Introduction
Brain gliomas are the most common primary
intracranial tumors. Malignant gliomas are often characterized by
invasive growth, a high rate of relapse and rich vascularity
(1). Despite advances in treatment
modalities and the meaningful benefits in regard to survival (the
median survival in glioblastoma increased from 12.1 to 14.6 months
with surgical resection followed by radiotherapy and temozolomide
chemotherapy) (2), the overall
outcome of gliomas remains extremely unsatisfactory. According to
the World Health Organization (WHO) classification of central
nervous system tumors, brain gliomas are classified into grades
I–IV, which partially indicate the degree of malignancy and
prognosis (3). However, even
gliomas with the same grade or histological type do not have
identical prognosis. For example, primary and secondary
glioblastoma are both included in WHO IV and they cannot be
distinguished by histopathology; however, the incidence, onset age,
survival rate and gene expression profiles are quite different
(4). Thus, it is critical to
illuminate the mechanisms underlying the development and
progression of gliomas and detect sensitive and specific biomarkers
for diagnosis, grading and prognosis of the disease.
MicroRNAs (miRNAs) are emerging as an important
class of endogenous small (~22 nts) non-coding RNAs. They have the
ability to negatively regulate gene expression at the
post-transcriptional level by binding to the complementary sequence
in the 3′-untranslated region (UTR) of target mRNAs (5). Aberrant expression of miRNAs has been
found in a wide variety of human tumors, including brain gliomas.
Most miRNAs have been shown to act as either oncogenes or tumor
suppressors (6). Since the first
identification of dysregulated miRNA-21 in human gliomas (7), a great number of dysregulated miRNAs
have been discovered in gliomas, which were also found to be
involved in multiple crucial biological processes such as
proliferation, apoptosis, invasion, differentiation and
angiogenesis (8). Importantly, some
miRNAs are associated with the grade, prognosis and sensitivity to
chemotherapy of gliomas (9). In the
present study, we aimed to identify miRNAs involved in
tumorigenesis and malignant progression and to conduct preliminary
prediction of the potential use of miRNAs in grading and survival
expectation. An miRNA microarray approach was used to unravel
aberrant miRNA expression in gliomas compared with matched normal
tissues.
Materials and methods
Clinical samples
The present study included 26 cases of gliomas
including 2, 6, 10 and 8 samples in grade of WHO I–IV,
respectively, according to the 2007 WHO criteria (Table I). Among them, 9 cases were used for
microarray, which had matched adjacent non-cancerous tissues, and
each grade of WHO II, III and IV had 3 patients. The non-neoplastic
brain tissues located >2 cm from the tumor tissues were obtained
(10). All of the patients had no
history of radiotherapy or chemotherapy. The samples were collected
at the time of surgery at the Department of Neurosurgery, Xinqiao
Hospital, Third Military Medical University (TMMU), Chongqing,
China, and then immediately stored at −80°C until use. Written
informed consents were obtained from all of the patients or their
relatives. The present study was approved by the Ethics Committee
of Xinqiao Hospital, TMMU. Clinical information of the patients
with tumors and adjacent tissues for microarray assay is summarized
in Table II.
 | Table IGeneral information regarding the
tumor samples for qRT-PCR. |
Table I
General information regarding the
tumor samples for qRT-PCR.
| Patientno. | Gender | Age (years) | Grade | Pathology |
|---|
| 1 | F | 24 | WHO I | DNT |
| 2 | F | 17 | WHO I | PA |
| 3 | F | 36 | WHO II | AO |
| 4 | M | 5 | WHO II | A |
| 5 | M | 38 | WHO II | A |
| 6 | M | 43 | WHO II | A |
| 7 | M | 71 | WHO II | A |
| 8 | M | 43 | WHO II | PrA |
| 9 | F | 38 | WHO III | AO |
| 10 | M | 34 | WHO III | AO |
| 11 | M | 49 | WHO III | A |
| 12 | M | 66 | WHO III | AO |
| 13 | M | 34 | WHO III | AO |
| 14 | M | 30 | WHO III | A |
| 15 | F | 41 | WHO III | A |
| 16 | F | 37 | WHO III | A |
| 17 | M | 49 | WHO III | A |
| 18 | F | 42 | WHO III | A |
| 19 | M | 57 | WHO IV | GBM |
| 20 | M | 67 | WHO IV | GBM |
| 21 | M | 63 | WHO IV | GBM |
| 22 | M | 58 | WHO IV | GBM |
| 23 | M | 62 | WHO IV | GBM |
| 24 | F | 62 | WHO IV | GBM |
| 25 | M | 66 | WHO IV | GBM |
| 26 | M | 67 | WHO IV | GBM |
 | Table IIGeneral information regarding the
tumor samples for miRNA microarray analysis. |
Table II
General information regarding the
tumor samples for miRNA microarray analysis.
