were identified using
nonparametric regression analysis. A survival curve was constructed
using the Kaplan-Meier method (23), and comparisons using the log rank
test were performed between patients with upregulated DElncRs and
those with downregulated DElncRs.
Construction of a ceRNA-regulated
network
The lncRNA targets of human miRNAs were downloaded
from the miRCode database (http://www.mircode.org/), a comprehensive searchable
map of putative miRNA target sites across the complete GENCODE
annotated transcriptome established by Jeggari et al
(24), and starBase v2.0
(http://starbase.sysu.edu.cn/), a
platform comprised of 111 CLIP-Seq data sets and focusing on
protein-lncRNA, protein-sncRNA, protein-mRNA and protein-pseudogene
interactions. The miRNA targets from the miRTarBase (http://mirtarbase.mbc.nctu.edu.tw), which is the
most updated collection of miRNA-target interactions, were
identified.
According to the identified miRNA-lncRNA
interactions and miRNA-mRNA interactions, a comprehensive
lncRNA-miRNA-mRNA network was constructed. Then, the interactions
in the lncRNA-miRNA-mRNA network were further identified by
calculating maximal information coefficients (MICs) with cut-off
values of MIC >0.15 and MIC-p2 >0.15.
Functional enrichment of DEGs in the
ceRNA-regulated network
The DEGs in the ceRNA-regulated network were
assigned to functional categories in the Kyoto Encyclopedia of
Genes and Genomes (KEGG) pathway database (25) (https://www.genome.jp/kegg/), and the pathways
significantly associated with ccRcc were obtained. Enrichment
P-values were calculated using Fisher's exact tests using the
equation below. Then, the functional pathways in the
ceRNA-regulated network in which the DEGs were enriched were
identified.
p=1-∑i=0x-1(Mi)(N-MK-i)(NK)
Here, N represents the number of genes in the whole
genome; M represents the number of genes enriched in KEGG pathways;
and K represents the number of differentially expressed genes. The
Fisher's score represents the probability that at least × genes of
K differentially expressed genes are enriched in KEGG pathways.
Results
Identification of DEGs, DelncRs, and
DEmiRs
lncRNAs and mRNAs were annotated and identified on
the HUGO gene nomenclature committee website. A total of 819
lncRNAs and 18,137 protein-coding transcripts were identified.
Subsequently, using an FDR statistic <0.05 and
|logFC2(fold change)>1|, the differential expression
analysis identified 1,573 DEGs and 37 DElncRs in the RNA-seq data
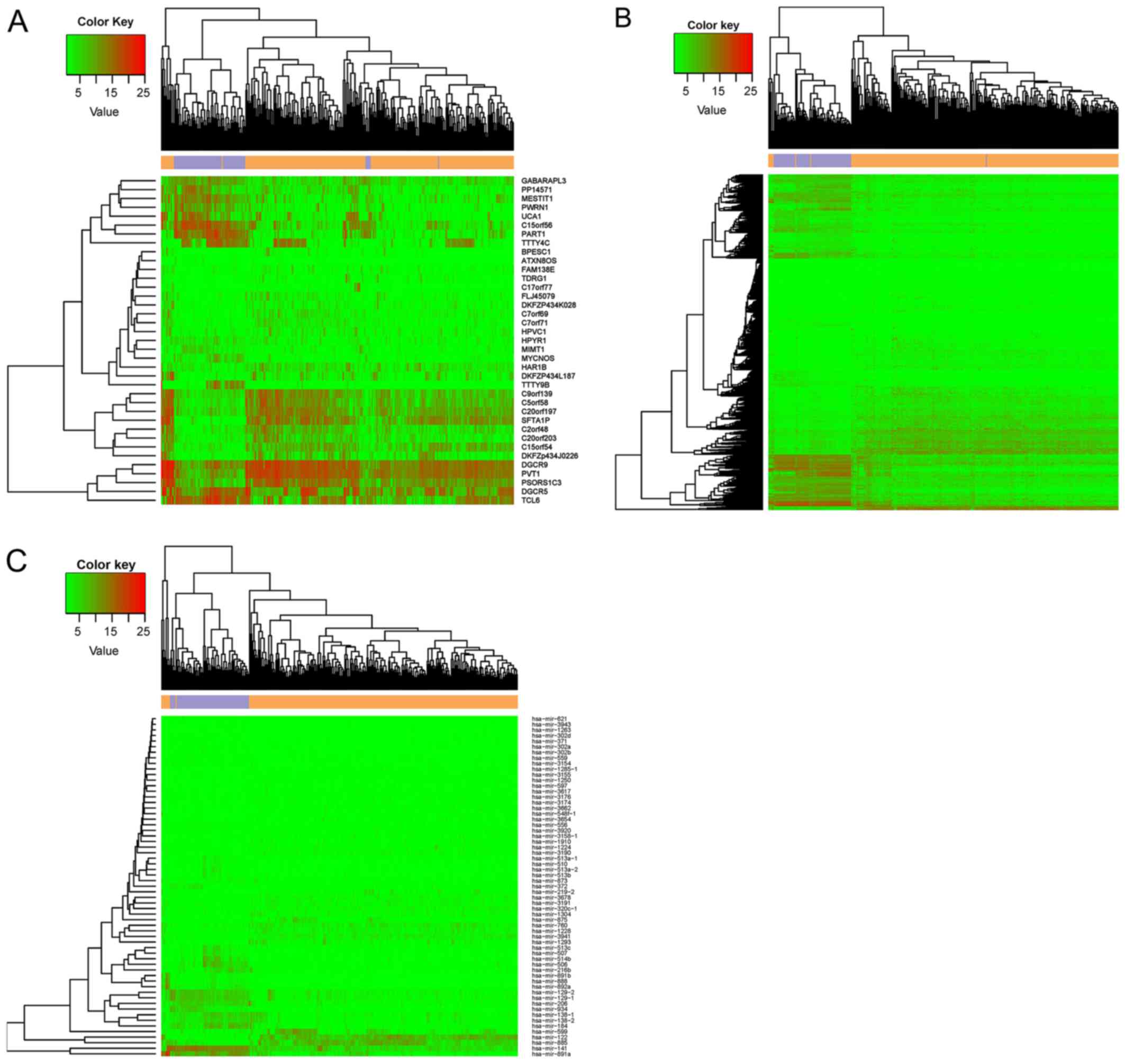
and 62 DEmiRs in the miRNA-seq data. A heatmap representing the
expression of the identified DElncRs, DEGs, and DEmiRs is presented
in Fig. 1. The ccRCC samples were
obviously separated from the control samples, suggesting that the
DEGs represent a large difference between the tumor and control
samples.
