Introduction
Lung cancer is one of the leading causes of
cancer-related death in the world (1). Human lung cancers can be classified
into two major histological types, non-small cell lung cancer
(NSCLC) and small cell lung cancer (SCLC), and the former one
accounts for more than 80% of all lung cancer cases (2). NSCLC is consisted of adenocarcinoma,
squamous carcinoma, adenosquamous carcinoma and large cell
carcinoma. Despite many advances achieved in the diagnosis and
management of NSCLC, the mechanisms underlying the pathogenesis of
NSCLC remain poorly understood and the overall 5-year survival rate
was no more than 15% (3). The
process of lung cancer is associated with alterations of both
oncogenes and tumor suppressors, as well some cell cycle regulators
related to cell proliferation and survival (4). Therefore, identifying new molecular
targets and mechanisms can provide novel strategies for the
diagnosis and treatment of NSCLC.
HES5 belongs to the basic helix-loop-helix (bHLH)
superfamily and is a DNA-binding transcription factor. It is a
downstream molecule and effector of mammalian Notch pathway, which
is mainly expressed in epithelia in the process of embryogenesis or
in neural stem cells (5). HES5 has
an important role in regulating mammalian neuronal differentiation
and the maintenance of neural stem cells (6) and positively regulates neuronal stem
cell self-renewal (7). Recent
studies have found that HES5 was involved in inducing cell
differentiation or promoting the tumor cellular proliferation in
many kinds of cancer, such as neuroblastoma cells, carcinoid tumor
cells and breast cancer cell lines (8–10).
This indicates that HES5 may be involved in the initiation and
process of cancers. Furthermore, it was reported that HES5 could
associate with JAK2 and STAT3, and promoted STAT3 phosphorylation
and activation, which protected hepatocytes from apoptosis after
I/R injury (11). Since many
studies have revealed that JAK2-STAT3 could regulate cell
proliferation and apoptosis in NSCLC (12,13),
HES5 may regulate the initiation and cell proliferation of NSCLC
via STAT3 pathway.
In the present study, we investigated the role of
HES5 in NSCLCs progression. Expression of HES5 in 8 paired tumor
and adjacent non-tumor tissues were detected by western blot
analysis. Immunohistochemistry (IHC) assay was performed in 114
NSCLC samples. Then, we also investigated the association of HES5
expression with clinical and pathological factors, as well as the
prognostic implications. Immunoprecipitation assay and western blot
analysis were used to detect the relation between HES5 and STAT3.
Moreover, we explored the potential involvement of HES5 in the
regulation of cell cycle progression and cell proliferation in
NSCLC cells using HES5 siRNA transfection.
Materials and methods
Patients and tissue samples
One hundred and fourteen lung cancer sections, 8
lung cancer tissue samples, and 8 normal tissue samples from
patients who underwent surgery from the period of 2005 to 2009 at
the Department of Pathology, Affiliated Hospital of Nantong
University. The Affiliated Hospital of Nantong University Hospital
provided formalin-fixed and paraffin-embedded tissues for
histopathological diagnosis and immunohistochemical study. The main
clinical and pathological variables are shown in Table I. Besides, 8 paired tumor and
adjacent non-tumor NSCLC fresh tissues were frozen in liquid
nitrogen immediately and stored at −80°C for western blot
analysis.
 | Table I.Expression of HES5 in 114 human lung
NSCLC tissues. |
Table I.
Expression of HES5 in 114 human lung
NSCLC tissues.
|
|
| HES5 expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Clinicopathological
features | Total | Low | High | P-value | χ2
value |
|---|
| Age (years) |
|
|
|
|
|
|
<60 | 51 | 27 | 24 | 0.953 | 0.004 |
| ≥60 | 63 | 33 | 30 |
|
|
| Gender |
|
|
|
|
|
|
Female | 55 | 25 | 30 | 0.138 | 2,196 |
| Male | 59 | 35 | 24 |
|
|
| Tumor size (cm) |
|
|
|
|
|
|
<3 | 50 | 38 | 12 | 0.046a | 19.508 |
| ≥3 | 64 | 22 | 42 |
|
|
| Smoking status |
|
|
|
|
|
|
Yes | 29 | 15 | 14 | 0.05 | 0.91 |
| No | 85 | 45 | 40 |
|
|
| Lymph node
status |
|
|
|
|
|
| 0 | 73 | 47 | 26 | 0.033a | 11.244 |
|
>0 | 41 | 13 | 28 |
|
|
| Clinical stage |
|
|
|
|
|
| I | 75 | 44 | 31 | 0.037a | 6.567 |
| II | 25 | 13 | 12 |
|
|
|
III | 14 | 3 | 11 |
|
|
| Histological
differentiation |
|
|
|
|
|
|
Well | 25 | 19 | 6 | 0.03a | 7.014 |
|
Moderate | 76 | 35 | 41 |
|
|
|
Poor | 13 | 6 | 7 |
|
|
| Ki-67
expression |
|
|
|
|
|
|
Low | 50 | 43 | 7 | 0.01a | 39.777 |
|
High | 64 | 17 | 47 |
Cell culture and transfection
The human NSCLC cell lines A549, H1299 and SPCA-1
were purchased from the China Academy of Science Cell Library
(Beijing, China). All the cells were cultured in RPMI-1640 medium
(Gibco-BRL, Grand Island, NY, USA) supplemented with 10% fetal
bovine serum (FBS) at 37°C and 5% CO2.
The HES5-siRNA and control-siRNA were chemically
synthesized (Shanghai GenePharma, Co., Ltd., Shanghai, China). The
HES5-specific siRNA target sequence was as follows:
5-AAGGCTACTCGTGGTGCCT-3 named as siRNA#1; 5-AGGACTACAGCGAAGGCTA-3
named as siRNA#2; and 5-TGTCAGCTACCTGAAGCAC-3 named as siRNA#3.
A549 cells were grown in dishes until they reached
70% confluence. The medium was replaced 6 h later with fresh medium
for transfection. A549 cells were transfected with HES5-siRNA or
control-siRNA according to the manufacturers instructions. Cells
were collected for western blot analysis, CCK-8 and flow cytometry
assays after transfection for 48 h (14).
