Introduction
Non-Hodgkin's lymphoma (NHL), a heterogeneous group
of lymphoid-derived malignancies, is the seventh most prevalent
type of cancer. According to the incidence trend, ~74,680 newly
diagnosed cases and 19,910 cases of mortality were estimated to
occur in 2016 in the USA (1).
According to its origination, NHL can be divided into three types:
B-cell origin, T-cell origin, and natural killer (NK)-cell origin
(2). Among cases, ~80-90% are
B-cell origin, namely B-NHL (3).
Specifically, diffuse B-cell lymphoma (DLBCL), mantle cell lymphoma
(MCL), follicular lymphoma (FL), chronic lymphocytic leukemia/small
lymphocytic lymphoma (CLL/SLL), and Burkitt lymphoma (BL) are the
most prevalent types of B-NHL (2).
Although there have been multiple trials in the treatment of B-NHL,
due to the diverse clinical and pathological performances, the
diagnosis and prognosis of B-NHL and its underlying mechanism
remain to be fully elucidated (4).
MicroRNAs (miRNAs) are a type of short, non-coding
RNA molecule (18–22 nt) which are mainly known to be involved in
the regulation of gene expression by binding to the 3′-untranslated
regions (3′-UTR) of target mRNAs (4). In previous years, miRNAs have been
found to be important in several types of cancer, including stomach
(5), colon (6) and ovarian cancer (7), in addition to B-NHL (8). Previous studies have reported that
aberrant high levels of miRNA (miR)-221, miR-21 and miR-155 are
significant indicators of a poor prognosis of DLBCL (9–11).
Akao et al (12) documented
that the expression levels of miR-143 and miR-145 were
downregulated in B-cell malignancies, and that increasing the
expression of both induced dose-dependent growth-inhibition in BL
cell lines. miR-150 and miR-16 have also been observed to have
elevated expression in CLL (13,14).
Taken together, this evidence indicates that miRNAs may be closely
correlated with the development of B-NHL.
The miR-17-92 gene cluster is a commonly gene
amplified loci in the non-protein-coding gene C13orf25 at 13q31.3
in several types of solid tumor and lymphoproliferative disorders
(15,16). In this region, six tandem loop
hairpin structures are contained and six mature miRNAs, including
miR-17, miR-18a, miR-19a, miR-19b, miR-20a and miR-92a, are
ultimately produced and involved in the regulation of cellular
processes (15). The miR-17-92
cluster is also crucial in the development of B-NHL. Tagawa et
al (17) documented that c-Myc
can not only promote the transcription of miR-17-92 cluster, but
can also act as a target of the miR-17-92 cluster. A high-level of
miR-17-92 cluster expression also results in the poor survival rate
of patients with MCL (18). With
the exception of MCL, the miR-17-92 gene cluster is also
upregulated in DLBCL and ALL, and correlated with a poorer
prognosis (19–21). Together, the above evidence
indicates that miR-17-92 may be crucial in B-NHL, however, the
detailed expression of mature miRNA in different types of B-NHL
remains to be elucidated. Therefore, the present study aimed to
examine the specific expression of the miR-17-92 gene cluster in
different types of B-NHL, in order to provide novel insights into
the treatment and prognostic prediction of B-NHL.
Materials and methods
Patients and enrolment criteria
Between January 2012 and October 2014, 71 patients,
who were first diagnosed with B-NHL in the Department of
Hematology, Harbin Medical University Cancer Hospital (Harbin,
China), were enrolled in the present study. The basic information
of patients, including gender, age, B symptoms, clinical stage,
international prognostic index (IPI) score and the level of lactic
dehydrogenase, were also collected. Patients were enrolled if they
met the following inclusive criteria: i) Patients were preliminary
diagnosed with B-NHL; ii) the diagnostic result was confirmed by
two pathologists; iii) the content of tumor cells was >80%. In
addition, patients were excluded if they had other types of tumor,
if the diagnostic results had no definite pathological
significance, or if certain types of tumor were not confirmed.
Additionally, five patients with reactive hyperplasia lymph nodes
were enrolled as controls. The follow-up was ended by recurrence or
the occurrence of patient mortality and the final follow-up time
was June 30th, 2016. The overall survival (OS) was defined as the
time from diagnosis to patient mortality from any cause, and
event-free survival (EFS) was designed as the time from diagnosis
to disease progression or mortality from any cause, whichever
occurred first. All patients were informed and provided signed
consent, and the clinical investigation was authorized by the
Ethics Committee of Harbin Medical University Cancer Hospital.
Cells and culture
Primary mouse lymphoma cells of wild-type (WT),
which had normal expression of the miR-17-92 gene cluster; knockout
(KO), which had deficient expression of the miR-17-92 gene cluster;
and TG, which had 3–4 times higher expression of the miR-17-92 gene
cluster than normal, were provided by the Ron Levy Laboratory of
Stanford University (Stanford, CA, USA) (22). All cell lines were maintained in
RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham,
MA, USA) containing 10% fetal bovine serum (Gibco; Thermo Fisher
Scientific, Inc.) at 37°C in a humidified incubator with a 5%
CO2 atmosphere.
RNA extraction
Following collection of the tumor and inflammatory
tissues, signal cell suspension was produced. According to the
manufacturer's protocol, total RNA of the cells was extracted using
an RNA extraction kit (Omega Bio-Tek, Inc., Norcross, GA, USA). In
addition, the concentration and purity of the RNA were measured
using a NanoDrop Nd-1000 spectrophotometer (Thermo Fisher
Scientific, Inc.).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Based on the concentration of RNA, the RNA in each
sample was adjusted to the same concentration and a reverse
transcription kit (TaqMan® MicroRNA assay; Applied
Biosystems; Thermo Fisher Scientific, Inc.) was utilized to
synthesize cDNA according to the manufacturer's protocol. The RT
primers were designed as follows: miR-17,
5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACCTACCTG-3′;
miR-18a,
5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACCTATCTG-3′;
miR-20a,
5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACCTACCTG-3′; and
U6, 5′-CGCTTCACGAATTTGCGTATCAT-3′. Subsequently, the synthesized
cDNA was used as template to detect the level of the miR-17-92 gene
cluster in the B-NHL and control groups via RT-qPCR analysis
(Quantifier Human DNA Quantification kit, Thermo Fisher Scientific,
Inc.) using the following conditions: 95°C for 10 min, and 40
cycles of 95°C for 10 min, 95°C for 15 sec, and 60°C for 31 sec.