| Patient ID. | Gender | Age (years) | Grade | Pathology |
|---|
| 04050361 | M | 71 | WHO II | A |
| 04784847 | M | 38 | WHO II | A |
| 04719577 | M | 43 | WHO II | PrA |
| 04103496 | M | 49 | WHO III | A |
| 03938319 | M | 34 | WHO III | AO |
| 03954349 | F | 38 | WHO III | AO |
| 04051023 | M | 67 | WHO IV | GBM |
| 04653022 | M | 62 | WHO IV | GBM |
| 04714692 | F | 62 | WHO IV | GBM |
miRNA microarray assay
Total RNA was isolated from tissues using the TRIzol
method (Life Technologies, USA), according to the manufacturer’s
protocol. NanoDrop® ND-1000 spectrophotometer (Thermo
Fisher Scientific Inc., Wilmington, De, USA) was used to determine
the purity and quantity of total RNA. In order to assess RNA
integrity, Agilent RNA 6000 Nano assay (Agilent Technologies, Inc.,
Santa Clara, CA, USA) and agarose gel electrophoresis, which also
checks for genomic DNA contamination, were used. DNase digestion
was performed to eliminate DNA found on the gel. Small RNA
molecules (<200 nt) were separated from large RNA molecules by
NanoSep® 100K, based on the filtrate and labeled with
cy3/cy5 by the platinum complex of the ULS molecule, which binds to
the N7 position of RNA guanine tightly. The labeled RNA was
hybridized to the Human miRNA OneArray microarray (Phalanx Biotech
Group, Taiwan) containing 1,926 miRNA probes, and the array was
later scanned by Axon 4000B (Axon Instruments, Union City, CA, USA)
from Molecular Devices.
Microarray data analysis
The data analysis of gene expression profiling,
including data filtering, normalization and statistical
calculations, was processed by R (R-Foundation for Statistical
Computing, Vienna, Austria; version 2.12.1). Arrays (including
technical replicates) of any compared sample set were normalized
together after filtering probes. The expression level of each miRNA
was calculated by pair-wise combination and error weighted average.
The significant differential expression of miRNAs between 9 tumors
and 9 matched adjacent noncancerous tissues was selected according
to fold-change and a P-value with the following criteria:
fold-change ≥1.5 or ≤0.67 and P-value ≤0.05 (11).
Quantitative RT-PCR
Quantitative reverse transcription-PCR (qRT-PCR) was
performed for verification of miRNA microarray results using 3
selected miRNAs (miR-20a, miR-21 and miR-4489). A total of 1
μg of RNA was reverse-transcribed into cDNA using the