Identification of DemiRs and DelncRs
that are significantly correlated with clinical
characteristics
The correlation analysis between the clinical
characteristics of patients and DemiRs indicated that the
expression level of hsa-mir-216b was associated with two clinical
variables, sex and tumor grade (Table
I).
 | Table I.The interactions between DEmiRs and
the clinical characteristics of patients. |
Table I.
The interactions between DEmiRs and
the clinical characteristics of patients.
|
| Related miRNAs |
|---|
|
|
|
|---|
| Comparisons | Upregulated | Downregulated |
|---|
| Sex (female vs.
male) | hsa-mir-1293,
hsa-mir-3654, hsa-mir-1285-1 | hsa-mir-875,
hsa-mir-216b, hsa-mir-599 |
| Age at diagnosis
(≥61 vs. <61) | – | hsa-mir-513c,
hsa-mir-1228, hsa-mir-3678, hsa-mir-514b, hsa-mir-513b |
| Tumor grade (G3 +
G4 vs. G1 + G2) | hsa-mir-3943,
hsa-mir-510 | hsa-mir-216b,
hsa-mir-891b |
| Tumor status (with
tumor vs. tumor-free) | hsa-mir-934 | – |
| AJCC pathological
stage (III + IV vs. I + II) | hsa-mir-891a,
hsa-mir-3174 | hsa-mir-507,
hsa-mir-371 |
| Metastatic (yes vs.
no) | hsa-mir-599,
hsa-mir-888, hsa-mir-1263 | hsa-mir-141 |
Moreover, the correlation between the clinical
characteristics of patients and the expression of identified
DelncRs was explored. The results are presented in Table II. The expression of DGCR9 and
TTTY4C was significantly correlated with three clinical variables,
sex, tumor status, and metastasis. TCL6 was revealed to be
significantly associated with tumor grade and AJCC pathological
stage. In addition, tumor metastasis was related to the upregulated
expression of BPESC1, C20orf203, C7orf71, C2orf48, DGCR9, DGCR5,
and DKFZp434J0226 and the downregulated expression of MYCNOS,
PP14571, TTTY4C, and PART1. These results revealed certain