The total volume of the reaction system was 20 µl, including 1.33
µl cDNA, 10 µl 2X TaqMan® GeneExpression Master Mix, 1
µl 20X Real time Primer and 7.67 µl dH2O. U6 was set as
internal control of this procedure. The primers were synthesized as
follows: miR-17, sense 5′-ATCCAGTGCGTGTCGTG-3′ and antisense
5′-TGCTTAAAGTGCTTACAGTG-3′; miR-18a, sense 5′-ATCCAGTGCGTGTCGTG-3′
and antisense 5′-TGCTTAAGGTGCATCTAGTG-3′; miR20, sense
5′-ATCCAGTGCGTGTCGTG-3′ and antisense 5′-TGCTTAAAGTGCTTAATAGTG-3′;
miR-17-92, sense 5′-CTGTCGCCCAATCAAACTG-3′ and antisense
5′-GTCACAATCCCCACCAAAC-3′; and U6, sense
5′-GCTTCGGCAGCACATATACTAAAAT-3′ and antisense
5′-CGCTTCACGAATTTGCGTATCAT-3′ (Beijing Aoke Biotechnology, Beijing,
China). The relative expression of the miRNAs was calculated by the
2−ΔΔCq method (23).
Tumor formation in nude mice
When sufficient numbers of the four types of cells,
including WT, KO and TG lymphoma cells obtained from mice and
reactive hyperplasia lymph cells obtained from mouse lymph nodes,
were obtained. A total of 18 female Balb/c nude mice (3–4 weeks
old; weighing 16–20 g) in SPC conditions purchased from Beijing
Vital River Laboratory Animal Technology Co., Ltd were used to
perform a tumor formation assay. The mice were housed at 22±2°C
with a 12-h light/dark cycle and had free access to regular diet
and purified water. Saline was used to adjust the cell
concentration. With the same concentration, 4×105 cells
of each type were injected into the subcutaneous tissue of the
shoulders. In the same mouse, the left shoulder was injected with
cells and the right shoulder was injected with the same volume of
saline. There were six mice for each cell type. Following
inoculation the lengths and diameters of the tumors were measured
persistently, and the volumes of the tumors were calculated using
the following formula: Volume=(π/6) × (length × diameter ×
diameter). After 6 weeks, the mice were sacrificed by cervical
dislocation and the weights of the tumors were estimated.
Statistical analysis
In the present study, SPSS 17.0 statistical software
(SPSS, Inc., Chicago, IL, USA) was utilized to conduct statistical
analyses. The correlations between the expression of the
miRNA-17-92 gene cluster and different clinical factors were
estimated using χ2 test. Continuous comparisons among
groups were estimated by two-way analysis of variance followed
Fisher's LSD test. Survival analyses of the mice were evaluated
using the Kaplan-Meier method, and comparisons between two groups
were assessed using the log-rank test. P<0.05 was considered to
indicate a statistically significant difference.
Results
Clinical information and expression of
the miR-17-92 gene cluster in patients
A total of 71 cases of B-NHL were confirmed with
assured pathological significance, and were enrolled in the present
study, including 48 cases of DLBCL (ABC, 33 cases; GBC, 15 cases),
five cases of MCL, two cases of BL, eight cases of CLL/SLL, and
eight cases of FL. The detailed clinical information of patients is
listed in Table I. Based on the
χ2 test, no significant correlations were identified
between the expression of the miR-17-92 gene cluster and gender,
age, lactate dehydrogenase (LDH), presence of B symptoms, clinical
stage or IPI score (Table
II).
 | Table I.Clinical information of the 71
enrolled patients with B-NHL. |
Table I.
Clinical information of the 71
enrolled patients with B-NHL.
|
| DLBCL |
|
|
|
|
|---|
|
|
|
|
|
|
|
|---|
| Factor | ABC | GCB | MCL | BL | CLL/SLL | FL |
|---|
| Gender |
|
|
|
|
|
|
| Male | 20 | 10 | 3 | 1 | 6 | 5 |
|
Female | 13 | 5 | 2 | 1 | 2 | 3 |
| Age (years) |
|
|
|
|
|
|
|
≥60 | 22 | 9 | 4 | 1 | 5 | 4 |
|
<60 | 11 | 6 | 1 | 1 | 3 | 4 |
| LDH |
|
|
|
|
|
|
|
Increased | 28 | 8 | 4 | 1 | 2 | 1 |
|
Normal | 5 | 7 | 1 | 1 | 6 | 7 |
| B symptoms |
|
|
|
|
|
|
|
Yes | 26 | 10 | 4 | 2 | 4 | 1 |
| No | 7 | 5 | 1 | 0 | 4 | 7 |
| Clinical stage |
|
|
|
|
|
|
|
I+II | 12 | 9 | 3 | 0 | 4 | 2 |
|
III+IV | 21 | 6 | 2 | 2 | 4 | 6 |
| IPI score |
|
|
|
|
|
|
|
High-risk + median
high-risk | 19 | 8 |
| 2 | 5 | 2 |
|
Low-risk + median
low-risk | 14 | 7 | 2 | 0 | 3 | 6 |
 | Table II.Correlations between microRNA-17-92
gene cluster and clinical information. |
Table II.
Correlations between microRNA-17-92
gene cluster and clinical information.
| Factor | Total | Downregulated | Upregulated | χ2 | P-value |
|---|
| Gender |
|
|
|
|
|
|
Male | 49 | 20 | 29 | 0.134 | 0.714 |
|
Female | 22 | 10 | 12 |
|
|
| Age (years) |
|
|
|
|
|
|
≥60 | 48 | 22 | 26 | 0.250 | 0.617 |
|
<60 | 32 | 12 | 11 |
|
|
| LDH |
|
|
|
|
|
|
Increased | 46 | 25 | 21 | 1.334 | 0.248 |
|
Normal | 25 | 10 | 15 |
|
|
| B symptoms |
|
|
|
|
|
|
Yes | 48 | 20 | 28 | 0.021 | 0.885 |
| No | 23 | 10 | 13 |
|
|
| Clinical stage |
|
|
|
|
|
|
I+II | 32 | 10 | 22 | 2.891 | 0.089 |
|
III+IV | 39 | 20 | 19 |
|
|
| IPI score |
|
|
|
|
|
|
High-risk + median
high-risk | 41 | 20 | 21 | 0.031 | 0.860 |
|
Low-risk + median
low-risk | 30 | 14 | 16 |
|
|
To obtain further insight into the development of
lymphoma, patients with reactive lymphoid hyperplasia were
collected as a control and six mature miRNAs (miR-17, miR-18a,
miR-19a, miR-19b, miR-20a and miR-92a) in the miR-17-92 cluster of
the control and lymphoma patients were evaluated with U6 as the
internal control. Compared with the control group, 19/71 (26.76%)
patients with lymphoma were identified with overexpression of the
miR-17-92 gene cluster (Fig. 1).