Western blot analysis
Tissue and cell protein were collected with two
lysis buffers containing 50 mM Tris-HCl, pH 7.5; 150 mM NaCl; 0.1%
NP-40; 5 mM EDTA; 60 mMb glycerophosphate; 0.1 Mm sodium
orthovanadate; 0.1 mM NaF; and complete protease inhibitor cocktail
(Roche Diagnostics, Indianapolis, IN, USA) and then incubated for
20 min at 4°C while rocking. Lysates were collected after
centrifugation (15 min at 12,000 rpm, 4°C). Protein concentrations
were measured with a Bio-Rad protein assay (Bio-Rad Laboratories,
Hercules, CA, USA). Then, the same total protein was separated by
SDS-PAGE and transferred to a polyvinylidene fluoride (PVDF)
membrane (Immobilon; Millipore, Billerica, MA, USA). The membranes
were first blocked with 5% non-fat milk in TBST (150 mM NaCl, 20 mM
Tris and 0.05% Tween-20), and the membranes were washed with TBST
three times after 2 h at room temperature and then incubated
overnight with the primary antibodies. Then, LumiGLO reagent and
peroxide (Cell Signaling Technology, Danvers, MA, USA) was used as
the secondary antibody. The band was then detected by enhanced
chemiluminescence (ECL) detection systems (Pierce, Rockford, IL,
USA). The band intensity was measured by an ImageJ analysis system
(Wayne Rasband; National Institutes of Health, Bethesda, MD, USA)
(15).
Antibodies
The antibodies used in the western blot analysis
included anti-HES5 (anti-rabbit, 1:500; Santa Cruz Biotechnology,
Santa Cruz, CA, USA), anti-STAT3 (anti-mouse, 1:500; Santa Cruz
Biotechnology), anti-p-STAT3 (anti-rabbit, 1:1,000; Abcam),
anti-PCNA (anti-mouse, 1:1,000; Santa Cruz Biotechnology) and
anti-GAPDH (anti-rabbit, 1:3,000; Sigma-Aldrich).
Immunohistochemistry
Surgically excised tissues were fixed with 10%
formalin and embedded in paraffin, and 4-µm-thick specimen sections
were prepared on glass slides. The sections were deparaffinized in
xylene and rehydrated with graded alcohol, and then, antigen
retrieval was performed by heating to 121°C for 3 min in 10 mmol/l
citrate buffer (pH 6.0) with an autoclave. Thereafter, endogenous
peroxidase activity was blocked by soaking in 0.3% hydrogen
peroxide for 20 min. After rinsing in phosphate-buffered saline
(PBS) (pH 7.2), the sections were then incubated with anti-HES5
antibody (diluted 1:100; Santa Cruz Biotechnology) for 2 h at room
temperature, and anti-Ki-67 antibody (diluted 1:400; Santa Cruz
Biotechnology) for 2 h at room temperature. All slides were
processed using the peroxidase-antiperoxidase method (Dako,
Hamburg, Germany). After being washed in PBS, the peroxidase
reaction was visualized by incubating the sections with DAB (0.1%
phosphate buffer solution, 0.02% diaminobenzidine
tetrahydrochloride and 3% H2O2). After
rinsing in water, the sections were counterstained with
hematoxylin, dehydrated and coverslipped.
Immunohistochemical evaluation
All of the immunostained sections were evaluated in
a blinded manner without knowledge of the clinical and pathological
parameters of the patients. For assessment of HES5 and Ki-67, five
views were chosen per slide, and at least 1,000 cells were counted
per view at high power fields. In more than one half of the
samples, staining was repeated three times to avoid technical
errors, and a consensus was achieved. Three independent
pathologists evaluated the immunostaining results. For statistical
analysis of HES5 stain, each slide was evaluated using a
semi-quantitative scoring system for both the intensity of the
stain and the percentage of positive malignant cells (16). The intensity of staining was coded
as follows: 0 (negative or poor staining), 1 (moderate staining),
and 2 (strong staining). The percentage of cells was scored as
follows: low-expression group (<50%) score 1,
moderate-expression group (50–75%) 2, and high expression group
(>75%) score 3. Then, we multiplied the two scores and divided
patients into two groups according to the average scores (3): high-expression group (>3) and low
expression group (≤3). In statistical analysis of Ki-67 stain, 50%
of malignant cells showing positive stain was used as a cut-off
value to distinguish tumors with a low (<50%) or high (≥50%)
level of expression (17).
Cell cycle analysis
A serum starvation and refeeding process was used to
imitate the cell cycle. First, we used RPMI-1640 medium without FBS
to incubate A549 cells for 72 h to synchronize cells, which was
then changed into complete medium. Then, cells were fixed in 70%
ethanol for 1 h at 4°C and incubated with 1 mg/ml RNaseA for 30 min
at 37°C. Subsequently, cells were stained with propidium iodide
(PI, 50 µg/ml PI) (Becton Dickinson, San Jose, CA) in PBS and 0.5%
Triton X-100, and analyzed using a Becton Dickinson flow cytometer
BD FACScan (Becton Dickinson) as well as CellQuest acquisition and
analysis programs.
Cell Counting kit-8 assays
Commercial Cell Counting kit-8 (CCK-8) assays
(Dojindo Laboratories, Kumamoto, Japan) were performed to evaluate
cell proliferation. A549 cells transfected with HES5-siRNA and
control-siRNA were seeded onto 96-well cell culture cluster plates
(Corning, Inc., Corning NY, USA) at a concentration of
2×104 cells/well in volumes of 100 µl and grown
overnight. CCK-8 reagents (Dojindo Laboratories) were added to each
well under different time-points, and the wells were incubated for
an additional 2 h at 37°C in the dark. The absorbency was measured
at a test wavelength of 450 nm and a reference wavelength of 650 nm
with a microplate reader (Bio-Rad Laboratories). The experiments
were repeated at least three times.
Statistical analysis
The SPSS 19.0 statistical program was used for
statistical analysis. The HES5 expression and clinicopathological
features were analyzed by the Chi-square (χ2) test. For
analysis of survival data, Kaplan-Meier curves were constructed and
log-rank test was performed. Multivariate analysis was performed
using Coxs proportional hazards model. The risk ratio and its 95%
confidence interval were recorded for each marker. P<0.05 was
considered statistically significant for all of the analysis. The
values were expressed as mean ± SEM (18). Each experiment consisted of at least
three replicates per condition.