candidate lncRNAs for ccRCC, such as C7orf71, C2orf48, DGCR9,
DGCR5, and TTTY4C, and also suggested a sex difference in the stage
distribution of ccRCC.
 | Table II.The correlation between DElncRNAs and
patient clinical features. |
Table II.
The correlation between DElncRNAs and
patient clinical features.
|
| Identified
DElncRNAs |
|---|
|
|
|
|---|
| Patient clinical
characteristics | Upregulated | Downregulated |
|---|
| Sex (female vs.
male) | C20orf203, HAR1B,
DGCR9, DGCR5, FAM138E | TTTY4C, TTTY9B |
| Age at diagnosis
(≥61 vs.<61 years) | C2orf48, DGCR9,
DGCR5 | ATXN8OS |
| Tumor grade (G3/G4
vs. G1/G2) | PSORS1C3, C7orf71,
DKFZP434K028, FAM138E | HPYR1, TCL6 |
| Tumor status
(tumor-burden state vs. tumor-free state) | C7orf71, HAR1B,
C2orf48, TDRG1 | TTTY4C |
| AJCC pathological
stage (III/IV vs. I/II) | C7orf71, PSORS1C3,
DKFZP434K028, FAM138E | MYCNOS, TCL6 |
| Metastasis (yes vs.
no) | BPESC1, C20orf203,
C7orf71, C2orf48, | MYCNOS, PP14571,
TTTY4C, |
|
| DGCR9, DGCR5,
DKFZp434J0226 | PART1 |
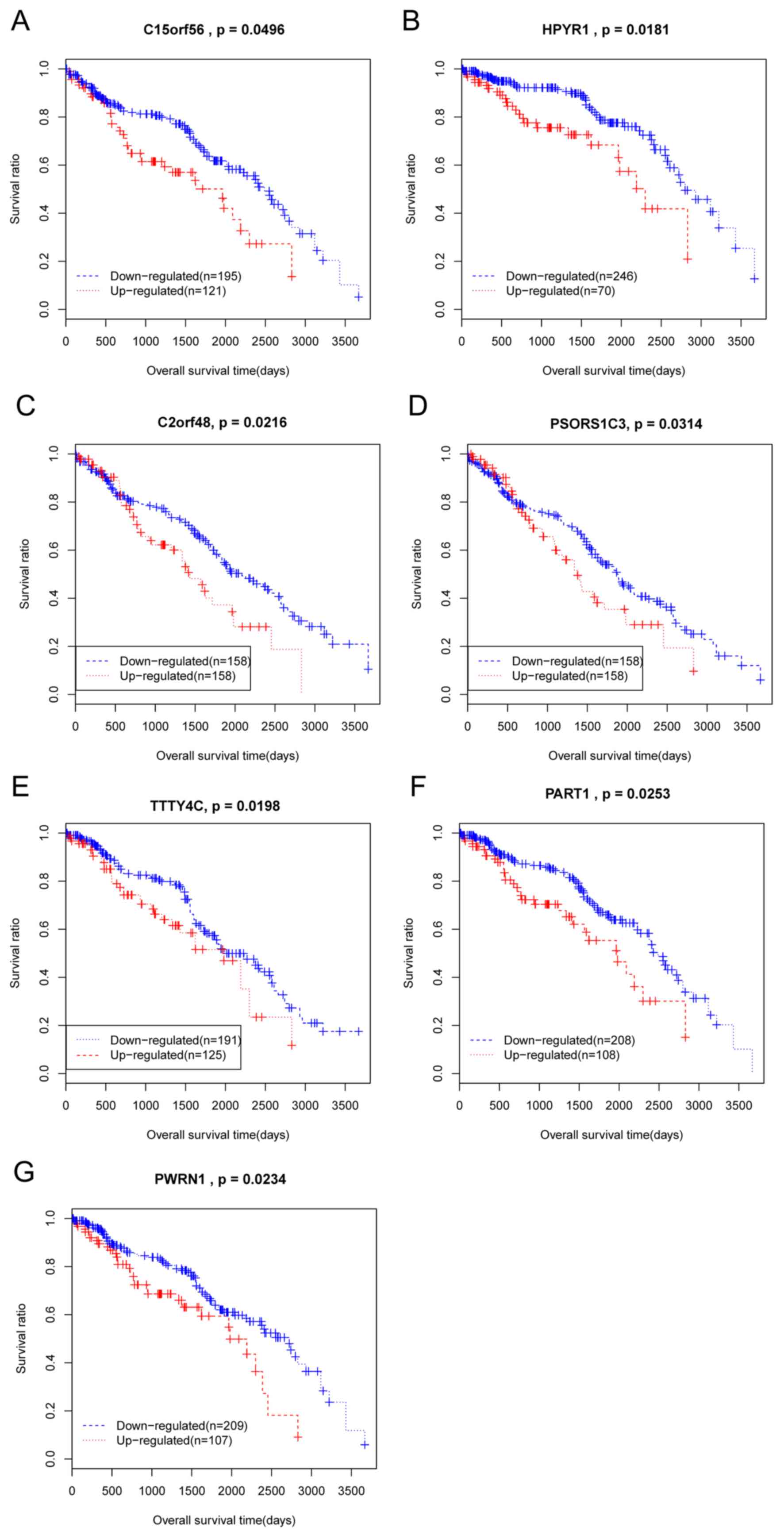
Kaplan Meier analysis was conducted on all the 37
DElncRs that were significantly associated with patient clinical
characteristics. As a result, 7 DelncRs that were significantly
associated with overall survival time of patients with ccRCC were
identified, TTTY4C, PSORS1C3, C2orf48, HPYR1, PWRN1, C15orf56, and
PART1. All of these DElncRs were negatively correlated with patient
overall survival, as patients with lower expression of these
DElncRs had longer survival times (Fig. 2).
Identification of DElncR-DEmiR
interaction pairs
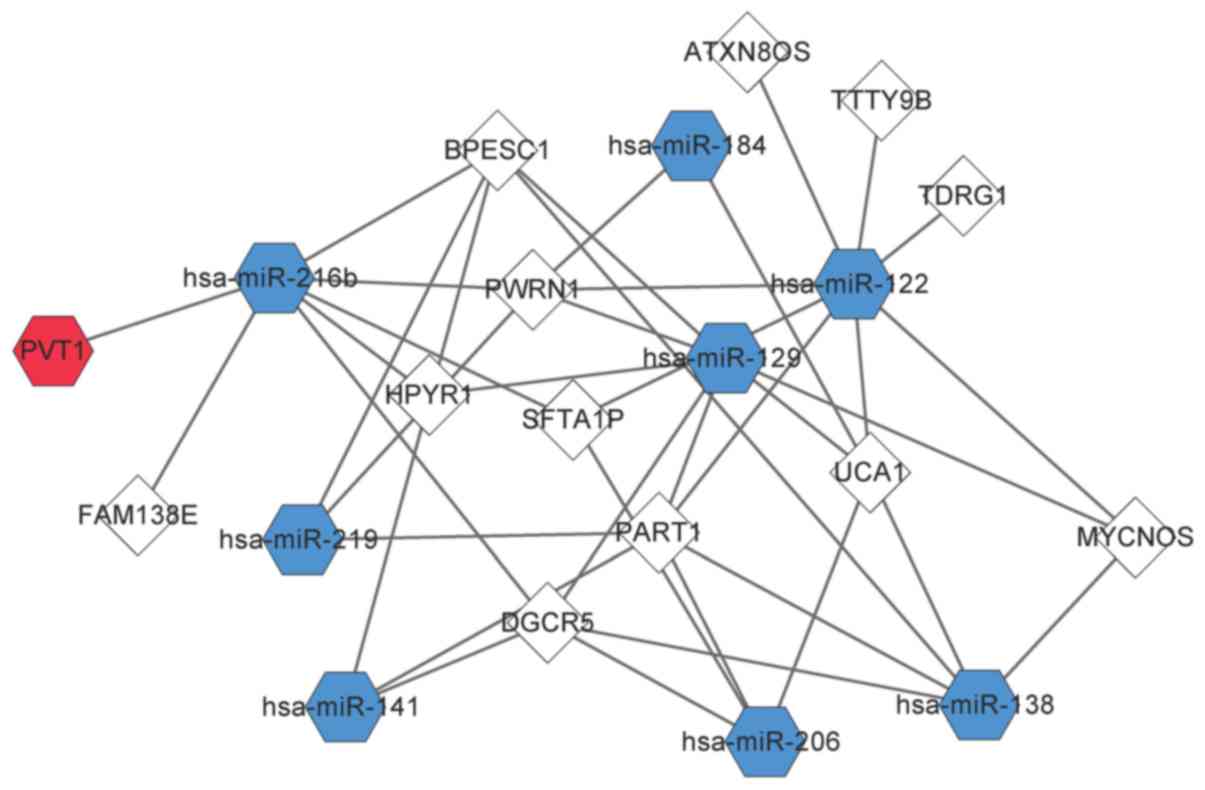
Predictions using miRcode and starBase revealed a
total of 353 pairs of DElncR-miRNA interactions and 38 pairs of
DElncR-DEmiR interactions. An interaction network of 13 DElncRs and