Following detailed analysis, 25% of the DLBCL cases had significant
upregulation of the miR-17-92 gene cluster, including miR-17,
miR-18, miR-19b and miR-92, compared with the control (P<0.05).
Significant upregulation of the miR-17-92 gene cluster was also
identified in two cases of BL and two cases of MCL, and this
upregulation was observed for all six members of the gene cluster
compared with the control (P<0.05). In addition, the
overexpression of miR-17-92 was observed in two cases of FL, and
marked increases in the levels of miR-18 and miR-20a were observed
(P<0.05). Only one case of CLL/SLL showed enhanced expression of
the miR-17-92 cluster, with specific upregulation identified in
miR-19a, miR-19b and miR-92, compared with the control
(P<0.05).
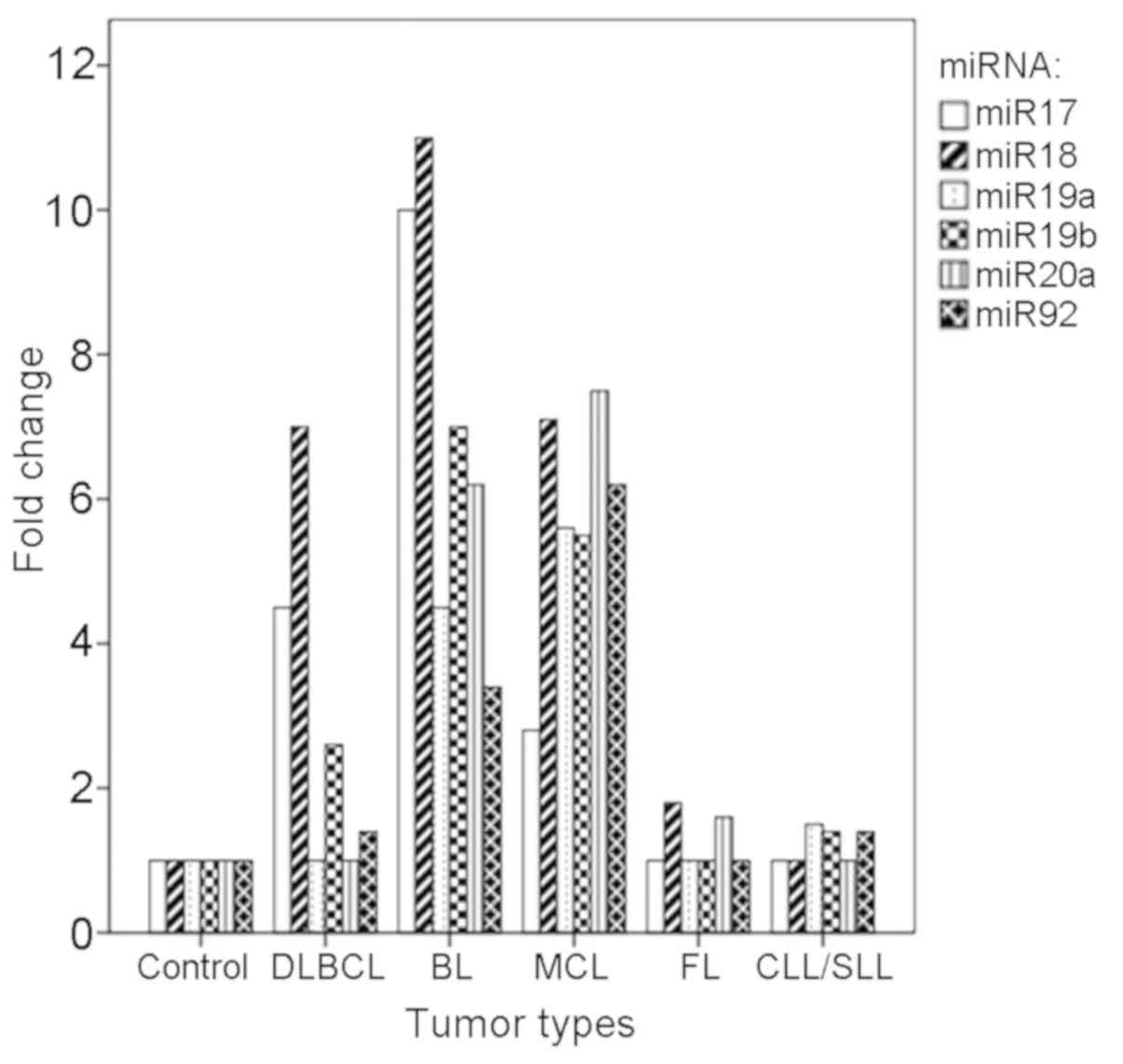 | Figure 1.Expression levels of miR-17, miR-18,
miR19a, miR-19b, miR-20a and miR-92 in different subtypes of B-NHL.
miR, microRNA; DLBCL, diffuse large B cell lymphoma; ABC, activated
B-cell like; GBC, germinal center B-cell like; MCL, mantle cell
lymphoma; BL, Burkitt lymphoma; CLL/SLL, chronic lymphocytic
leukemia/small lymphocytic lymphoma; FL, follicular lymphoma;
B-NHL, B-cell non-Hodgkin's lymphoma. |
Expression of the miR-17-92 gene
cluster in different types of B-NHL
Based on the χ2 test, comparisons of the
miR-17-92 gene cluster between different types of B-NHL were made
(Table III). Compared with ABC,
significantly lower expression levels of miR-17-92 gene cluster
members, with the exception of miR-18, were identified in patients
with GCB (P<0.05), however, no significant difference was
detected in patients with FL (P>0.05). Compared with GCB,
significantly higher expression was observed in all members of the
miR-17-92 gene cluster in patients with FL (P<0.05), however,
these differences disappeared when specifically compared with stage
III FL, with the exception of miR-20a (P>0.05). Overexpressed
levels of miR-17, miR-18b, miR-19b, miR-20a and miR-92a were also
identified in 80% of patients with DLBCL compared with
non-transformed FL, although differences were only significant for
the expression of miR-17 and miR-20a (P<0.05). Patients with
DLBCL derived from non-transformed FL also had higher expression
levels of miR-18, miR-19b and miR-92a, compared with patients with
DLBCL derived from transformed FL (P<0.05). No significant
difference was identified between MCL and CLL/SLL (P>0.05).
 | Table III.Comparisons of the miR-17-92 gene
cluster among different types of B-NHL. |
Table III.