Results
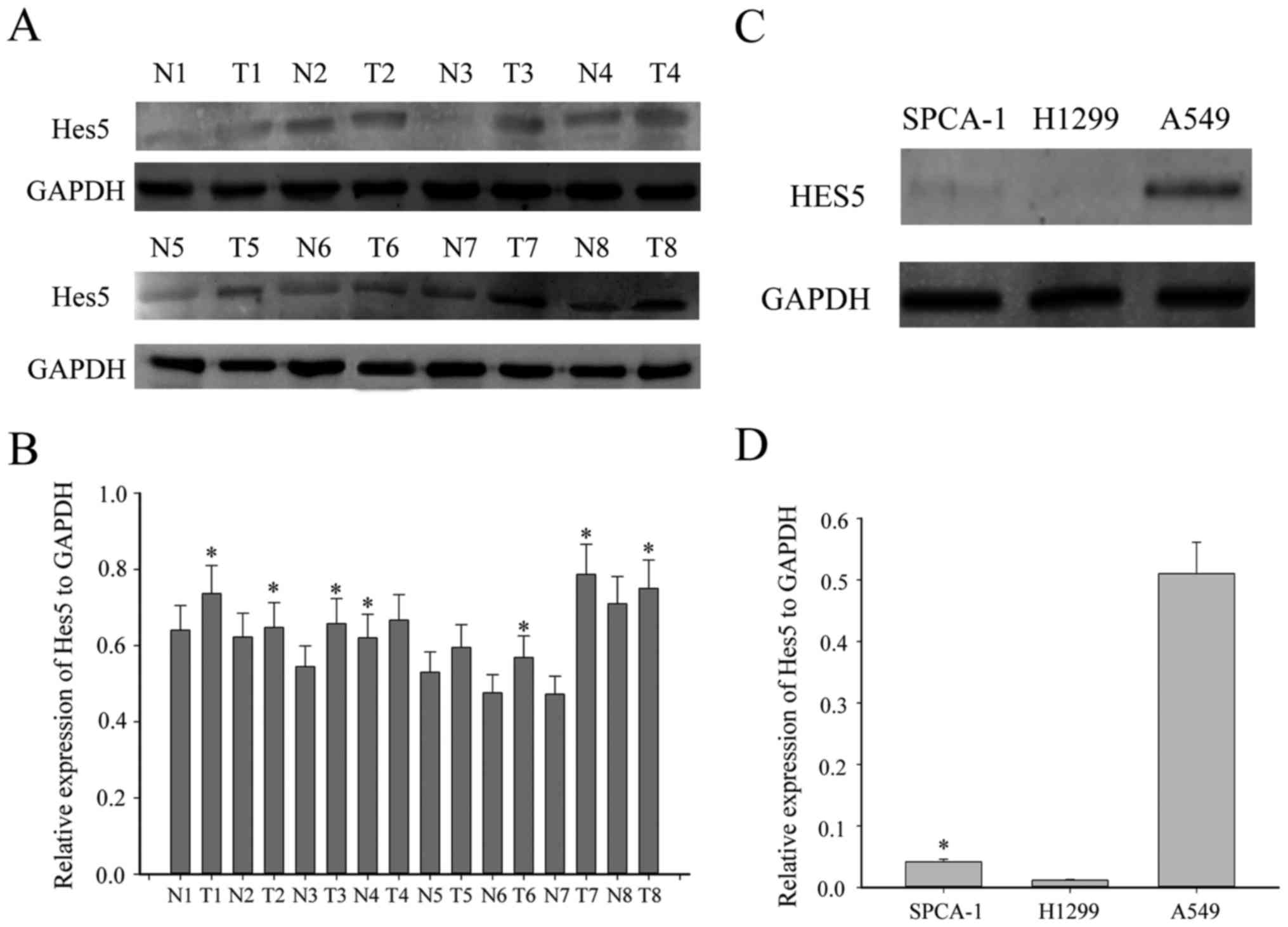
HES5 was increased in NSCLC
tissues
To explore whether abnormal expression of HES5 is
associated with the progression of NSCLC, first, we investigated
the expression of HES5 between eight paired NSCLC tissues and the
adjacent non-tumorous tissues by western blotting. As shown in
Fig. 1A, HES5 was significantly
upregulated in most NSCLC tissues compared to the adjacent
non-tumor tissues. Moreover, we examined the expression profile of
HES5 in three human NSCLC cell lines, SPCA1, H1299 and A549. As
expected, HES5 was highly expressed in A549 cell line, while there
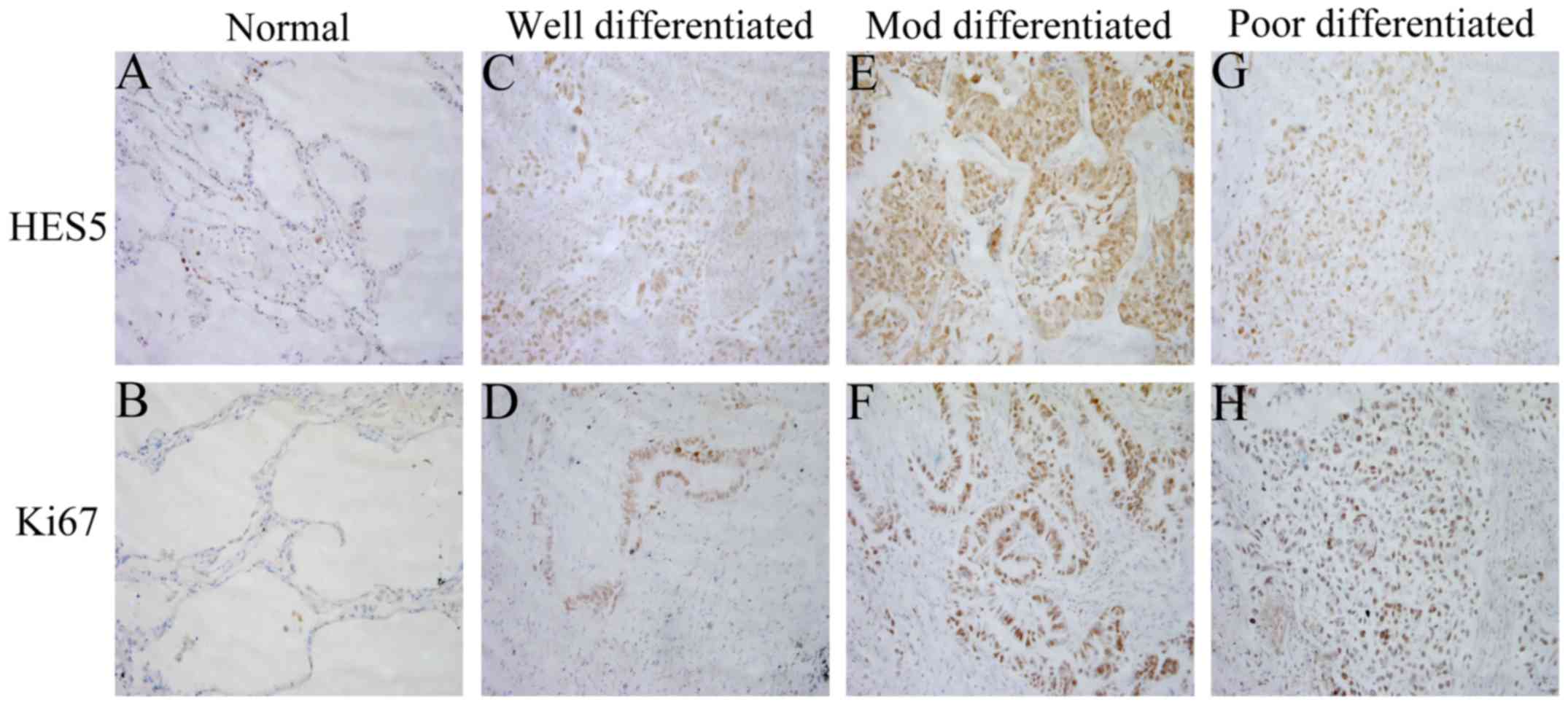
was no expression in the other two cell lines (Fig. 1C). To further investigate the
expression of HES5 in clinicopathological specimens, 114 samples
from patients with NSCLC were analyzed using immunohistochemical
assay. We found immunoreactivity of HES5 was seen mainly in the
nucleus (Fig. 2). While Ki-67,
which is a cell proliferation index, was also expressed
predominantly in the nucleus (Fig.