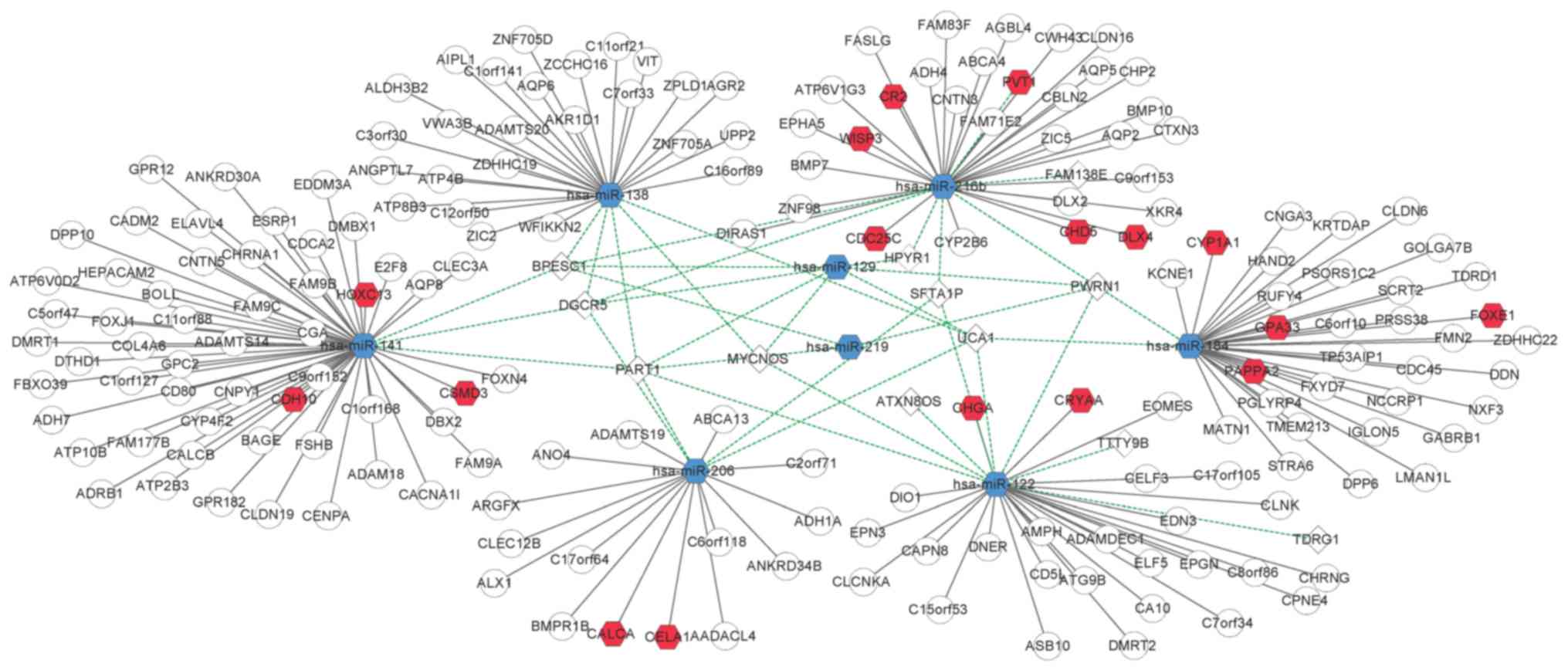
8 DEmiRs was constructed (Fig. 3).
miR-122, miR-129, and miR-216b targeted more lncRNAs than the other
miRNAs. miR-122 had a relationship with 8 DElncRNAs, including
MYCNOS, PART1, and PWRN1. miR-129 targeted 7 DElncRNAs, including
DGCR5, HPYR1, MYCNOS, PART1, and PWRN1. miR-216b had 7 targets,
BPESC1, DGCR5, FAM138E, HPYR1, PVT1, PWRN1, and SFTA1P.
Identification of DEmiR-DEG
interaction pairs
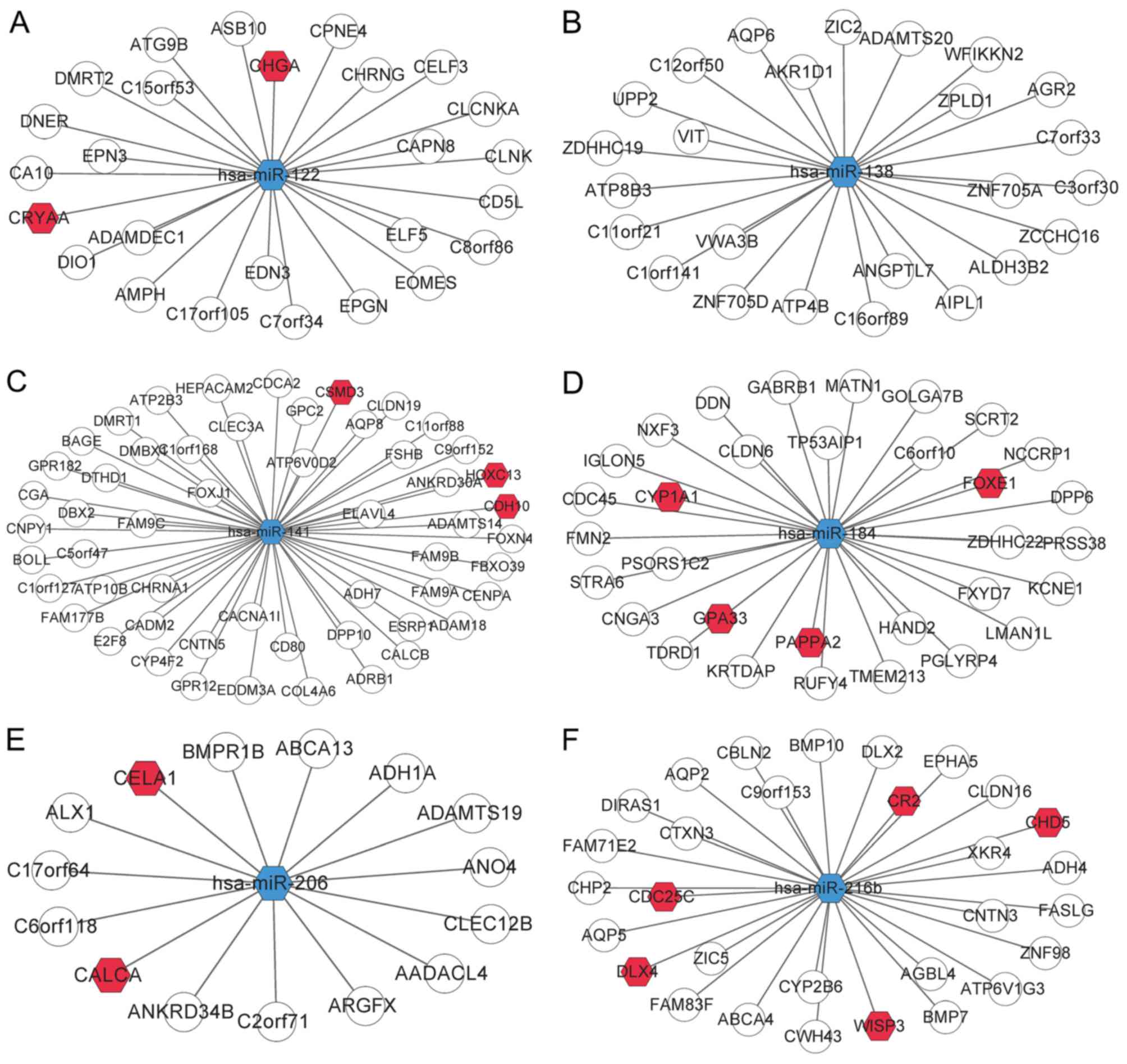
As aforementioned, a total of 8 DEmiRs were revealed
to be DElncR-targeting miRNAs. Then, the DEmiR-mRNA interactions
were collected from the miRTarBase database, and 6 out of 8 DEmiRs
were revealed to target DEGs: hsa-miR-122 targeted 26 DEGs,
hsa-miR-216b targeted 30 DEGs, hsa-miR-141 targeted 54 DEGs,
hsa-miR-206 targeted 15 DEGs, hsa-miR-138 targeted 25 DEGs, and
hsa-miR-184 targeted 32 DEGs (Table
III). Collectively, these data indicated that hsa-miR-122 and
hsa-miR-216b may be important in ccRCC. In addition, hsa-miR-141
had the most targets, followed by hsa-miR-184, also indicating
their crucial role in ccRCC. Among the 182 DEmiR targets, 130 DEGs
were identified as cancer-related genes after comparison with the
allOnco database (http://www.bushmanlab.org/links/genelists), a
collection of 2,125 cancer-related genes. The DEmiR-DEG interaction
network is presented in Fig.
4.
 | Table III.The DEmiRs-DEGs interaction pairs
extracted from miRTarBase. |
Table III.
The DEmiRs-DEGs interaction pairs
extracted from miRTarBase.
| DEmiRs | No. of targeting
DEGs | DEmiRs targeting
DEGs |
|---|
| has-miR-122 | 26 | ADAMDEC1, AMPH,
ASB10, ATG9B, C15orf53, C17orf105, C7orf34, C8orf86, CA10, CAPN8,
CD5L, CELF3, CHGA, CHRNG, CLCNKA, CLNK, CPNE4, CRYAA, DIO1, DMRT2,
DNER, EDN3, ELF5, EOMES, EPGN, EPN3 |