Comparisons of the miR-17-92 gene
cluster among different types of B-NHL.
| Type | miR-17 (FC) | miR-18 (FC) | miR-19a (FC) | miR-19b (FC) | miR-20a (FC) | miR-92 (FC) | Increase (%) |
|---|
| ABC vs. GCB | S | NS | S | S | S | S | 80 |
| FL vs. ABC | NS |
| NS | NS | NS | NS | 70 |
| FL vs. GCB | S | S | S | S | S | S | 100 |
| GCB vs. FL III |
| NS |
| NS | S | NS | 100 |
| DLBCL vs.
nt-FL | S | NS |
| NS | S | NS | 80 |
| DLBCL-nt vs.
DLBCL-t |
| S |
| S |
| S | 85 |
| MCL vs.
CLL/SLL |
|
|
|
| NS |
| 90 |
Influence of overexpression of the
miR-17-92 gene cluster on the prognosis of patients with B-NHL
Influence of overexpression of the
miR-17-92 gene cluster on OS
Compared with the average expression levels of
miRNAs in the reactive hyperplasia lymph node group, changes in the
expression of miRNAs in B-NHL were determined, and the OS of
patients was analyzed. According to the analytical results, the OS
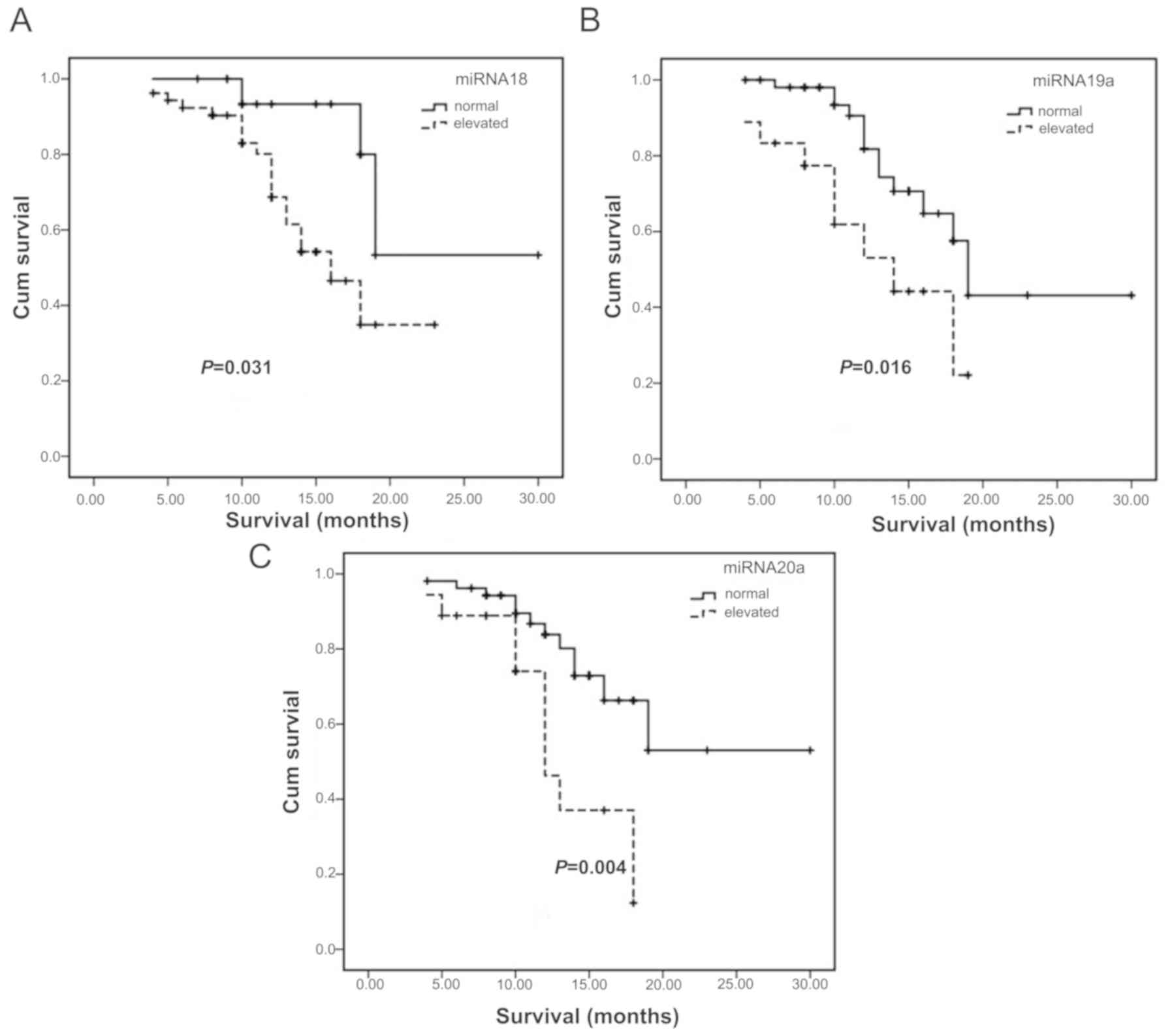
of patients with overexpressed miR-18 (Fig. 2A), miR-19a (Fig. 2B) and miR-20a (Fig. 2C) were shortened compared with that
of patients with normal expression levels (Table IV).
 | Table IV.Median OS of patients with differing
expression of miRNAs. |
Table IV.