All-in-One™ miRNA First-strand cDNA synthesis kit (AMRT-0600;
GeneCopoeia, Rockville, MD, USA) according to the manufacturer’s
instructions. Next, the resulting cDNA (1:5 dilution) was amplified
and assayed by qPCR, with the 7500 Real-Time PCR system (Applied
Biosystems). Specific primers of the miRNAs (HmiRQP0312,
HmiRQP0316, HmiRQP2129 and HmiRQP9001) were provided by
GeneCopoeia. All PCR reactions were analyzed in triplicate.
Relative quantification for each sample was calculated using the
2−ΔΔCt method, with the universally expressed small
nuclear RNA U6 used as the endogenous control. A pooled human brain
cerebral cortex total RNA (Clontech, USA) was used as the normal
control.
miRNA target prediction and functional
analysis
Some online software programs were used for target
gene prediction of miRNAs, including miRanda (http://www.microrna.org/microrna/home.do), PICTAR
(http://www.ncrna.org/KnowledgeBase/link-database/mirna_target_database)
and TargetScan (http://www.targetscan.org). To reduce the number of
false positives, only miRNA target genes identified by all of these
programs were retained. Predicted target genes were imported into
DAVID v6.7, a web-based tool for annotation, visualization and
integrated discovery, for functional analysis. We performed
biological process of Gene Ontology (GO) and Pathway analysis on
target genes of the miRNAs with differential expression based on
the GO database and KEGG database. The statistical significance of
GO and pathways was determined on the basis of a P-value
<0.01.
Statistical analysis
The significance of differences between the compared
groups, gliomas and the matched adjacent noncancerous tissues
performed by qRT-PCR, was determined by a paired samples t-test
using SPSS 16.0 (IBM Corporation, USA) with a P-value of <0.05
considered to indicate a statistically significant result.
Results
miRNA microarray analysis
In the microarray data analysis, we investigated
1,926 miRNAs. Thirteen human miRNAs were significantly
differentially expressed between 9 gliomas and the adjacent tissues
(fold-change ≥1.5 or ≤0.67; P≤0.05). Among these, 12 miRNAs were
upregulated and 1 was downregulated. Seven miRNAs (hsa-miR-92a-3p,
hsa-miR-106b-5p, hsa-miR-93-5p, hsa-miR-20a-5p, hsa-miR-17-5p,
hsa-miR-25-3p and hsa-miR-19b-3p) are members of the miR-17-92
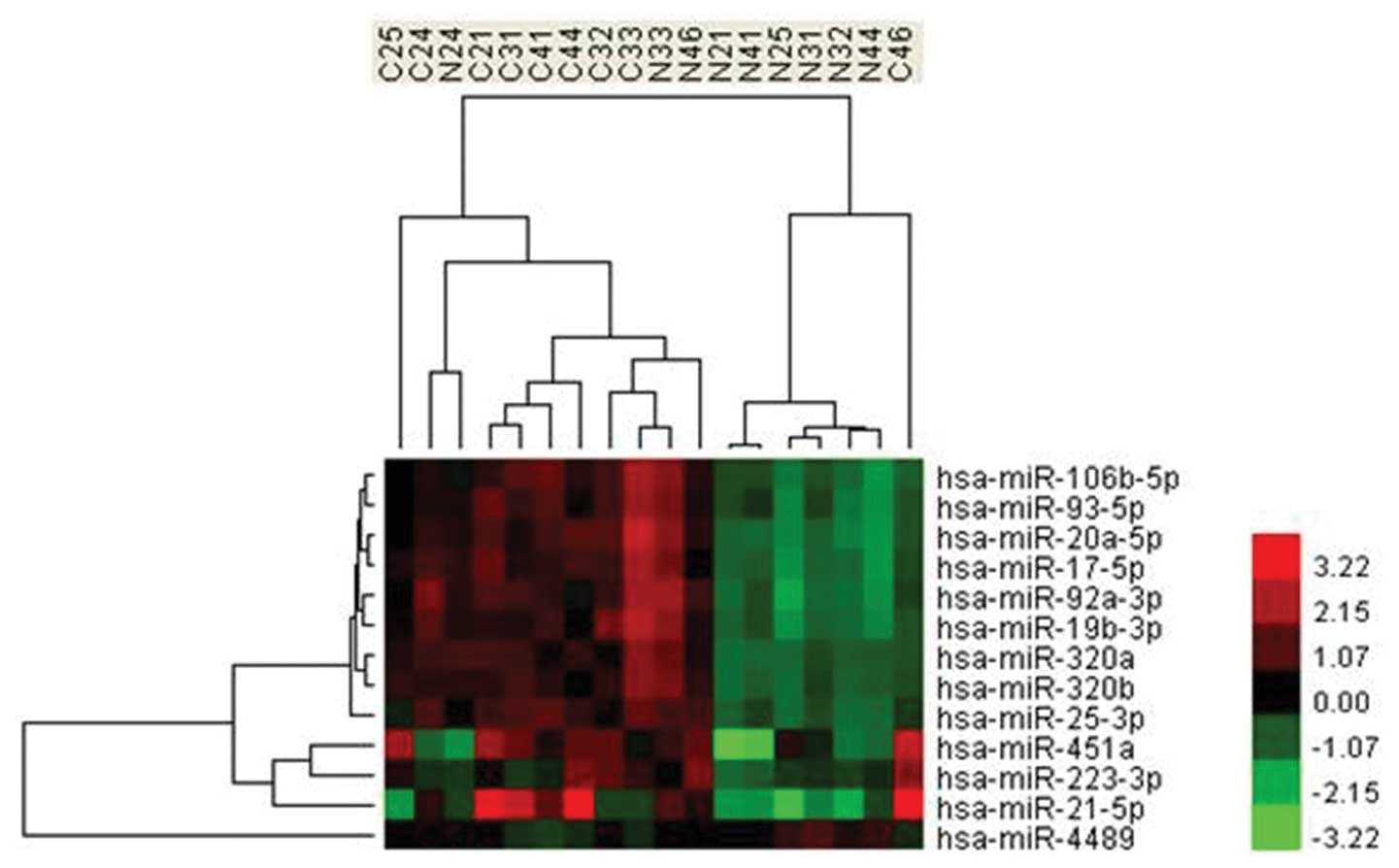