| has-miR-138 | 25 | ADAMTS20, AGR2,
AIPL1, AKR1D1, ALDH3B2, ANGPTL7, AQP6, ATP4B, ATP8B3, C11orf21,
C12orf50, C16orf89, C1orf141, C3orf30, C7orf33, UPP2, VIT, VWA3B,
WFIKKN2, ZCCHC16, ZDHHC19, ZIC2, ZNF705A, ZNF705D, ZPLD1 |
| has-miR-141 | 54 | ADAM18, ADAMTS14,
ADH7, ADRB1, ANKRD30A, AQP8, ATP10B, ATP2B3, ATP6V0D2, BAGE, BOLL,
C11orf88, C1orf127, C1orf168, C5orf47, C9orf152, CACNA1I, CADM2,
CALCB, CD80, CDCA2, CENPA, CGA, CHRNA1, CLDN19, CLEC3A, CNPY1,
CNTN5, COL4A6, CSMD3, CYP4F2, DBX2, DMBX1, DMRT1, DPP10, DTHD1,
E2F8, EDDM3A, ELAVL4, ESRP1, FAM177B, FAM9A, FAM9B, FAM9C, FBXO39,
FOXJ1, FOXN4, FSHB, GPC2, GPR12, GPR182, HEPACAM2, HOXC13 |
| has-miR-184 | 32 | C6orf10, CDC45,
CLDN6, CNGA3, CYP1A1, DDN, DPP6, FMN2, FOXE1, FXYD7, GABRB1,
GOLGA7B, GPA33, HAND2, IGLON5, KCNE1, KRTDAP, LMAN1L, MATN1,
NCCRP1, NXF3, PAPPA2, PGLYRP4, PRSS38, PSORS1C2, RUFY4, SCRT2,
STRA6, TDRD1, TMEM213, TP53AIP1, ZDHHC22 |
| has-miR-206 | 15 | AADACL4, ABCA13,
ADAMTS19, ADH1A, ALX1, ANKRD34B, ANO4, ARGFX, BMPR1B, C17orf64,
C2orf71, C6orf118, CALCA, CELA1, CLEC12B |
| has-miR-216b | 30 | ABCA4, ADH4, AGBL4,
AQP2, AQP5, ATP6V1G3, BMP10, BMP7, C9orf153, CBLN2, CDC25C, CHD5,
CHP2, CLDN16, CNTN3, CR2, CTXN3, CWH43, CYP2B6, DIRAS1, DLX2, DLX4,
EPHA5, FAM71E2, FAM83F, FASLG, WISP3, XKR4, ZIC5, ZNF98 |
Construction and functional annotation
of a ceRNA-regulated network
A comprehensive lncRNA-miRNA-mRNA network was
established based on the identified DElncR-DEmiR interactions and
DEmiR-DEG interactions. By calculating MICs, with MIC >0.15 and
MIC-p2 >0.15, a ceRNA-regulated network comprised of
203 nodes and 221 edges was constructed (Fig. 5). This network indicated the roles
of hsa-miR-141, hsa-miR-184, hsa-miR-122, hsa-miR-216b,
hsa-miR-138, and hsa-miR-206 in the ceRNA-regulated network in
ccRCC.