Median OS of patients with differing
expression of miRNAs.
| miRNA | Variation | Patients (n) | OS (months) | P-value |
|---|
| miRNA-17 | Increase | 16 | 18 | 0.057 |
|
| Normal | 55 | 19 |
|
| miRNA-18 | Increase | 18 | 16 | 0.031 |
|
| Normal | 53 | 20 |
|
| miRNA-19a | Increase | 18 | 14 | 0.016 |
|
| Normal | 53 | 19 |
|
| miRNA-19b | Increase | 16 | 18 | 0.321 |
|
| Normal | 55 | 19 |
|
| miRNA-20a | Increase | 18 | 12 | 0.004 |
|
| Normal | 53 | 25 |
|
| miRNA-92a | Increase | 16 | 18 | 0.087 |
|
| Normal | 55 | 19 |
|
Influence of overexpression of the
miR-17-92 gene cluster on EFS
Similar to the analysis of OS, the influence of
overexpression of the miR-17-92 gene cluster on ESF was
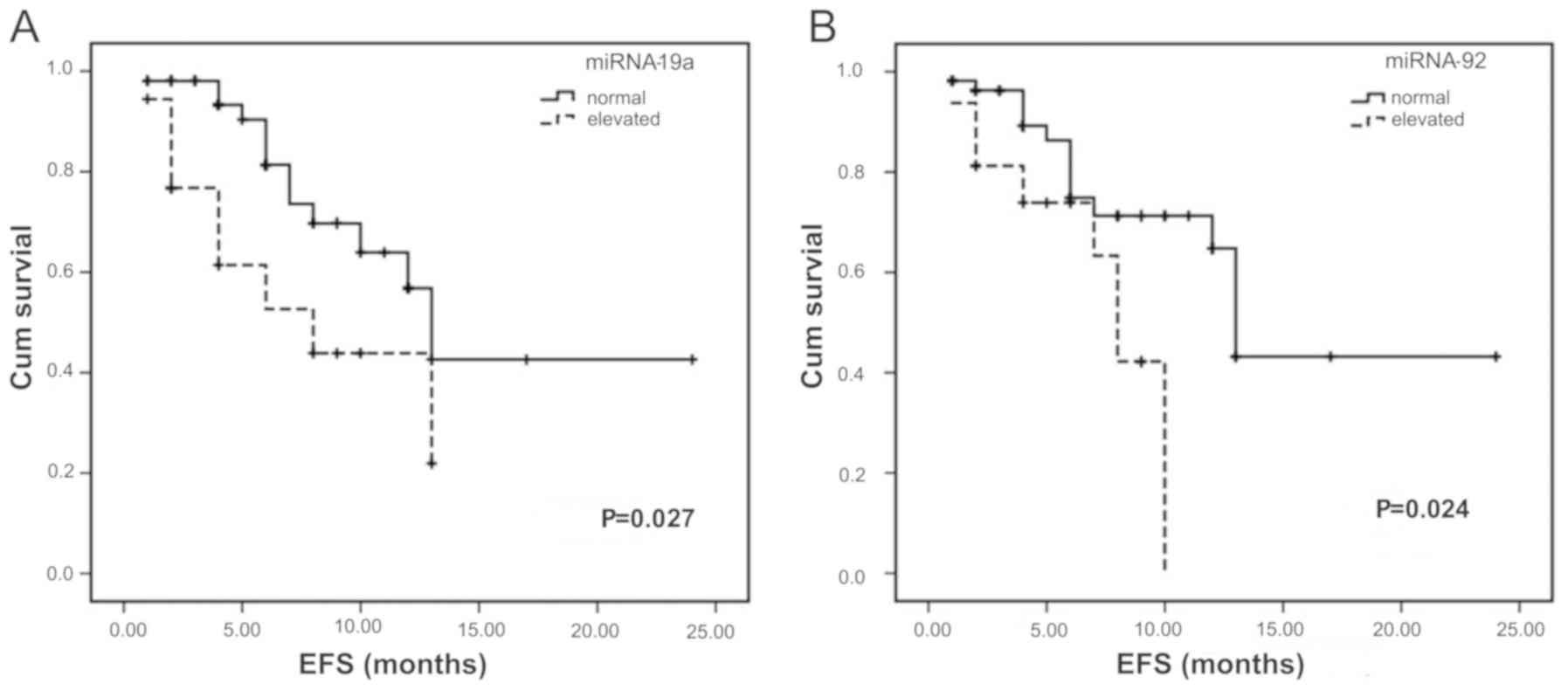
investigated. Following analysis, marked reductions EFS were
identified in patients with overexpressed miR-19a and miR-92a
(Fig. 3A and B) compared with the
patients with normal levels (Table
V).
 | Table V.Median EFS of patients with differing
expression of miRNAs. |
Table V.
Median EFS of patients with differing
expression of miRNAs.