2). These findings together revealed that HES5 was highly
expressed in NSCLC tissues.
HES5 expression is correlated with
Ki-67 in NSCLC - relationship to clinicopathological variables
Next, we evaluated the clinicopathological
significance of HES5 expression and the physiological or
pathologicalal association between HES5 and Ki-67 in
clinicopathological variables of NSCLC. As shown in Table I, we found that the expression of
HES5 was significantly related with tumor size (P=0.046), lymph
node metastasis (P=0.033), clinical stage (P=0.037), histological
differentiation (P=0.03) and Ki-67 expression (P=0.01), while there
was no correlation with other prognostic factors such as age
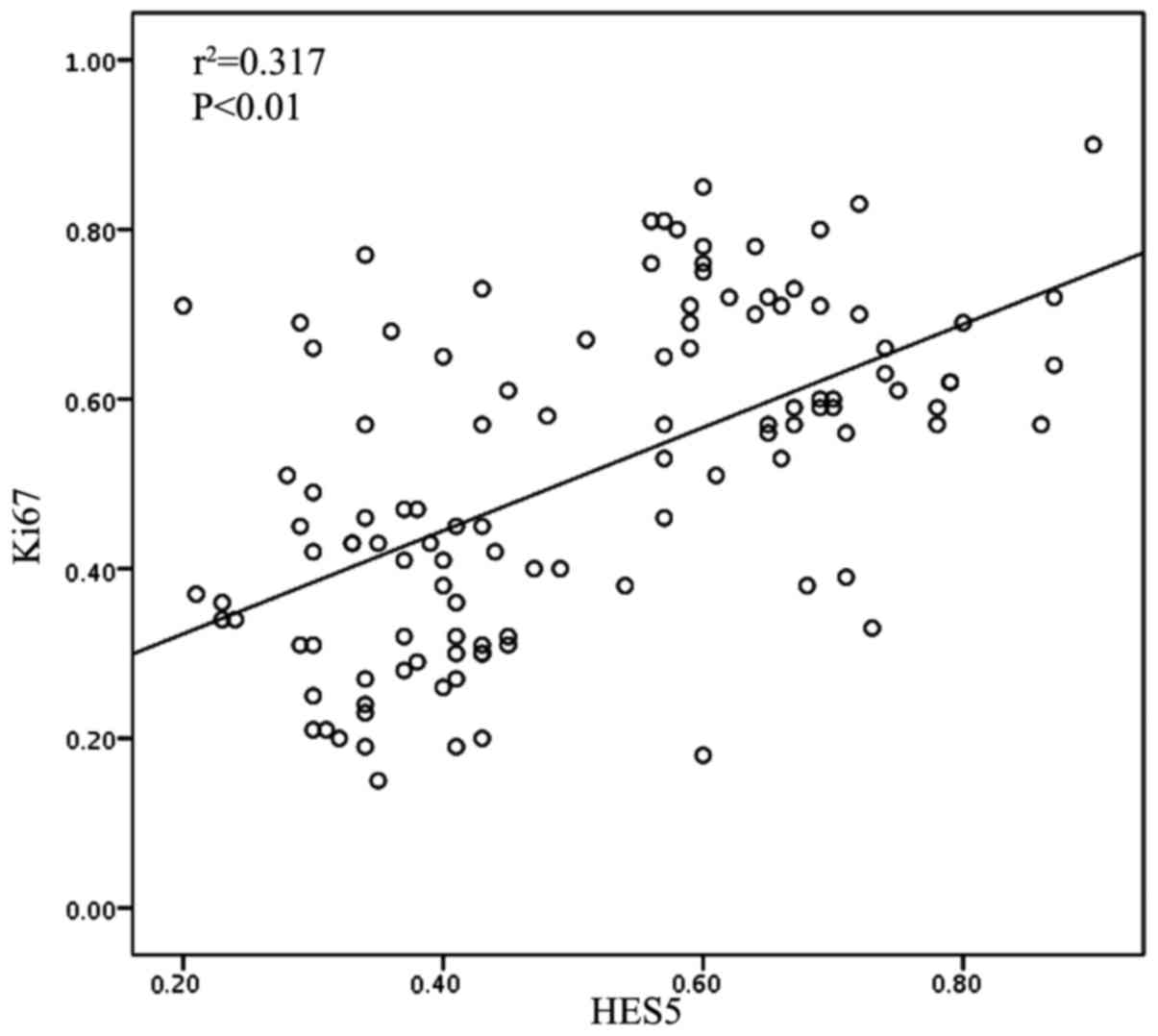
(P=0.953) and gender (P=0.138). Furthermore, we found that there
was a positive relation between the expression HES5 expression and
Ki-67 using Spearmans rank correlation test (P<0.01,
r2=0.317; Fig. 3). Taken
together, upregulated expression of HES5 could be a strong
determinant of poor prognosis in NSCLC.