cluster and its paralogs, miR-106b-25 cluster (Fig. 1). miRNAs encoded by the miR-17-92
cluster show widespread enforced expression and are known as
oncogenes in different types of cancers in previous studies
(12,13). The cluster analyses revealed
complete separation of the patient and control groups based on the
expression profiles of the differentially expressed miRNAs
(Fig. 2). In addition, by analysis
of aberrant expression of miRNAs in WHO II, III and IV compared
with the matched adjacent non-cancerous tissues, each grade
containing 3 glioma samples, we found 7 upregulated and 1
downregulated miRNA in WHO II, 6 upregulated and 3 downregulated
miRNAs in WHO III and 6 upregulated and 9 downregulated miRNAs in
WHO IV (Table III).
 | Table IIIAberrantly expressed miRNAs in
different grades of gliomas. |
Table III
Aberrantly expressed miRNAs in
different grades of gliomas.
| miRNAs | Ratio (T/N) | P-value |
|---|
| WHO II |
|
hsa-miR-92a-3p | 2.962 | 0.044 |
|
hsa-miR-335-5p | 3.634 | 0.042 |
|
hsa-miR-18a-5p | 1.812 | 0.032 |
|
hsa-miR-19b-3p | 2.335 | 0.001 |
| hsa-miR-3653 | 1.684 | 0.050 |
|
hsa-miR-1247-3p | 1.505 | 0.041 |
|
hsa-miR-4632-5p | 1.766 | 0.034 |
|
hsa-miR-204-3p | 0.437 | 0.014 |
| WHO III |
|
hsa-miR-125b-5p | 1.772 | 0.041 |
|
hsa-miR-106b-5p | 1.958 | 0.018 |
|
hsa-miR-99a-3p | 1.582 | 0.017 |
| hsa-miR-93-5p | 1.946 | 0.013 |
| hsa-miR-17-5p | 1.962 | 0.015 |
| hsa-miR-320a | 1.760 | 0.024 |
|
hsa-miR-29b-3p | 0.473 | 0.043 |
|
hsa-miR-29c-3p | 0.403 | 0.044 |
|
hsa-miR-4433-5p | 0.604 | 0.031 |
| WHO IV |
| hsa-miR-32-3p | 1.639 | 0.021 |
| hsa-miR-1275 | 1.931 | 0.035 |
| hsa-miR-3178 | 1.861 | 0.004 |
|
hsa-miR-3940-5p | 1.631 | 0.036 |
|
hsa-miR-4725-3p | 2.073 | 0.018 |
|
hsa-miR-642b-3p | 1.580 | 0.022 |
|
hsa-miR-138-5p | 0.396 | 0.019 |
|
hsa-miR-29b-2-5p | 0.658 | 0.010 |
|
hsa-miR-139-5p | 0.353 | 0.037 |
|
hsa-miR-3144-5p | 0.587 | 0.016 |
| hsa-miR-3161 | 0.622 | 0.024 |
| hsa-miR-4526 | 0.631 | 0.040 |
| hsa-miR-4525 | 0.496 | 0.022 |
|
hsa-miR-4694-3p | 0.665 | 0.032 |
|
hsa-miR-6501-3p | 0.605 | 0.012 |
miRNA expression validation
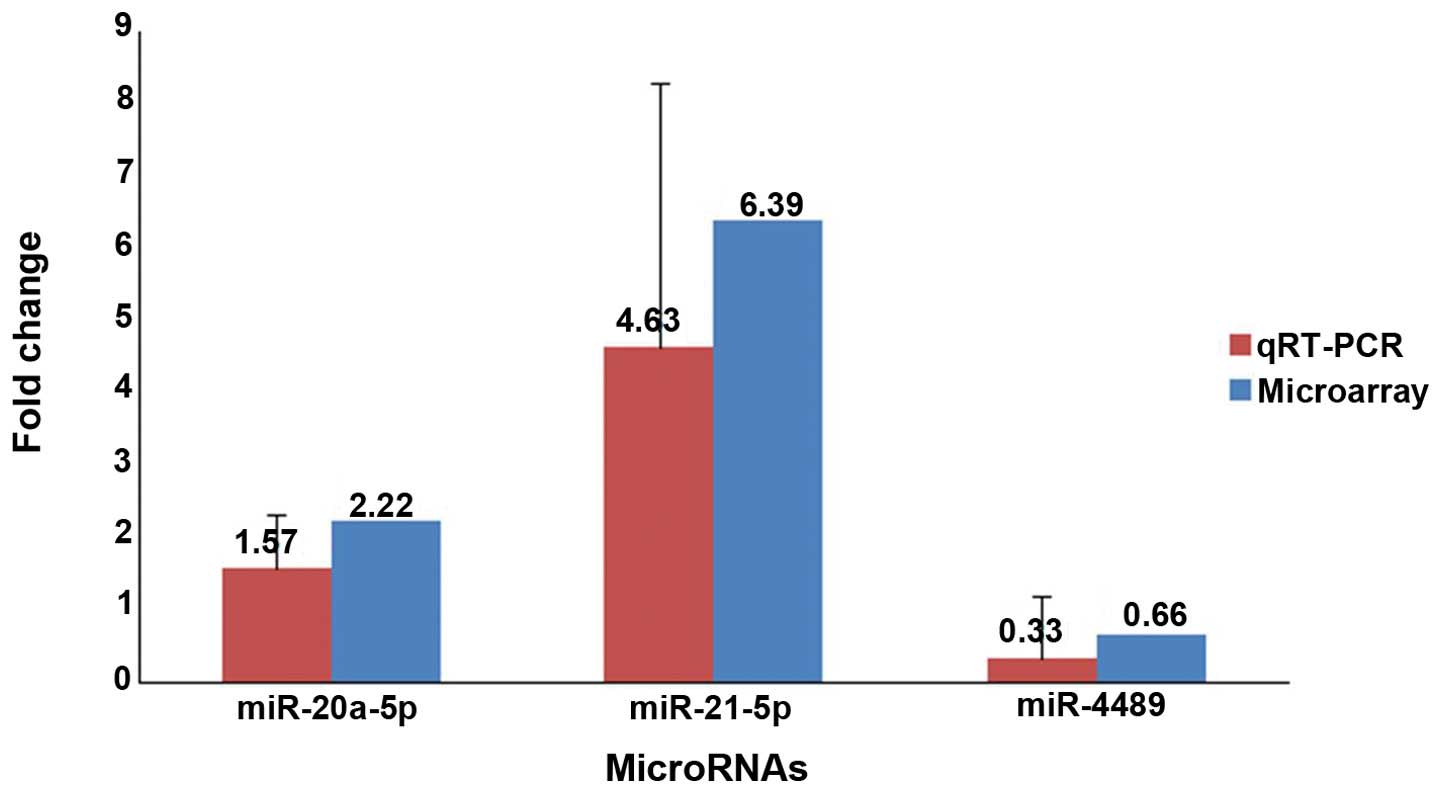
To improve sample representativeness, 3 candidate
miRNAs, miR-20a-5p (randomly selected), miR-21-5p (the common
upregulated) and miR-4489 (the only one downregulated), were
selected to validate the microarray data by quantitative RT-PCR
assay using the same RNA samples that were used for the