| miRNA | Variation | Patients (n) | EFS (months) | P-value |
|---|
| miRNA-17 | Increase | 16 | 10 | 0.052 |
|
| Normal | 55 | 13 |
|
| miRNA-18 | Increase | 18 | 12 | 0.058 |
|
| Normal | 53 | 13 |
|
| miRNA-19a | Increase | 18 | 8 | 0.027 |
|
| Normal | 53 | 13 |
|
| miRNA-19b | Increase | 16 | 13 | 0.407 |
|
| Normal | 55 | 13 |
|
| miRNA-20a | Increase | 18 | 10 | 0.106 |
|
| Normal | 53 | 13 |
|
| miRNA-92a | Increase | 16 | 8 | 0.024 |
|
| Normal | 55 | 13 |
|
Tumor formation assay in nude
mice
According to the results described above, the
miR-17-92 gene cluster may have a potential effect in promoting the
development of B-NHL. In order to evaluate the biofunctions of the
miR-17-92 gene cluster in vivo, a tumor formation assay in
nude mice was performed. A total of 24 samples in four groups (WT,
KO, TG and reactive hyperplasia lymph node) were used for this
assay, and all animals in the WT and TG groups exhibited tumor
formation as a result, with the exception of two cases in the TG
group, in which the mice died 10 days following injection (Fig. 4). However, no tumor nodes were
produced in the KO or reactive hyperplasia lymph node groups
following injection with the same concentration of KO and reactive
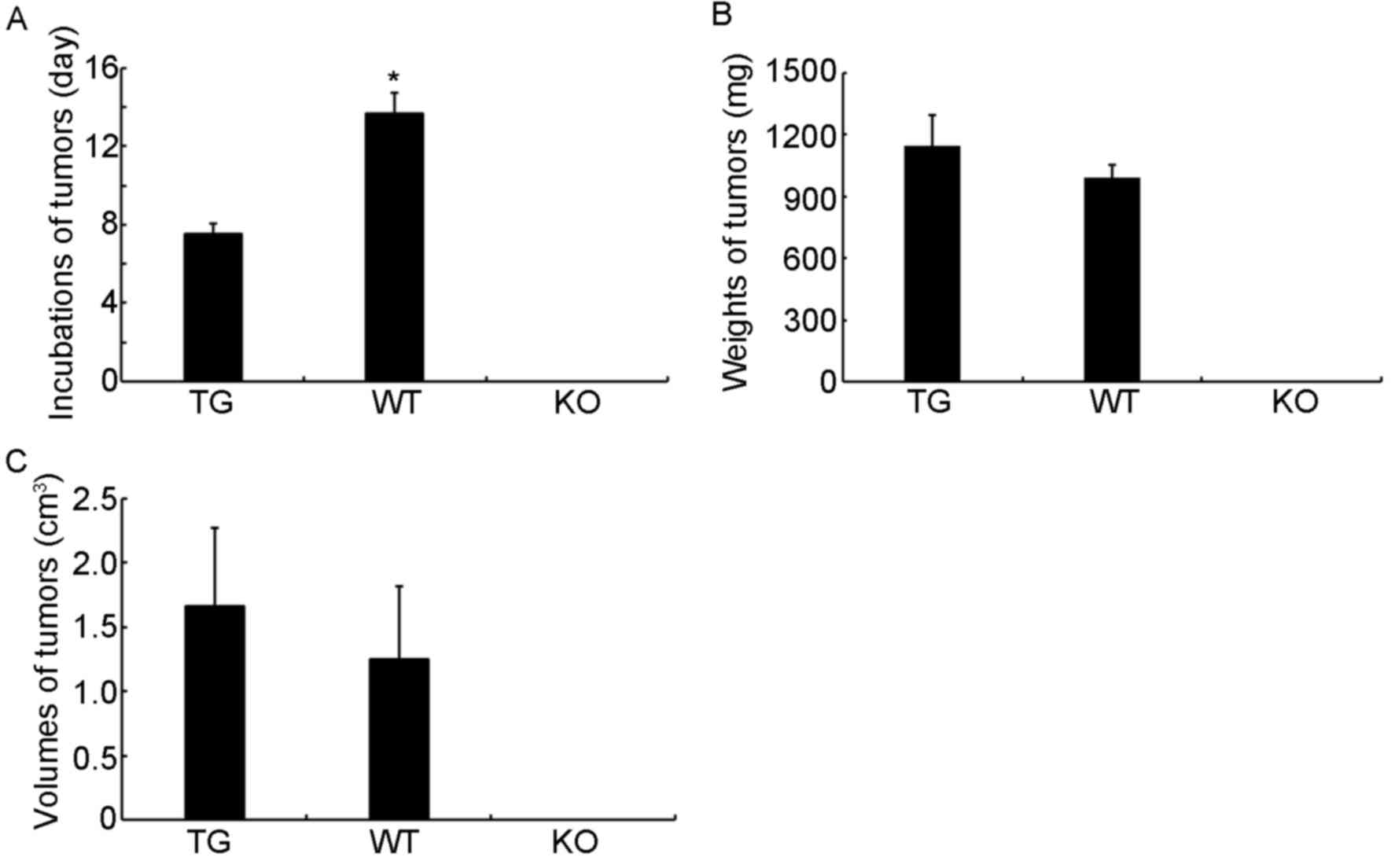
hyperplasia lymph node cells. In terms of the formation of tumors,
the duration of incubation required until tumor visualization was
significantly shorter in the TG group than in the WT group
(P<0.05; Fig. 5A). The tumor
weights and volumes in the TG group were also higher than those in
the WT group, although no significant difference was identified
(P>0.05; Fig. 5B and C). These
results indicate that the miR-17-92 gene cluster may have a
potential carcinogenic characteristic, and this feature may
correlate with the occurrence and development of lymphoma.
Discussion
In the present study, the six mature miRNAs in the
miR-17-92 gene cluster were detected in several types of B-NHL. The
results showed that ~27% of B-NHL cases presented with a high level
of the miR-17-92 gene cluster, including DLBCL, BL, MCL, FL and
CLL/SLL, although the levels of the six mature miRNAs derived from
miR-17-92, (miR-17, miR-18, miR-19a, miR-19b, miR-20a and miR-92)
differed between these types. The investigation of OS and EFS
demonstrated that overexpressed miRNA18, miR-19a and miR-20a have a
positive effect on the OS of patients with B-NHL, and elevated
expression levels of miR-19a and miR-92a led to a poor EFS for
patients with B-NHL. In addition, the tumor formation assay
indicated that overexpression of the miR-17-92 gene cluster led to
acceleration in the occurrence of tumors in nude mice, however, the
tumor weights did not differ.
miR-17-92 is encoded by C13orf25, which is a unique
non-coding RNA in the aberrant amplification of 13q31-32 in B-NHL.
Therefore, it is useful to investigate the mechanism of B-NHL via
examining the associated biofunction of the miR-17-92 gene cluster
(15). Among previously published
literature, c-Myc is the most referred to molecule, which is key in
cell apoptosis (15). A mouse
model indicates that c-Myc can activate the expression of the
miR-17-92 gene cluster, and can also be regulated by the miR-17-92
gene cluster. This indicates that there may be negative regulation
between c-Myc and the miR-17-92 gene cluster, and loss of control
of this regulation is important for the development of B-NHL
(17). miR-17-92 also can inhibit
the expression levels of phosphatase and tensin homolog (PTEN),
P21, and Bcl-2-like 11 (Bim) to promote proliferation and suppress
the apoptosis of cancer cells (24). The xenograft tumor assay confirmed
that the incubation duration of mice with the overexpressed
miR-17-92 gene cluster was shorter than that with normal levels,
however, the deletion of miR-17-92 resulted in failure of tumor
formation. This indicates that miR-17-92 may serve as an oncogene
in the occurrence and development of B-NHL.