High expression of HES5 correlates
with poor survival of NSCLC patients
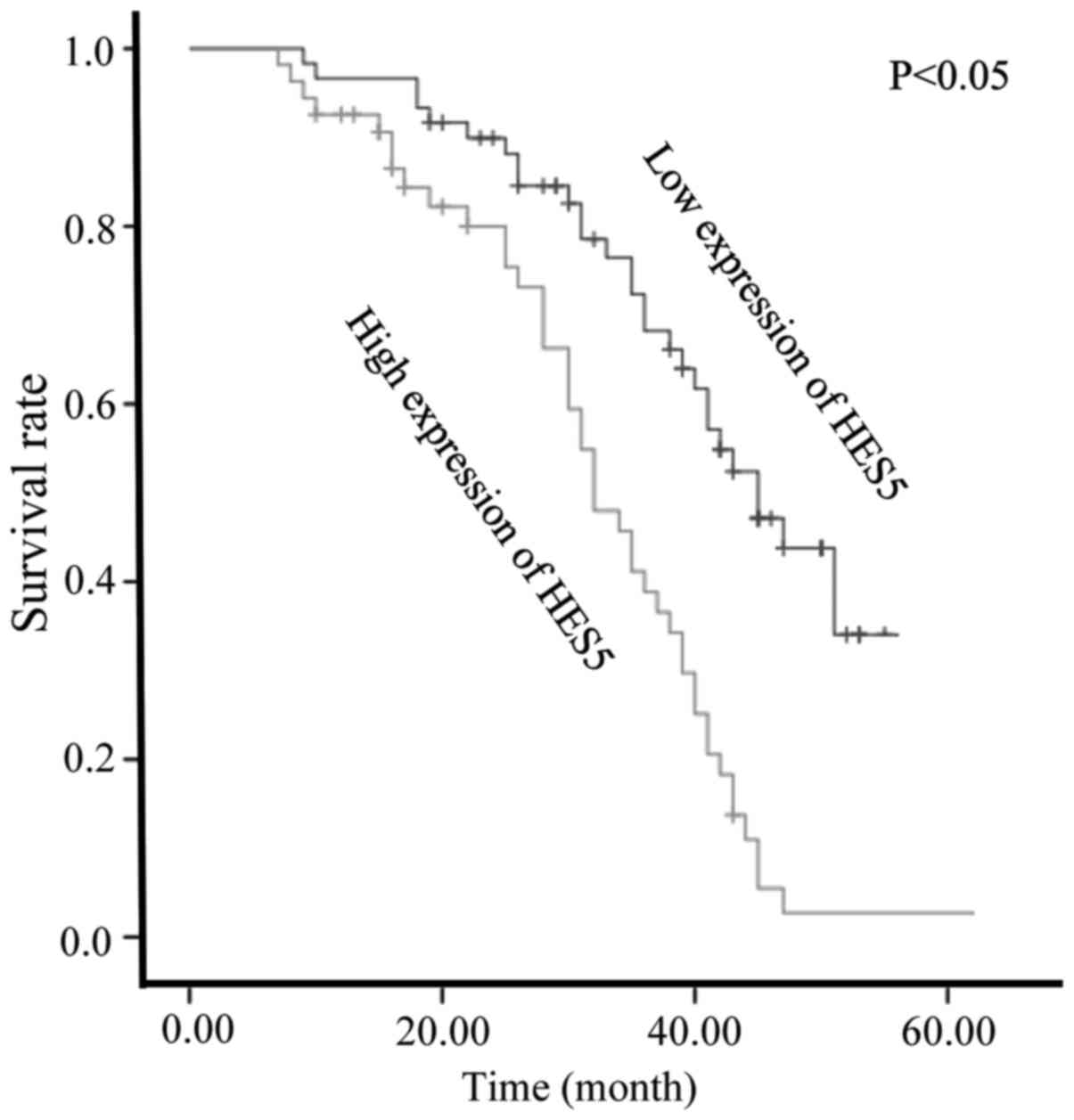
We used Kaplan-Meier analysis to calculate the
association between the expression of HES5 and patients' survival
(Fig. 4). The survival curves
showed that NSCLC patients with high HES5 expression had
significantly decreased overall survival, compared with those with
low HES5 expression. Moreover, Cox proportional hazards model
indicated that HES5 expression (P=0.027), as well as Ki-67
(P=0.018) and histological differentiation (P=0.048), were
independent prognostic indicators for overall survival of the
patients (Table II).
 | Table II.Contribution of various potential
prognostic factors to survival by Cox regression analysis in 114
NSCLC specimens. |
Table II.
Contribution of various potential
prognostic factors to survival by Cox regression analysis in 114
NSCLC specimens.
|
| Hazard ratio | 95.0% CI | P-value |
|---|
| Age | 1.245 | 0.703–2.206 | 0.453 |
| Gender | 0.542 | 0.532–2.705 | 0.660 |
| Clinical stage | 1.055 | 0.686–1.623 | 0.406 |
| Tumor size | 1.448 | 0.653–3.212 | 0.363 |
| Histological
differentiation | 0.587 | 0.345–0.996 | 0.048a |
| Lymph node
status | 0.987 | 0.534–1.822 | 0.966 |
| Ki-67
expression | 1.241 | 0.326–0.901 | 0.018a |
| HES5
expression | 1.922 | 1.077–3.430 | 0.027a |
Expression of HES5 promotes
proliferation of NSCLC cells
Based on the the fact that HES5 expression was
positively correlated with the expression of Ki-67 and higher
histological grade, we speculated that HES5 might play a role in
cell cycle progression of NSCLC cells. To verify this hypothesis,
we chose A549 cells for the serum starvation and refeeding process.
A549 cells were serum starved for 72 h and then recovered by serum
refeeding. We analyzed the cell cycle progression after serum
deprivation for 48 h using flow cytometry. As shown in Fig. 5A and B, A549 cells were arrested in
G1 phase. Upon serum addition, A549 cells were released from the
G0/G1 phase and gradually entered into S and G2/M phases. To
further confirm the results, western blot assays were performed to
analyze proliferating cell nuclear antigen (PCNA) and the