microarrays. The adjacent non-cancerous tissues acted as the
control in the same groups for both experiments. Compared with the
adjacent non-cancerous tissues, the expression levels of miR-20a-5p
(P=0.049, n=9) and miR-21-5p (P=0.017, n=9) were present in higher
abundance, whereas miR-4489 (P=0.045, n=9) was present in lower
abundance in the gliomas. The results of quantitative RT-PCR were
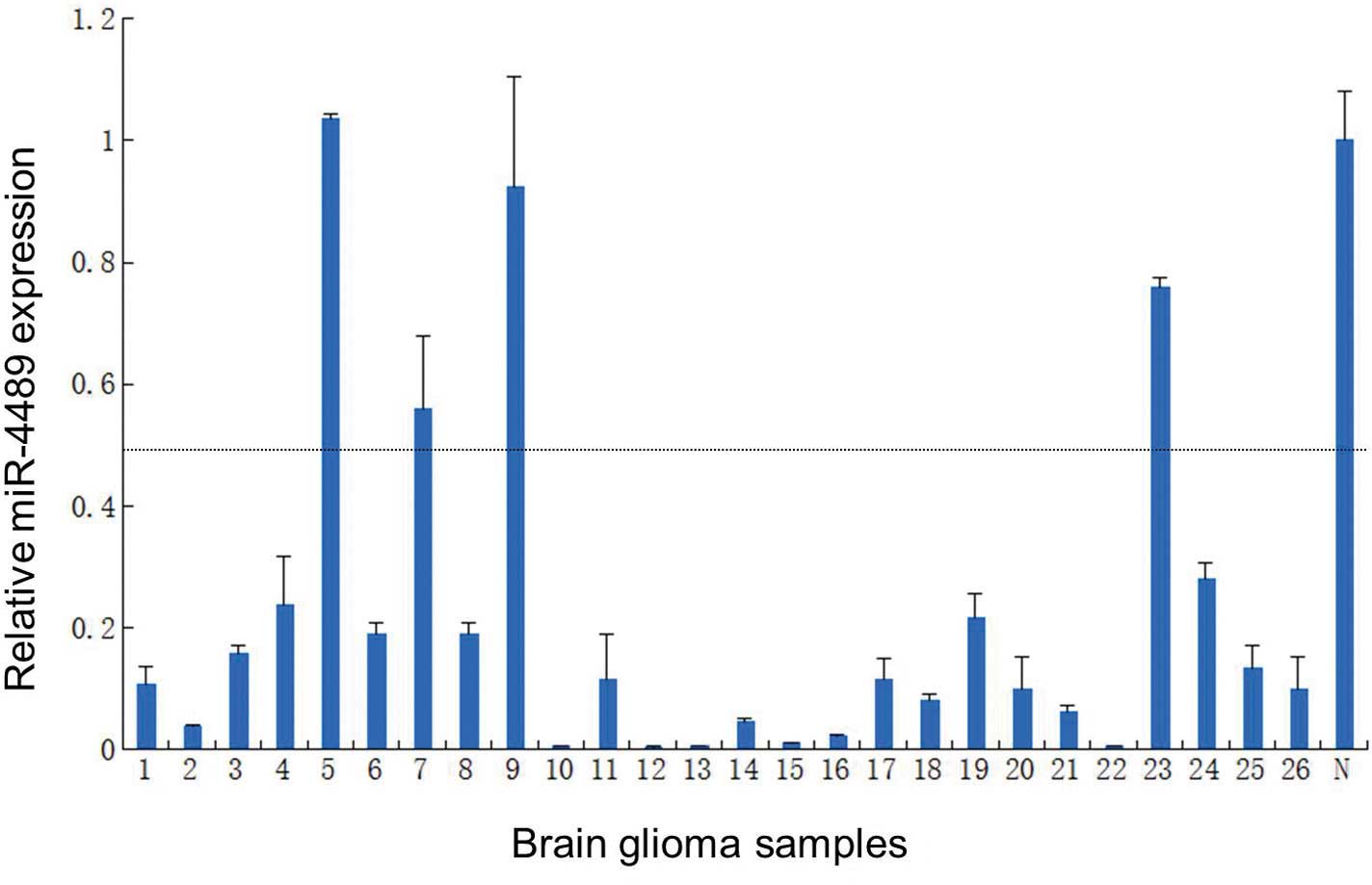
in accordance with the miRNA microarray results (Fig. 3). Furthermore, we investigated the
expression of miR-4489 by qRT-PCR in 26 glioma samples. The results
presented in Fig. 4 revealed that
84.62% (22/26) of gliomas had a reduced miR-4489 expression
compared to the normal human cerebral cortex total RNA
(Clontech).
miRNA target gene prediction, GO and
Pathway analysis
The potential target genes of the 13 dysregulated
miRNAs were predicted by different algorithms, including
TargetScan, PICTAR and miRanda. Only miRNA target genes identified
by all of these algorithms were considered. Some of the miRNAs, for
example, miR-4489 were only included in the database of TargetScan;
hence, target genes of miR-4489 in TargetScan were selected as a
part of the target gene set for further analysis. A total of 926
target genes were predicted. Target genes (720) and 162 significant
categories were found in Gene Ontology Biological Process when GO
analysis was performed in the DAVID database (DAVID Bioinformatics
Resources 6.7). A large number of target genes were involved in the
enriched GO categories notably genes involved in the regulation of
cell proliferation, the cell cycle, apoptosis and neuronal
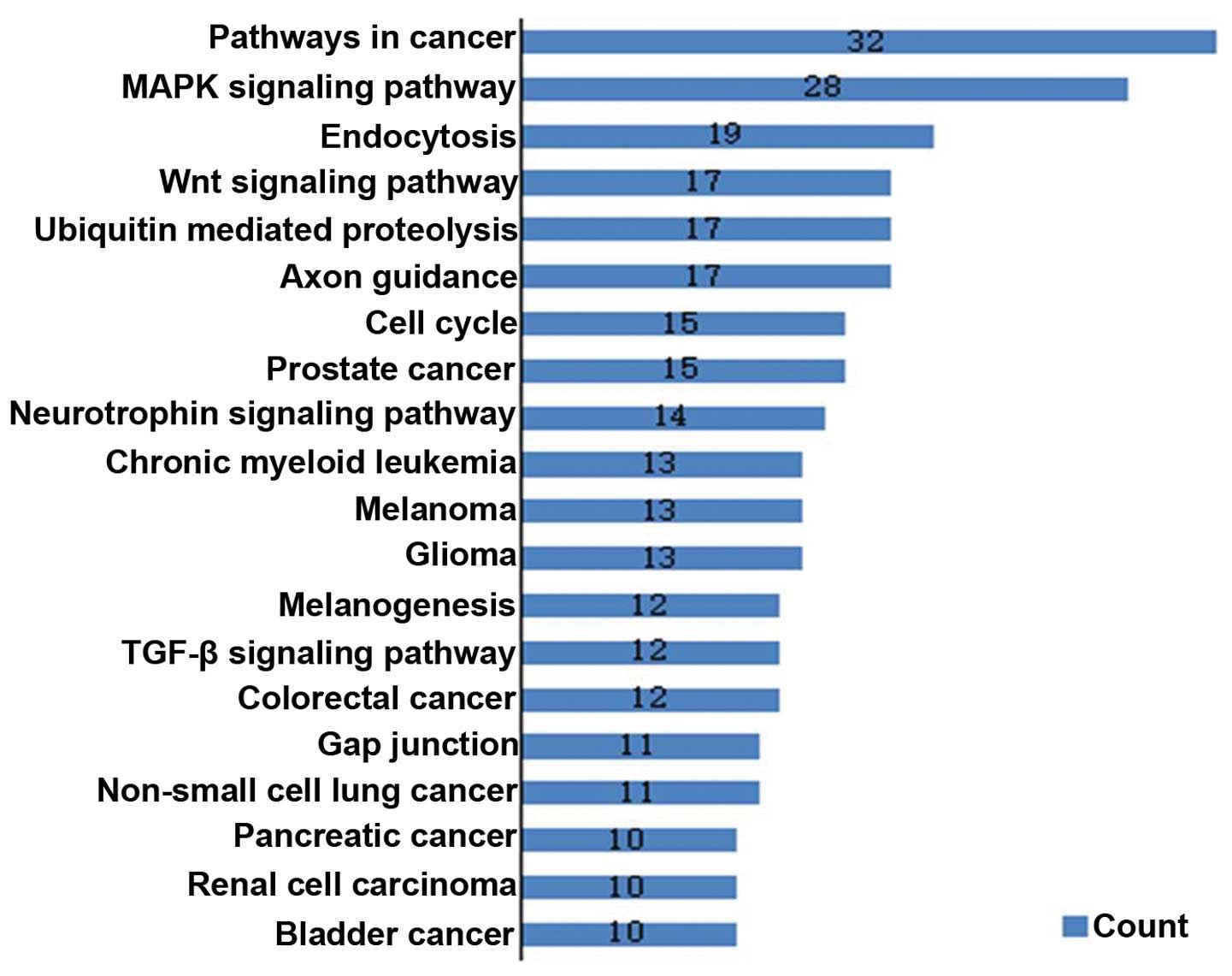
differentiation. Pathway analysis based on the KEGG pathway
database identified 239 target genes and 20 enriched pathways
(P<0.01) (Fig. 5).
Discussion
In the past 9 years, extensive research has
indicated that miRNAs contribute to the development and progression
of gliomas (8). However, the
expression patterns of grade-specific miRNAs that may act as
reliable biomarkers for early diagnosis, progression detection and
prediction of survival are largely unknown. In the present study,
using an miRNA microarray approach, we uncovered the aberrant
expression of several miRNAs in gliomas of different grades
compared with the adjacent tissues. In previous studies using
microarrays, normal tissues and brain tumors were selected from