DLBCL is the most common type of B-NHL and accounts
for ~33% (15). A previous study
showed that >18% of patients with DLBCL had 2–36-fold higher
expression of the miR-17-92 gene cluster (25). However, due to the high
heterogeneity of DLBCL, the levels of members of the miR-17-92
cluster are not consistent in ABC and GCB. Robertus et al
(26) confirmed that the
expression of miR-19b is only elevated in ABC. In the present
study, >27% of DLBCL cases had elevated expression of the
miR-17-92 gene cluster. With the exception of miR-18, the levels of
the remaining miRNAs in the miR-17-92 gene cluster were
significantly higher than those in the GCB subtype. Significant
differences were also identified between transformed and
untransformed DLBCL in the expression levels of miR-18, miR-19b and
miR-92. Controversially, Lenz et al (27) reported that 13q31 amplification
frequently occurs in GCB but not ABC, and that the overexpression
of miR-17-92 is always correlated with high levels of MYC and its
target genes. The above evidence indicates that expression of the
miR-17-92 gene cluster is not coincident in different DLBCL
subtypes, however, the particular expression levels in these
subtypes require further elucidation.
FL is the most frequent indolent tumor type of
B-NHL, and 90% of this type can transform into DLBCL 31. Zhao et
al (28) reported that the
increased expression of miR-17 can act as an important marker in
the invasive transformation and prognosis of FL. Studies by Lawrie
et al (29) and Fassina
et al (30) also suggested
that miR-17-92 profiling can be utilized to distinguish
morphologically similar disease between grade III of FL, and de
novo and transformed DLBCL. In the present study, a marked
difference in expression of the miR-17-92 gene cluster was also
observed between FL and GCB, but not ABC. This suggests that
miR-17-92 may be a useful biomarker in the distinction of FL and
GCB. In addition, the distinction of non-transformed FL and DLBCL
was only detected in miR-17 and miR-20a, and the difference between
grade III FL and GCB was only identified in miR-20a. Taken
together, these findings suggest that the expression of miR-17-92
may serve as a biomarker to distinguish FL and GCB, although the
differences in the different states of FL and the subtype of DLBCL
remain to be fully elucidated, of which further investigation is
required.
The miR-17-92 gene cluster is not only involved in
the development of B-NHL, but is also affects the prognosis of
patients with B-NHL. In the present study, the estimations of OS
and EFS showed that the OS rates were significantly lower in
patients with overexpressed miR-18, miR-19a and miR-20a, and that
the EFS rates were markedly decreased in patients with high levels
of miR-19a and miR-92a. This indicates that miR-19a may be
important in the OS and EFS rates of patients with B-NHL. It is
documented that miR-19a and miR-19b can downregulate suppressor of
cytokine signaling 1, a negative regulator of the interleukin 6
pathway, inducing constitutive activation of the Janus
kinase/signal transducer and activator of transcription 3 (STAT3)
signaling pathway and contributing to myelomagenesis (31). The pharmacological inhibition of
STAT3 induces a dose-dependent reduction of the miR-17-92 cluster
(32). This may be a potential
mechanism for miR-19a in B-NHL and may serve as a biomarker in the
prognosis of B-NHL. miR-19 can also negatively regulate the
phosphatidylinositol-3-kinase pathway via silencing the genes PTEN,
protein phosphatase 2, Bim and protein kinase AMP-activated α1 in
acute leukemia (33). As a
lymphoid malignancy, this mechanism may also be involved in B-NHL,
although confirmation is required.
In conclusion, it was made apparent that the
miR-17-92 gene cluster is important in the development and
prognosis of B-NHL. Among the subtypes of B-NHL, the expression
levels of the six members of miR-17-92 varied, and the distinction
between FL and GCB was most evident. Based on this evidence, it can
be inferred that miR-19a may be important in the prognosis of
B-NHL, although the detailed mechanism requires further
elucidation. However, compared with existing evidence,
controversies remain, and more detailed investigation based on a
large sample is required.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contributions
SY and CJ were responsible for the conception and
design of the research, and drafting the manuscript. LQ performed
the data acquisition. LZ performed the data analysis and
interpretation. YT and AL participated in the design of the study
and performed the statistical analysis. All authors have read and
approved the manuscript.
Ethics approval and consent to
participate
All patients were informed and provided signed
consent, and the clinical investigation was authorized by the
Ethics Committee of Harbin Medical University Cancer Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clinicians. 68:7–30. 2018. View Article : Google Scholar
|
|
2
|
Song W, Liu MG, Zhang JB, Zhang JJ, Sun MM
and Yu QK: Mechanism of action of EBV, Bcl-2, p53, c-Myc and Rb in
non-Hodgkin's lymphoma. Eur Rev Med Pharmacol Sci. 20:1093–1097.
2016.PubMed/NCBI
|
|
3
|
Datta S, Chatterjee S, Policegoudra RS,
Gogoi HK and Singh L: Hepatitis viruses and non-Hodgkin's lymphoma:
A review. World J Virol. 1:162–173. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sun CM and Luan CF: Overexpression of
microRNA-21 in peripheral blood mononuclear cells of patients with
B-cell non-Hodgkin's lymphoma is associated with disease stage and
treatment outcome. Eur Rev Med Pharmacol Sci. 19:3397–3402.
2015.PubMed/NCBI
|
|
5
|
Zhu C, Ren C, Han J, Ding Y, Du J, Dai N,
Dai J, Ma H, Hu Z, Shen H, et al: A five-microRNA panel in plasma
was identified as potential biomarker for early detection of
gastric cancer. Br J Cancer. 110:2291. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xu L, Zhang Y, Wang H, Zhang G, Ding Y and
Zhao L: Tumor suppressor miR-1 restrains epithelial-mesenchymal
transition and metastasis of colorectal carcinoma via the MAPK and
PI3K/AKT pathway. J Transl Med. 12:2442014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lu L, Katsaros D, Risch HA, Canuto EM,
Biglia N and Yu H: MicroRNA let-7a modifies the effect of
self-renewal gene HIWI on patient survival of epithelial ovarian
cancer. Mol Carcinog. 55:357–365. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bruni R, Marcantonio C, Pulsoni A, Tataseo
P, De Angelis F, Spada E, Marcucci F, Panfilio S, Bianco P,
Riminucci M, et al: microRNA levels in paraffin-embedded indolent
B-cell non-Hodgkin lymphoma tissues from patients chronically
infected with hepatitis B or C virus. BMC Infect Dis. 5:S62014.