expression of HES5. We found that the expression of PCNA and cell
cycle regulator cyclin D1 were increased after serum stimulation in
A549 cells. The protein level of HES5 was also upregulated
(Fig. 5C and D). Thus, this result
suggested that HES5 might have a function as a positive regulator
of NSCLC cells in a cell cycle-dependent manner.
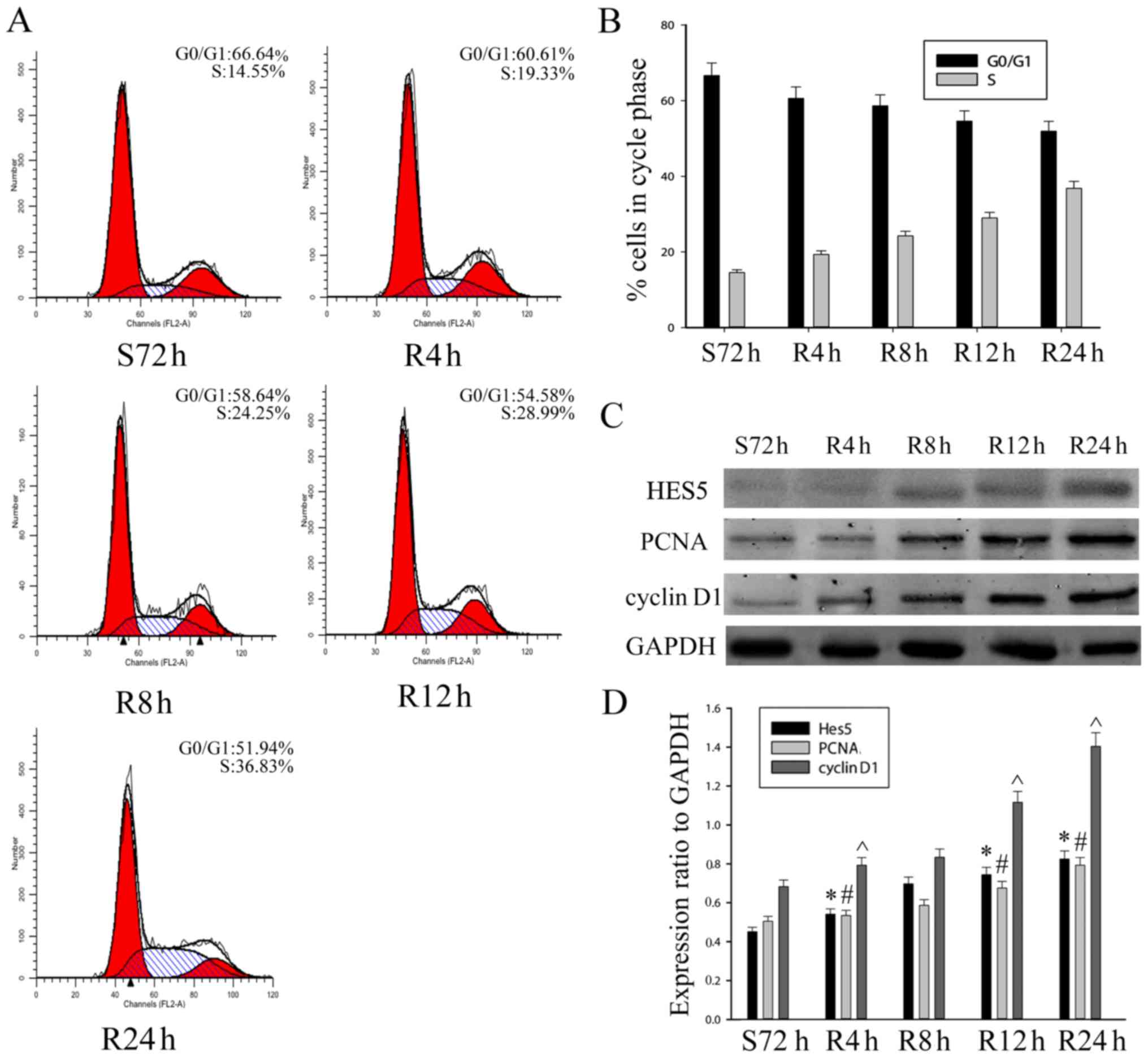 | Figure 5.Expression of HES5 promotes
proliferation of NSCLC cells. (A and B) Cells were synchronized at
G0/G1 and progressed into the cell cycle when serum was added for
S72 h, R6 h, R12 h, R24 h and R48 h. The experiment was conducted
by flow cytometry. (C and D) The S72 h A549 cells were released by
refeeding with serum, and cell lysates were prepared and analyzed
by western blot analysis using antibodies against HES5, cyclin D1,
PCNA and GAPDH (loading control). The bar chart demonstrates the
ratio of HES5, cyclin D1 and PCNA to GAPDH by densitometry. The
data are means ± SEM. *,#,^P<0.05, compared with
control cells serum starved for 48 h (S72 h). S, serum starvation;
R, serum release. |
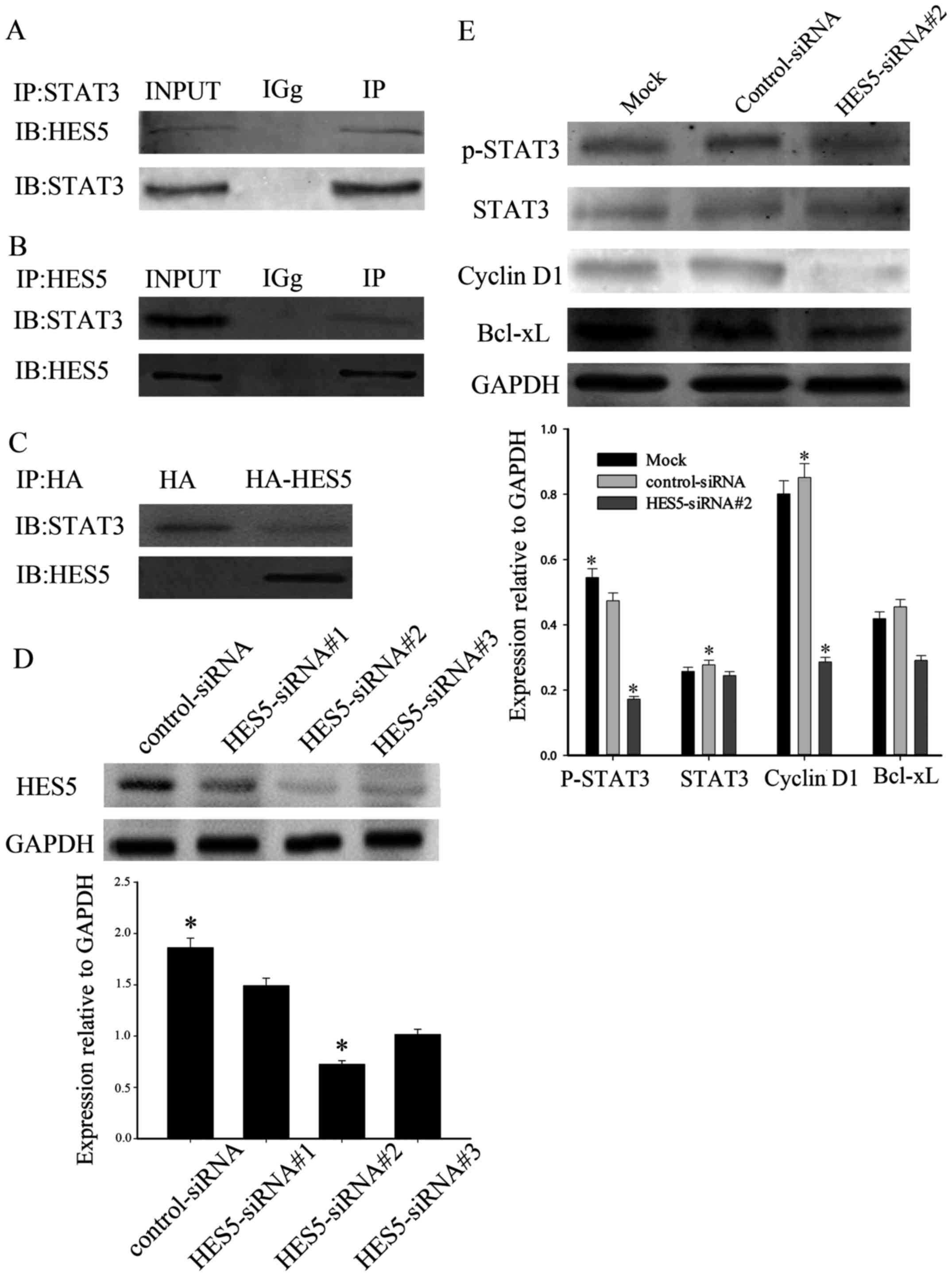
HES5 interacts with STAT3 and affects
activation of STAT3
Taken together, the results above demonstrated that
HES5 was obviously higher expressed in NSCLC cells. Cross-talk has
been reported between HES5 and STAT3 in hepatocytes and
hepatocellular carcinoma, which provided a reason to research the
mechanism of HES5 involved in NSCLC. There are many studies
suggesting that STAT3 was closely related with the proliferation
and growth of NSCLC. As a result, we performed immunoprecipitation
to further investigate the mechanism of HES5 on cellular
proliferation. Co-immunoprecipitation experiments were on
non-transfected A549 cell lysates with anti-HES5 or anti-STAT3
antibody. As shown in Fig. 6A and
B, HES5 could co-immunoprecipitate with the anti-STAT3
antibody, but not in control precipitation, indicating a naturally
occurring interaction between endogenous HES5 and STAT3 in
vivo, and vice versa. Furthermore, we explored whether the
HES5/STAT3 interaction can be detected exogenously. We transfected
A549 cells with HA-HES5 and anti-HA was used to precipitate STAT3.