To study the function of DEGs in the ceRNA-regulated
network, KEGG pathway and gene ontology (GO) catalogue annotation
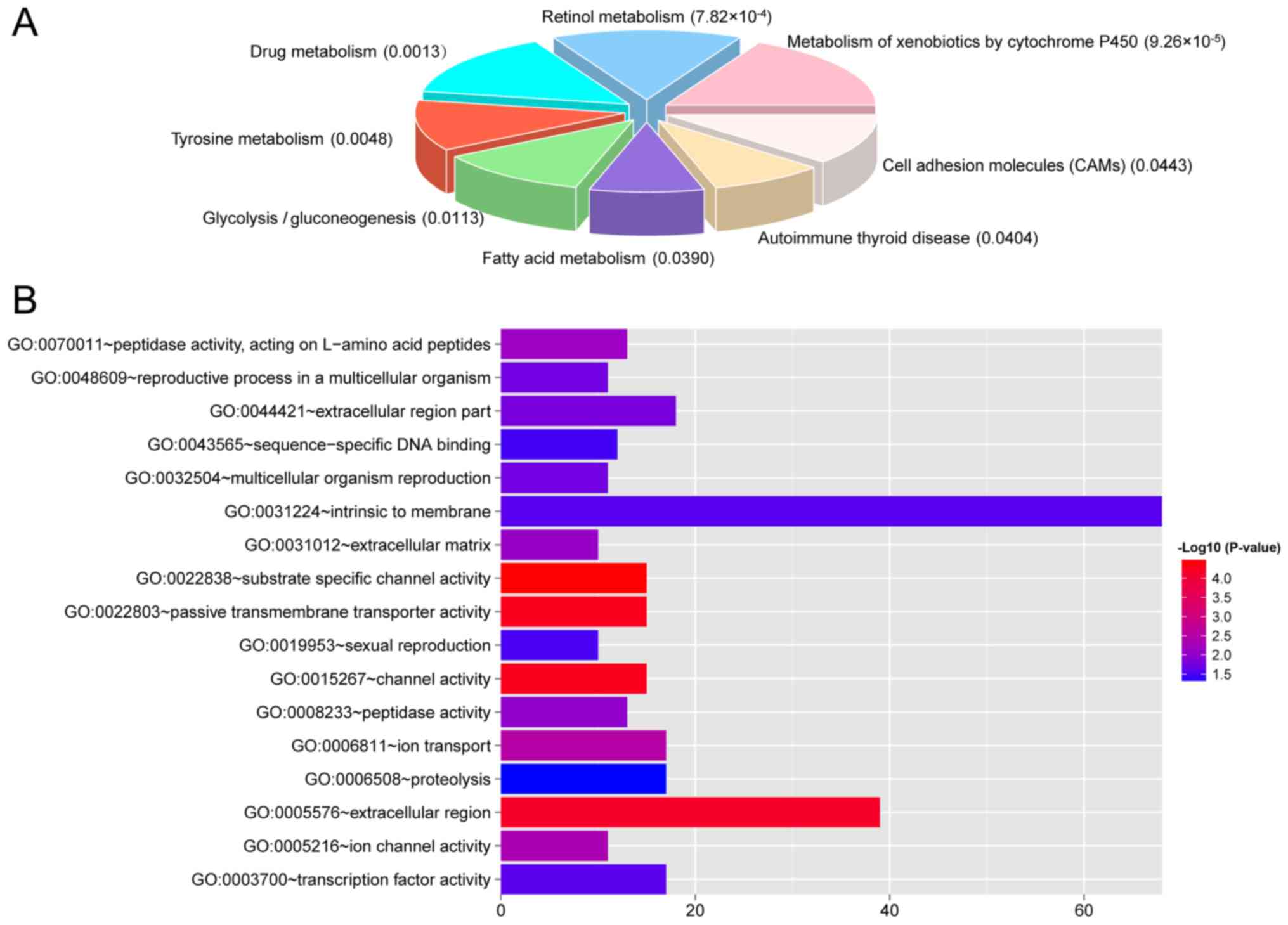
were performed. These DEGs were enriched in 8 KEGG pathways
(Fig. 6A), such as metabolism of
xenobiotics by cytochrome P450, retinol metabolism, and drug
metabolism, and 17 GO terms, such as intrinsic to membrane,
extracellular region, extracellular region part, transcription
factor activity, ion transport, and proteolysis (Fig. 6B).
Discussion
Human lncRNAs have been revealed to act as
competitors of mRNAs for miRNA binding in RNA-RNA cross-talk
networks and regulate mRNA expression at the post-transcriptional
level. Although certain differentially expressed lncRNAs and mRNAs
have been identified in ccRCC by expression profiling and a branch
of the MALAT1/miR-200s/ZEB2 pathway, and the functional triplets
consisting of lncRNAs, mRNAs, and miRNAs, have been proposed to
regulate the progression of ccRCC, a systematic analysis of the
ceRNA-regulated network in ccRCC has not been performed. To
understand the roles of the ceRNA-regulated network in the
pathology of ccRCC, a transcriptome-wide identification and
characterization of differentially expressed RNAs, including
lncRNAs, miRNAs, and mRNAs was performed, and a comprehensive
lncRNA-miRNA-mRNA network was constructed based on identified
lncRNA-miRNA interactions and miRNA-mRNA interactions downloaded
from an online data portal. The present study provides new insights
that will aid in understanding the molecular mechanism involved in
ccRCC.
To the best of our knowledge, the present study
represents the first comprehensive description of the
ceRNA-regulated network in ccRCC, as identified through the
bioinformatics analysis of RNAseq data in a large patient cohort
that encompasses 254 ccRCC tumor samples and 71 normal renal tissue
samples. Several novel findings were obtained from our in-depth
genomic analysis.