different patients (14). However,
in the present study, the gliomas and non-neoplasms were selected
from the same patients concurrently, and this effectively reduced
the impact of individual differences. We selected 3 pairs of
samples in each grade of WHO II, III and IV for miRNA microarray
analysis. Totals of 8, 9 and 15 dysregulated miRNAs were detected
in WHO II, III and IV gliomas, respectively, and the miRNAs in each
grade of gliomas were entirely different from those in the other
grades. Aberrantly expressed miRNAs only appeared in a certain
grade but were absent in the lower grades, indicating that these
miRNAs result in the malignant progression of gliomas. These may
also be used as valuable biomarkers for glioma grading. As a single
mRNA can be regulated by multiple miRNAs, an association would be
found in the level of protein or mRNA despite the apparent
unrelatedness of miRNAs in each grade. In addition, a majority of
our miRNAs, such as miR-4433-5p and miR-4725-3p, have not been
involved in other studies. These miRNAs may serve as important
resources for future functional study in gliomas.
Thirteen human miRNAs were found to be significantly
differentially expressed between gliomas and the adjacent tissues
in an integrated analysis. A total of 926 predicted target genes of
the 13 miRNAs were obtained by bioinformatics. GO and Pathway
analysis showed that a large number of target genes were
significantly enriched in biological processes and signaling
pathways associated with the nervous system and tumors. Several
cancer-related pathways, including the MAPK, p53, Wnt signaling
pathways and cell cycle-related pathway, were included in the
enriched pathways of the target genes. Aberrant activation of Wnt
signaling, which increases cell proliferation and enhances
invasiveness, is involved in various types of cancers, including
gliomas (15). MAPK signaling has
been considered to contribute to the pathogenesis of gliomas by
supporting the migration and invasion of malignant glioma cells
(16). TP53 signaling is important
in apoptosis and cell cycle arrest in cellular responses to DNA
damage and aberrant pathway activation (17,18).
Approximately 87% of glioblastomas harbor at least one component of
TP53 signaling aberrations (19).
This indicates that aberrant expression of miRNAs in our results
contributes to the pathogenesis of gliomas.