It was shown that STAT3 co-immunoprecipitated specifically with
anti-HA antibody, which demonstrated that HES5 could interact with
STAT3 in vitro (Fig. 6C). To
further investigate the role of HES5 in cellular proliferation, we
analyzed HES5 in A549 cells using lentivirus-mediated RNA
interference. We transiently transfected A549 cells with
HES5-siRNA#1, HES5-siRNA#2, HES5-siRNA#3 or control siRNA. After 36
h, western blot analysis was used to evaluate the efficiency of
transfection. We found that HES5 protein levels decreased most
significantly in A549 cells infected with HES5-siRNA#2, which
compared with cells treated with other si-HES5 (Fig. 6D). Thus, we used HES5-siRNA#2 to
perform the following experiments. To evaluate whether loss of
STAT3 activation was involved in the effects induced by HES5
knockdown, we detected STAT3 and the expression of its target
genes. We observed that knockdown of HES5 inhibited the activation
of STAT3 and decreased the expression of cyclin D1 and Bcl-xL in
A549 cells transfected with HES5-siRNA#2 (Fig. 6E). Thus, these results suggest that
HES5 is responsible for tumor oncogenicity and regulate
proliferation in NSCLC via STAT3 pathway.
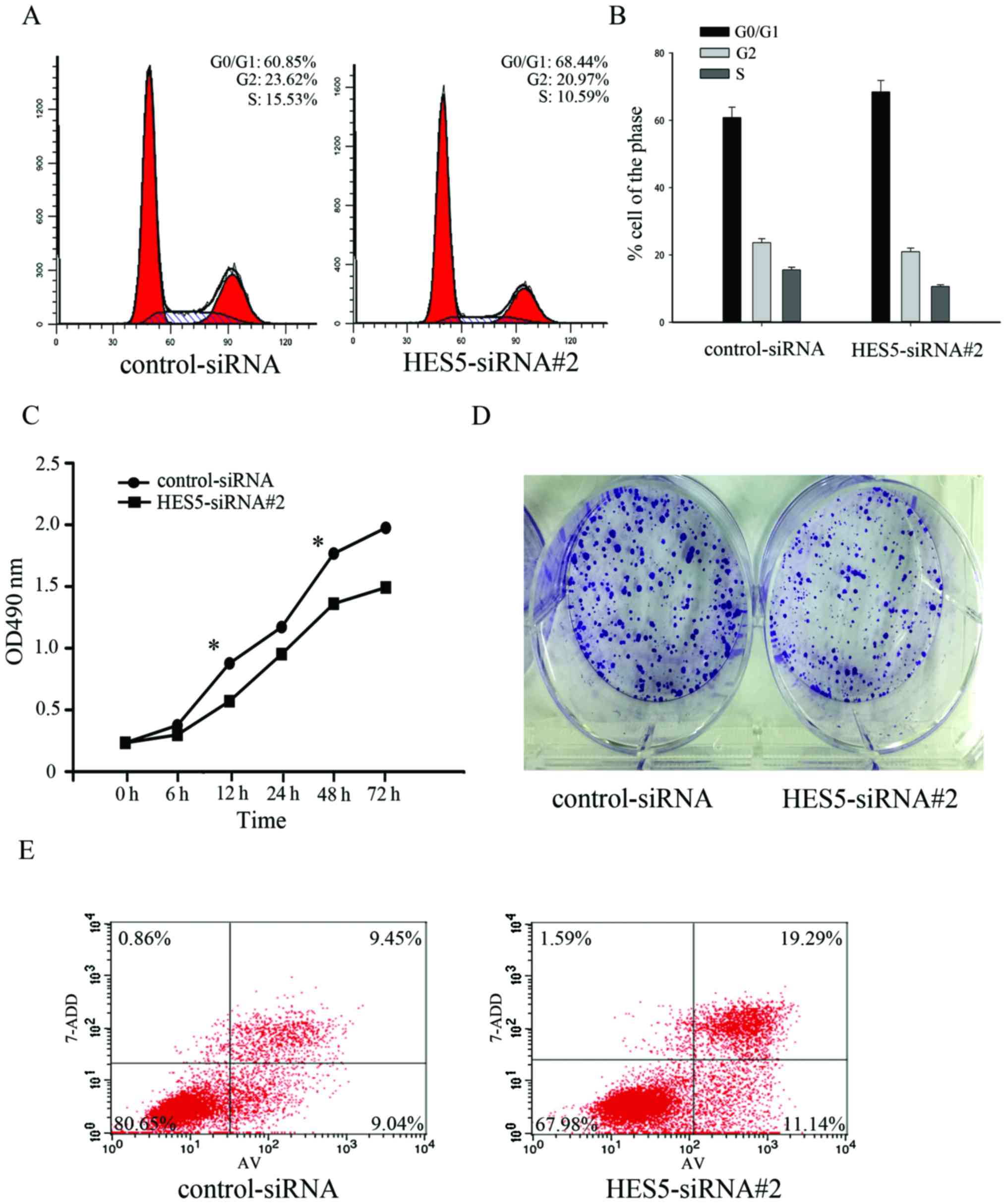
Knockdown of HES5 inhibits cell
proliferation in NSCLC cells
Next, we investigated the role of HES5 on cell
proliferation in NSCLC. We transfected A549 cells with
control-siRNA and HES5-siRNA#2, and the flow cytometric analyses of
the cell cycle showed an increase of cell number in the G0/G1 phase
from 60.85 to 68.44% and a decrease of cell number in the S phase
from 15.53 to 10.59% (Fig. 7A and
B). It indicates that downregulation of HES5 could slow down
the NSCLC cell cycle. In addition, a CCK-8 assay was used to
confirm the effect of HES5. Knockdown of HES5 can decrease the cell
proliferation (Fig. 7C). By colony
formation assay, we found that the rate of colony formation was
significantly attenuated in knockdown of HES5 (Fig. 7D). Furthermore, the Annexin
V-FITC/7-ADD assay also showed significant increase of cell death
after HES5 knockdown (Fig. 7E). In
conclusion, these results suggested that knockdown of HES5 might
inhibit G0/G1-S transition and delay the proliferation of A549
cells.