First, the expression of certain differentially
expressed lncRNAs, such as C2orf48, DGCR9, DGCR5, DKFZp434J0226,
MYCNOS, PP14571, TTTY4C, and PART1, was correlated with tumor
metastasis; moreover, the expression of TTTY4C, PSORS1C3, C2orf48,
HPYR1, PWRN1, C15orf56, and PART1 was negatively correlated with
patient overall survival. The expression level of miR-216b was
revealed to be associated with sex and tumor grade in patients with
ccRCC (26). Notably, TTTY4C and
certain other lncRNAs were also revealed to be related to sex,
which is consistent with the sex-dependent manner of ccRCC
(27). All these identified
lncRNAs have not been previously reported to be associated with
ccRCC. DGCR5 has been revealed to be involved in the regulation of
the proliferation, migration, and invasion of lung cancer (28) and lung adenocarcinoma (29). MYCNOS has been reported to
cooperate with a crucial transcription factor, CCCTC-binding
factor, to promote neuroblastoma progression through MYCN
expression (30). PSORS1C3 has
been demonstrated to be expressed in pluripotent and tumor cell
lines (31). Therefore, further
studies are required to clarify the role of these identified
lncRNAs in ccRCC.
Second, a comprehensive lncRNA-miRNA-mRNA network in
ccRCC was constructed, which revealed the roles of hsa-miR-141,
hsa-miR-184, hsa-miR-122, hsa-miR-216b, hsa-miR-138, and
hsa-miR-206 in the ceRNA-regulated network in ccRCC. The identified
miRNAs in the ceRNA-regulated network, including miR-141 (32,33),
miR-138 (32–34), miR-206c (35), miR-122 (36), miR-184 (37), and miR-216b (26), have been reported to play important
roles in tumor progression in human ccRCC. In addition to these
mRNAs, lncRNAs were also identified in the ceRNA-regulated network.
As aforementioned, DGCR5 plays an important role in lung cancer
(28,29) and was revealed to target miR-216b,
miR-129, miR-206, miR-141, and miR-138 in the present study. MYCNOS
was revealed to target miR-129, miR-122, and miR-138. PART1 has
also been revealed to target miR-129, miR-206, miR-219, miR-206,
miR-141, and miR-138. Based on our results, the ceRNA-regulated
network revealed how these competing modules (lncRNA-miRNA-mRNA)
play a role in the pathogenesis of ccRCC and indicated the roles
played by lncRNAs in the disease.
Moreover, the transcripts in the ceRNA network of
ccRCC were revealed to be enriched in several pathways such as
metabolism of xenobiotics by cytochrome P450, retinol metabolism,
and drug metabolism and critical biological processes such as
intrinsic to membrane, extracellular region, extracellular region
part, transcription factor activity, ion transport, and
proteolysis. These pathways share certain genes, such as ADH4 and
ALDH3B2. Previous studies have reported that xenobiotic metabolism
by cytochrome P450 and drug activation are involved in
tumorigenesis in RCC (38,39).
Collectively, the use of bioinformatic methods can
significantly improve our understanding of RNA communication in
ccRCC at the transcriptome level. However, these functional modules
obtained from bioinformatic analyses require experimental
confirmation.
In conclusion, our study represents the first
comprehensive analysis of a lncRNA-miRNA-mRNA network in ccRCC. The
results revealed certain novel lncRNAs involved in ccRcc, such as
DGCR5, MYCNOS, and PART1, which may be involved in the progression
of ccRCC through cytochrome P450-mediated drug metabolism. These
findings expand the existing knowledge on lncRNA characteristics
and provide new tools for the identification of lcRNAs in ccRCC,
which can help to elucidate the disease processes and identify new
targets for therapy.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81702495), the
Natural Science Foundation of Anhui Province (grant no.
KJ2018A0214), and the Anhui Natural Science Foundation (grant no.
1808085MH293).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
BL and TM performed the data analyses and wrote the
manuscript. QL, SW, WS and WL contributed significantly in data
analyses and manuscript revision. JL and YG conceived and designed
the study. All authors read and approved the final manuscript and
agree to be accountable for all aspects of the research in ensuring
that the accuracy or integrity of any part of the work are
appropriately investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gago-Dominguez M, Castelao JE, Yuan JM,
Ross RK and Yu MC: Increased risk of renal cell carcinoma
subsequent to hysterectomy. Cancer Epidemiol Biomarkers Prev.
8:999–1003. 1999.PubMed/NCBI
|
|
3
|
Linehan WM: Genetic basis of kidney
cancer: Role of genomics for the development of disease-based
therapeutics. Genome Res. 22:2089–2100. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kapranov P, Cheng J, Dike S, Nix DA,
Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J,
Hofacker IL, et al: RNA maps reveal new RNA classes and a possible
function for pervasive transcription. Science. 316:1484–1488. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Guttman M, Donaghey J, Carey BW, Garber M,
Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al:
lincRNAs act in the circuitry controlling pluripotency and
differentiation. Nature. 477:295–300. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wilusz JE, Sunwoo H and Spector DL: Long
noncoding RNAs: Functional surprises from the RNA world. Genes Dev.
23:1494–1504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Farazi TA, Spitzer JI, Morozov P and
Tuschl T: miRNAs in human cancer. J Pathol. 223:102–115. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kwok ZH and Tay Y: Long noncoding RNAs:
Lincs between human health and disease. Biochem Soc Trans.
45:805–812. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Crea F, Watahiki A, Quagliata L, Xue H,
Pikor L, Parolia A, Wang Y, Lin D, Lam WL, Farrar WL, et al:
Identification of a long non-coding RNA as a novel biomarker and
potential therapeutic target for metastatic prostate cancer.