Several of the differentially expressed miRNAs, such
as miR-21, in our results had previously been demonstrated to be
associated with gliomas and to participate in disease development
and progression. Chan et al (7) initially found high expression of
miR-21 in gliomas by miRNA microarray. In the present study, a
4.63- and 6.39-fold miR-21 expression level in gliomas was detected
by qRT-PCR analysis (P=0.017, n=9) and microarray (P=0.028, n=9),
respectively, compared with the adjacent non-cancerous tissues
(Fig. 3). Moreover, our results are
in accordance with Chan et al. The upregulation of miR-21
was found to increase cell growth by inhibiting PDCD4 (20) and to enhance invasiveness by
inhibiting SPYR2 (21). Moreover,
Han et al (22) identified
that the miR-21 expression level increased with an increasing
histological grade of gliomas. A novel dysregulated miRNA,
miR-4489, was found to be related to human gliomas in the present
study. However, the significance of dysregulated expression of
miR-4489 was not found in WHO II, III and IV in the microarray
(Table III). To further detect
the expression of miRNA-4489, 26 samples were used for qRT-PCR
method compared with human normal cerebral cortex (Clontech). In
the present study, 84.62% (22/26) of glioma samples showed
downregulation of miR-4489 by qRT-PCR. A total of 89 potential
target genes for miR-4489 were obtained from the TargetScan
database. According to the result of the GO analysis, a number of
target genes were enriched in the process of cell proliferation.
Among these, several genes such as CDK6 were verified to be
correlated with gliomas (23).
Analyses of clinical samples from GBM patients identified a higher
CDK6 antigen expression in tumors than in adjacent normal brain
tissues. Knockdown of CDK6 by siRNAs induced cell cycle arrest at
the G1/S transition and inhibited cell proliferation. In addition,
TGF-β2, a predicted target of miR-4489, is a key factor in the
pathogenesis of gliomas, and is involved in tumor growth, invasion
and metastasis (24).
Bioinformatics software provided a primary analysis of miR-4489
functions and target genes, and this contributes to new clues for
research on gliomas.
The miR-17-92 cluster (miR-17, miR-18a, miR-19a,
miR-20a, miR-19b-1 and miR-92-1) is a typical case known as
oncogenes in diverse tumor subtypes. However, as a paralog of the
miR-17-92 cluster, the function of the miR-106b-25 cluster,
including 3 miRNAs (miR-106b, miR-93 and miR-25), remains largely
unclear in carcinogenesis. In the present study, all members of the
miR-106b-25 cluster and 4 miRNAs in the miR-17-92 cluster were
included in the aberrantly expressed miRNAs. Ventura et al
(25) showed that the functions of
the miR-106b-25 cluster are seemingly dispensable in normal mouse
embryonic development and is the only study in the context of
miR-17-92 cluster loss. In our data, the aberrant expression of
members in the miR-17-92 cluster (miR-92a-3p, miR-18a-5p and
miR-19b-3p) was found in WHO II gliomas, while dysregulated
expression of miRNAs in the miR-106b-25 cluster (miR-106b-5p and
miR-93-5p) was detected in WHO III gliomas. Two members of the
miR-106b-25 cluster have an identical seed sequence (nucleotides
2–7), which is also the seed sequence of two members of the
miR-17-92 cluster (Fig. 1). The
highly conserved feature of this region indicates the functional
importance of these miRNAs. However, according to the results of
prevailing target prediction models, it is also reasonable to
suppose that these miRNAs should be functionally redundant. As
observed during normal development of mice, enhanced expression of
the miR-106b-25 cluster plays an assistant role in glioma
progression and results in greater proliferation ability and
anti-apoptotic effects in tumors with upregulated expression levels
of the miR-17-92 cluster. There is reason to believe that the
miR-106b-25 cluster plays an indispensable role in the malignant
process of gliomas by promoting cell cycle progression, enhancing
cell proliferation, inhibiting cell apoptosis and inducing blood
vessel formation (26–29). Despite the great progress made in
understanding the cluster’s roles, the expression pattern and the
precise role of each miRNA in gliomas need to be deeply elucidated.
Further study of the function of the miR-106b-25 cluster may
provide new insight into the role of miRNAs in the development and
progression of human gliomas.
To summarize, we described the differential
expression of miRNAs and explored the grade-related miRNA
expression profiles in brain gliomas. These miRNAs could disrupt
crucial pathophysiological processes. These results have
significant importance in uncovering a relationship between
specific miRNAs and glioma tumorigenesis, malignant progression and
different glioma grades.
Acknowledgments
We thank Dr Kaifeng Shen and Mrs. Zhenle Zang for
their technical assistance. The present study was supported by the
National Natural Science Foundation of China (NSFC: 81272783), The
Natural Science Foundation Project of CQ (CSTC: 2010BB5028) and The
National Key Technology Research and Development Program of the
Ministry of Science and Technology of China (2014BAI04B02).
References
|
1
|
Kanzawa T, Germano IM, Komata T, Ito H,
Kondo Y and Kondo S: Role of autophagy in temozolomide-induced
cytotoxicity for malignant glioma cells. Cell Death Differ.
11:448–457. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Stupp R, Mason WP, van den Bent MJ, Weller
M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn
U, et al European Organisation for Research and Treatment of Cancer
Brain Tumor and Radiotherapy Groups; National Cancer Institute of
Canada Clinical Trials Group: Radiotherapy plus concomitant and
adjuvant temozolomide for glioblastoma. N Engl J Med. 352:987–996.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervous system. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ohgaki H and Kleihues P: The definition of
primary and secondary glioblastoma. Clin Cancer Res. 19:764–772.
2013. View Article : Google Scholar
|
|
5
|
Ambros V: microRNAs: Tiny regulators with
great potential. Cell. 107:823–826. 2001. View Article : Google Scholar
|
|
6
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chan JA, Krichevsky AM and Kosik KS:
MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells.