View Article : Google Scholar
|
|
9
|
Medina PP, Nolde M and Slack FJ: OncomiR
addiction in an in vivo model of microRNA-21-induced pre-B-cell
lymphoma. Nature. 467:86–90. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lawrie CH, Soneji S, Marafioti T, Cooper
CD, Palazzo S, Paterson JC, Cattan H, Enver T, Mager R, Boultwood
J, et al: MicroRNA expression distinguishes between germinal center
B cell-like and activated B cell-like subtypes of diffuse large B
cell lymphoma. Int J Cancer. 121:1156–1161. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Eis PS, Tam W, Sun L, Chadburn A, Li Z,
Gomez MF, Lund E and Dahlberg JE: Accumulation of miR-155 and BIC
RNA in human B cell lymphomas. Proc Nati Acad Sci USA.
102:3627–3632. 2005. View Article : Google Scholar
|
|
12
|
Akao Y, Nakagawa Y, Kitade Y, Kinoshita T
and Naoe T: Downregulation of microRNAs-143 and −145 in B-cell
malignancies. Cancer Sci. 98:1914–1920. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ferrajoli A, Shanafelt TD, Ivan C, Shimizu
M, Rabe KG, Nouraee N, Ikuo M, Ghosh AK, Lerner S, Rassenti LZ, et
al: Prognostic value of miR-155 in individuals with monoclonal
B-cell lymphocytosis and patients with B chronic lymphocytic
leukemia. Blood. 122:1891–1899. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Döhner H, Stilgenbauer S, Benner A,
Leupolt E, Kröber A, Bullinger L, Döhner K, Bentz M and Lichter P:
Genomic aberrations and survival in chronic lymphocytic leukemia. N
Engl J Med. 343:1910–1916. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ota A, Tagawa H, Karnan S, Tsuzuki S,
Karpas A, Kira S, Yoshida Y and Seto M: Identification and
characterization of a Novel Gene, C13orf25, as a Target for
13q31-q32 amplification in malignant lymphoma. Cancer Res.
64:3087–3095. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mendell JT: miRiad roles for the miR-17-92
cluster in development and disease. Cell. 133:217–222. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tagawa H, Karube K, Tsuzuki S, Ohshima K
and Seto M: Synergistic action of the microRNA-17 polycistron and
Myc in aggressive cancer development. Cancer Sci. 98:1482–1490.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zheng RL, Jiang YJ and Wang X: Role of
microRNAs on therapy resistance in non-Hodgkin's lymphoma. Int J
Clin ExpMed. 7:3818–3832. 2014.
|
|
19
|
Zanette DL, Rivadavia F, Molfetta GA,
Barbuzano FG, Proto-Siqueira R, Silva WA Jr, Falcão RP and Zago MA:
miRNA expression profiles in chronic lymphocytic and acute
lymphocytic leukemia. Braz J Med Biol Res. 40:1435–1440. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
He L, Thomson JM, Hemann MT,
Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe
SW, Hannon GJ and Hammond SM: A microRNA polycistron as a potential
human oncogene. Nature. 435:828–833. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ventura A Young AG, Winslow MM, Lintault
L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone
JR, et al: Targeted deletion reveals essential and overlapping
functions of the miR-17~92 Family of miRNA clusters. Cell.
132:875–886. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jin HY, Oda H, Lai M, Skalsky RL, Bethel
K, Shepherd J, Kang SG, Liu WH, Sabouri-Ghomi M, Cullen BR, et al:
MicroRNA-17~92 plays a causative role in lymphomagenesis by
coordinating multiple oncogenic pathways. Embo J. 32:2377–2391.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔ C T method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
O'Donnell KA, Wentzel EA, Zeller KI, Dang
CV and Mendell JT: c-Myc-regulated microRNAs modulate E2F1
expression. Nature. 435:839–843. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jin HY, Lai M and Xiao C: MicroRNA-17~92
is a powerful cancer driver and a therapeutic target. Cell Cycle.
13:495–496. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Robertus JL, Harms G, Blokzijl T, Booman
M, de Jong D, van Imhoff G, Rosati S, Schuuring E, Kluin P and van
den Berg A: Specific expression of miR-17-5p and miR-127 in
testicular and central nervous system diffuse large B-cell
lymphoma. Mod Pathol. 22:547–555. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lenz G, Wright GW, Emre NC, Kohlhammer H,
Dave SS, Davis RE, Carty S, Lam LT, Shaffer AL, Xiao W, et al:
Molecular subtypes of diffuse large B-cell lymphoma arise by
distinct genetic pathways. Proc Nati Acad Sci USA. 105:13520–13525.
2008. View Article : Google Scholar
|
|
28
|
Zhao JJ, Lin J, Lwin T, Yang H, Guo J,
Kong W, Dessureault S, Moscinski LC, Rezania D, Dalton WS, et al:
microRNA expression profile and identification of miR-29 as a
prognostic marker and pathogenetic factor by targeting CDK6 in
mantle cell lymphoma. Blood. 115:2630–2639. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lawrie CH, Chi J, Taylor S, Tramonti D,
Ballabio E, Palazzo S, Saunders NJ, Pezzella F, Boultwood J,
Wainscoat JS and Hatton CS: Expression of microRNAs in diffuse
large B cell lymphoma is associated with immunophenotype, survival
and transformation from follicular lymphoma. J Cell Mol Med.
13:1248–1260. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fassina A, Marino F, Siri M, Zambello R,
Ventura L, Fassan M, Simonato F and Cappellesso R: The miR-17-92
microRNA cluster: A novel diagnostic tool in large B-cell
malignancies. Lab Invest. 92:1574–1582. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Benetatos L and Vartholomatos G:
Deregulated microRNAs in multiple myeloma. Cancer. 118:878–887.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Spaccarotella E, Pellegrino E, Ferracin M,
Ferreri C, Cuccuru G, Liu C, Iqbal J, Cantarella D, Taulli R,
Provero P, et al: STAT3-mediated activation of microRNA cluster
17~92 promotes proliferation and survival of ALK-positive
anaplastic large cell lymphoma. Haematologica. 99:116–124. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Schotte D, Pieters R and Boer MLD:
MicroRNAs in acute leukemia: From biological players to clinical
contributors. Leukemia. 26:1–12. 2012. View Article : Google Scholar : PubMed/NCBI
|