Oncotarget. 5:764–774. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ebert MS and Sharp PA: Emerging roles for
natural microRNA sponges. Curr Biol. 20:R858–R861. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Arvey A, Larsson E, Sander C, Leslie CS
and Marks DS: Target mRNA abundance dilutes microRNA and siRNA
activity. Mol Syst Biol. 6:3632010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Panzitt K, Tschernatsch MM, Guelly C,
Moustafa T, Stradner M, Strohmaier HM, Buck CR, Denk H, Schroeder
R, Trauner M and Zatloukal K: Characterization of HULC, a novel
gene with striking up-regulation in hepatocellular carcinoma, as
noncoding RNA. Gastroenterology. 132:330–342. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cesana M, Cacchiarelli D, Legnini I,
Santini T, Sthandier O, Chinappi M, Tramontano A and Bozzoni I: A
long noncoding RNA controls muscle differentiation by functioning
as a competing endogenous RNA. Cell. 147:358–369. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bertozzi D, Iurlaro R, Sordet O, Marinello
J, Zaffaroni N and Capranico G: Characterization of novel antisense
HIF-1α transcripts in human cancers. Cell Cycle. 10:3189–3197.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Frevel MA, Sowerby SJ, Petersen GB and
Reeve AE: Methylation sequencing analysis refines the region of H19
epimutation in Wilms tumor. J Biol Chem. 274:29331–29340. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chiesa N, De Crescenzo A, Mishra K, Perone
L, Carella M, Palumbo O, Mussa A, Sparago A, Cerrato F, Russo S, et
al: The KCNQ1OT1 imprinting control region and non-coding RNA: New
properties derived from the study of Beckwith-Wiedemann syndrome
and Silver-Russell syndrome cases. Hum Mol Genet. 21:10–25. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cameron AC and Trivedi PK:
Regression-based tests for overdispersion in the Poisson model. J
Econ. 46:347–364. 1990. View Article : Google Scholar
|
|
21
|
Maritz JS and Lwin T: Empirical bayes
methods with applications (Chapman & Hall/CRC Monographs on
Statistics & Applied Probability). (2nd). 2018.
|
|
22
|
Solari A and Goeman JJ: Minimally adaptive
BH: A tiny but uniform improvement of the procedure of Benjamini
and Hochberg. Biom J. 59:776–780. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Koletsi D and Pandis N: Survival analysis,
part 2: Kaplan-Meier method and the log-rank test. Am J Orthod
Dentofacial Orthop. 152:569–571. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jeggari A, Marks DS and Larsson E:
miRcode: A map of putative microRNA target sites in the long
non-coding transcriptome. Bioinformatics. 28:2062–2063. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kanehisa M, Furumichi M, Tanabe M, Sato Y
and Morishima K: KEGG: New perspectives on genomes, pathways,
diseases and drugs. Nucleic Acids Res. 45:D353–D361. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang W, Zhao E, Yu Y, Geng B, Zhang W and
Li X: MiR-216a exerts tumor-suppressing functions in renal cell
carcinoma by targeting TLR4. Am J Cancer Res. 8:476–488.
2018.PubMed/NCBI
|
|
27
|
Ricketts CJ and Linehan WM: Gender
specific mutation incidence and survival associations in clear cell
renal cell carcinoma (CCRCC). PLoS One. 10:e01402572015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen EG, Zhang JS, Xu S, Zhu XJ and Hu HH:
Long non-coding RNA DGCR5 is involved in the regulation of
proliferation, migration and invasion of lung cancer by targeting
miR-1180. Am J Cancer Res. 7:1463–1475. 2017.PubMed/NCBI
|
|
29
|
Dong HX, Wang R, Zeng J and Pan J: LncRNA
DGCR5 promotes lung adenocarcinoma (LUAD) progression via
inhibiting hsa-mir-22-3p. J Cell Physiol. 233:4126–4136. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhao X, Li D, Pu J, Mei H, Yang D, Xiang
X, Qu H, Huang K, Zheng L and Tong Q: CTCF cooperates with
noncoding RNA MYCNOS to promote neuroblastoma progression through
facilitating MYCN expression. Oncogene. 35:3565–3576. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Malakootian M, Mirzadeh-Azad F, Naeli P,
Fouani Y and Mowla SJ: A long non-coding RNA, PSORS1C3, located
upstream of the human Oct4 gene is expressed in pluripotent and
tumor cell lines. Modares J Med Sci Pathobiol. 17:105–117.
2014.
|
|
32
|
Huang Y, Murakami T, Sano F, Kondo K,
Nakaigawa N, Kishida T, Kubota Y, Nagashima Y and Yao M: Expression
of aquaporin 1 in primary renal tumors: A prognostic indicator for
clear-cell renal cell carcinoma. Eur Urol. 56:690–698. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ma J, Zhou C, Yang J, Ding X, Zhu Y and
Chen X: Expression of AQP6 and AQP8 in epithelial ovarian tumor. J
Mol Histol. 47:129–134. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu F, Wu L, Wang A, Xu Y, Luo X, Liu X,
Hua Y, Zhang D, Wu S, Lin T, et al: MicroRNA-138 attenuates
epithelial-to-mesenchymal transition by targeting SOX4 in clear
cell renal cell carcinoma. Am J Transl Res. 9:3611–3622.
2017.PubMed/NCBI
|
|
35
|
Xiao H, Xiao W, Cao J, Li H, Guan W, Guo
X, Chen K, Zheng T, Ye Z, Wang J and Xu H: miR-206 functions as a
novel cell cycle regulator and tumor suppressor in clear-cell renal
cell carcinoma. Cancer Lett. 374:107–116. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Fan Y, Ma X, Li H, Gao Y, Huang Q, Zhang
Y, Bao X, Du Q, Luo G, Liu K, et al: miR-122 promotes metastasis of
clear-cell renal cell carcinoma by downregulating Dicer. Int J
Cancer. 142:547–560. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Huang J, Kong W, Zhang J, Chen Y, Xue W,
Liu D and Huang Y: c-Myc modulates glucose metabolism via
regulation of miR-184/PKM2 pathway in clear-cell renal cell
carcinoma. Int J Oncol. 49:1569–1575. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Mitsui Y, Chang I, Fukuhara S, Hiraki M,
Arichi N, Yasumoto H, Hirata H, Yamamura S, Shahryari V, Deng G, et
al: CYP1B1 promotes tumorigenesis via altered expression of CDC20
and DAPK1 genes in renal cell carcinoma. BMC Cancer. 15:9422015.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zaravinos A, Pieri M, Mourmouras N,
Anastasiadou N, Zouvani I, Delakas D and Deltas C: Altered
metabolic pathways in clear cell renal cell carcinoma: A
meta-analysis and validation study focused on the deregulated genes
and their associated networks. Oncoscience. 1:117–131. 2014.
View Article : Google Scholar : PubMed/NCBI
|