Cancer Res. 65:6029–6033. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Davis-Dusenbery BN and Hata A: MicroRNA in
cancer: The involvement of aberrant microRNA biogenesis regulatory
pathways. Genes Cancer. 1:1100–1114. 2010. View Article : Google Scholar
|
|
9
|
He J, Deng Y, Yang G and Xie W:
MicroRNA-203 down-regulation is associated with unfavorable
prognosis in human glioma. J Surg Oncol. 108:121–125. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yan LX, Huang XF, Shao Q, Huang MY, Deng
L, Wu QL, Zeng YX and Shao JY: MicroRNA miR-21 overexpression in
human breast cancer is associated with advanced clinical stage,
lymph node metastasis and patient poor prognosis. RNA.
14:2348–2360. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
He HC, Han ZD, Dai QS, Ling XH, Fu X, Lin
ZY, Deng YH, Qin GQ, Cai C, Chen JH, et al: Global analysis of the
differentially expressed miRNAs of prostate cancer in Chinese
patients. BMC Genomics. 14:7572013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cho WC: OncomiRs: The discovery and
progress of microRNAs in cancers. Mol Cancer. 6:602007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Diosdado B, van de Wiel MA, Terhaar Sive
Droste JS, Mongera S, Postma C, Meijerink WJ, Carvalho B and Meijer
GA: MiR-17-92 cluster is associated with 13q gain and c-myc
expression during colorectal adenoma to adenocarcinoma progression.
Br J Cancer. 101:707–714. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li Y, Xu J, Chen H, Bai J, Li S, Zhao Z,
Shao T, Jiang T, Ren H, Kang C, et al: Comprehensive analysis of
the functional microRNA-mRNA regulatory network identifies miRNA
signatures associated with glioma malignant progression. Nucleic
Acids Res. 41:e2032013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang K, Wang X, Zou J, Zhang A, Wan Y, Pu
P, Song Z, Qian C, Chen Y, Yang S, et al: miR-92b controls glioma
proliferation and invasion through regulating Wnt/beta-catenin
signaling via Nemo-like kinase. Neurooncol. 15:578–588. 2013.
|
|
16
|
Guo G, Yao W, Zhang Q and Bo Y: Oleanolic
acid suppresses migration and invasion of malignant glioma cells by
inactivating MAPK/ERK signaling pathway. PLoS One. 8:e720792013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kruse JP and Gu W: Modes of p53
regulation. Cell. 137:609–622. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Farnebo M, Bykov VJ and Wiman KG: The p53
tumor suppressor: A master regulator of diverse cellular processes
and therapeutic target in cancer. Biochem Biophys Res Commun.
396:85–89. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cancer Genome Atlas Research Network:
Comprehensive genomic characterization defines human glioblastoma
genes and core pathways. Nature. 455:1061–1068. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gaur AB, Holbeck SL, Colburn NH and Israel
MA: Down-regulation of Pdcd4 by mir-21 facilitates glioblastoma
proliferation in vivo. Neurooncol. 13:580–590. 2011.
|
|
21
|
Kwak HJ, Kim YJ, Chun KR, Woo YM, Park SJ,
Jeong JA, Jo SH, Kim TH, Min HS, Chae JS, et al: Downregulation of
Spry2 by miR-21 triggers malignancy in human gliomas. Oncogene.
30:2433–2442. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Han L, Yue X, Zhou X, Lan FM, You G, Zhang
W, Zhang KL, Zhang CZ, Cheng JQ, Yu SZ, et al: MicroRNA-21
expression is regulated by β-catenin/STAT3 pathway and promotes
glioma cell invasion by direct targeting RECK. CNS Neurosci Ther.
18:573–583. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen SM, Chen HC, Chen SJ, Huang CY, Chen
PY, Wu TW, Feng LY, Tsai HC, Lui TN, Hsueh C, et al: MicroRNA-495
inhibits proliferation of glioblastoma multiforme cells by
down-regulating cyclin-dependent kinase 6. World J Surg Oncol.
11:872013. View Article : Google Scholar
|
|
24
|
Wick W, Naumann U and Weller M:
Transforming growth factor-beta: A molecular target for the future
therapy of glioblastoma. Curr Pharm Des. 12:341–349. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ventura A, Young AG, Winslow MM, Lintault
L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone
JR, et al: Targeted deletion reveals essential and overlapping
functions of the miR-17 through 92 family of miRNA clusters. Cell.
132:875–886. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kan T, Sato F, Ito T, Matsumura N, David
S, Cheng Y, Agarwal R, Paun BC, Jin Z, Olaru AV, et al: The
miR-106b-25 polycistron, activated by genomic amplification,
functions as an oncogene by suppressing p21 and Bim.
Gastroenterology. 136:1689–1700. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ivanovska I, Ball AS, Diaz RL, Magnus JF,
Kibukawa M, Schelter JM, Kobayashi SV, Lim L, Burchard J, Jackson
AL, et al: MicroRNAs in the miR-106b family regulate p21/CDKN1A and
promote cell cycle progression. Mol Cell Biol. 28:2167–2174. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu F, Gong J, Huang W, Wang Z, Wang M,
Yang J, Wu C, Wu Z and Han B: MicroRNA-106b-5p boosts glioma
tumorigensis by targeting multiple tumor suppressor genes.
Oncogene. 33:4813–4822. 2014. View Article : Google Scholar
|
|
29
|
Fang L, Deng Z, Shatseva T, Yang J, Peng
C, Du WW, Yee AJ, Ang LC, He C, Shan SW, et al: MicroRNA miR-93
promotes tumor growth and angiogenesis by targeting integrin-β8.
Oncogene. 30:806–821. 2011. View Article : Google Scholar
|