Discussion
The initiation and progression of NSCLC is
comprehensive, where complex alterations of oncogenes and tumor
suppressor genes are involved. Despite significant progress in
diagnostic and therapeutic strategy, the prognosis of NSCLC
patients remained unsatisfactory due to high incidence of tumor
recurrence, invasion and metastasis. Therefore, it is urgent to
identify novel therapeutic targets and develop new anticancer
therapies such as molecular-targeted drugs or antibodies. HES5,
together with HES1, is an important transcription factor that
involved in neural stem cells. HES1 has similar sequence with HES5,
which suggests that they are probably functionally related genes.
Previous studies showed that HES1 is involved in oncogenesis. HES1
is highly expressed in many cancer types, including colorectal
cancer cells, cervical carcinoma cells and colon cancer (19–21). A
study in human lung cancers also showed that HES1 expression was at
abundant level in several non-small cell lung cancer cell lines
without neuroendocrine features (22), but the roles of HES5 in NSCLC remain
unknown.
In the present study, we confirmed that HES5 might
be an important regulator in cell proliferation of NSCLC. Firstly,
we analyzed the HES5 expression in NSCLC tissues and the cell lines
using western blot analysis. The results revealed that HES5 was
upregulated in NSCLC tumor tissues and NSCLC cell line compared
with adjacent non-tumor ones (Fig.
1). Secondly, immunohistochemistry analysis of 114 NSCLC
samples were performed and showed that HES5 expression was
associated with tumor size, histological differentiation, clinical
stage, lymph node status and Ki-67 expression (Table I). Ki-67 is a useful marker of tumor
proliferative activity and only expressed during the active phases
of the cell cycle (23,24). In addition, multivariate analysis
indicated that HES5 could be an independent prognostic factor for
the survival of NSCLC patients (Table
II). Furthermore, Kaplan-Meier analysis showed that
overexpression of HES5 predicted poor survival (Fig. 4). We found that HES5 expression was
positively correlated with cell proliferation by serum starvation
and release assay (Fig. 5).
Knockdown of HES5 resulted in decreased rate of cell growth, colony
formation and alleviated cellular apoptosis. In addition, we
performed cell cycle analysis in NSCLC cells, and we found that
HES5 expression was increasingly upregulated during G1 to S phase,
while the proportion of cells in S phase was decreased in cells low
expressed of HES5 (Fig. 7).
Therefore, HES5 may be involved in the process of NSCLC
tumorigenesis.
Previous studies revealed that HES5 proteins
associated with STAT3 and JAK2, and facilitated complex formation
between STAT3 and JAK2, thus, promoting STAT3 phosphorylation and
activation (11,25). We demonstrated that HES5 interacted
with STAT3 in NSCLC cells, and knockdown of HES5 could inhibit the
activity of STAT3 and decrease the expression of the downstream
targets (Fig. 6). This suggested
that HES5 might affect the proliferation through STAT3 pathway.
However, further studies are needed to clarify the molecular
mechanisms of HES5 in NSCLC pathogenesis.
In summary, all these results showed that HES5 was
upregulated in NSCLC and promoted cell proliferation process
through the activation of STAT3. Therefore, HES5 might be a novel
molecular target for the diagnosis and therapy of NSCLC.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (no. 81472185).
References
|
1
|
Wang J, Wei H, Zhao B, Li M, Lv W, Lv L,
Song B and Lv S: The reverse effect of X-ray irradiation on
acquired gefitinib resistance in non-small cell lung cancer cell
line NCI-H1975 in vitro. J Mol Histol. 45:641–652. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu SV and Giaccone G: Lung cancer in
2013: Refining standard practice and admitting uncertainty. Nat Rev
Clin Oncol. 11:69–70. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Osada H and Takahashi T: Genetic
alterations of multiple tumor suppressors and oncogenes in the
carcinogenesis and progression of lung cancer. Oncogene.
21:7421–7434. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ohtsuka T, Sakamoto M, Guillemot F and
Kageyama R: Roles of the basic helix-loop-helix genes Hes1 and Hes5
in expansion of neural stem cells of the developing brain. J Biol
Chem. 276:30467–30474. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ohtsuka T, Ishibashi M, Gradwohl G,
Nakanishi S, Guillemot F and Kageyama R: Hes1 and Hes5 as notch
effectors in mammalian neuronal differentiation. EMBO J.
18:2196–2207. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hatakeyama J and Kageyama R: Notch1
expression is spatiotemporally correlated with neurogenesis and
negatively regulated by Notch1-independent Hes genes in the
developing nervous system. Cereb Cortex. 16:(Suppl 1). i132–i137.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Axelson H: The Notch signaling cascade in
neuroblastoma: Role of the basic helix-loop-helix proteins HASH-1
and HES-1. Cancer Lett. 204:171–178. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kunnimalaiyaan M, Yan S, Wong F, Zhang YW
and Chen H: Hairy enhancer of Split-1 (HES-1), a Notch1 effector,
inhibits the growth of carcinoid tumor cells. Surgery.
138:1137–1142; discussion 1142. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ström A, Arai N, Leers J and Gustafsson
JA: The hairy and enhancer of Split homologue-1 (HES-1) mediates
the proliferative effect of 17beta-estradiol on breast cancer cell
lines. Oncogene. 19:5951–5953. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yu HC, Qin HY, He F, Wang L, Fu W, Liu D,
Guo FC, Liang L, Dou KF and Han H: Canonical notch pathway protects
hepatocytes from ischemia/reperfusion injury in mice by repressing
reactive oxygen species production through JAK2/STAT3 signaling.
Hepatology. 54:979–988. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Haura EB, Zheng Z, Song L, Cantor A and
Bepler G: Activated epidermal growth factor receptor-Stat-3
signaling promotes tumor survival in vivo in non-small cell lung
cancer. Clin Cancer Res. 11:8288–8294. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Malanga D, De Marco C, Guerriero I,
Colelli F, Rinaldo N, Scrima M, Mirante T, De Vitis C, Zoppoli P,
Ceccarelli M, et al: The Akt1/IL-6/STAT3 pathway regulates growth
of lung tumor initiating cells. Oncotarget. 6:42667–42686.
2015.PubMed/NCBI
|
|
14
|
Xue Q, Zhou Y, Wan C, Lv L, Chen B, Cao X,
Ju G, Huang Y, Ni R and Mao G: Epithelial membrane protein 3 is
frequently shown as promoter methylation and functions as a tumor
suppressor gene in non-small cell lung cancer. Exp Mol Pathol.
95:313–318. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ji L, Ni T, Shen Y, Xue Q, Liu Y, Chen B,
Cui X, Lv L, Yu X, Cui Y, et al: Transformer 2β (Tra2β/SFRS10)
positively regulates the progression of NSCLC via promoting cell
proliferation. J Mol Histol. 45:573–582. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ni T, Mao G, Xue Q, Liu Y, Chen B, Cui X,
Lv L, Jia L, Wang Y and Ji L: Upregulated expression of ILF2 in
non-small cell lung cancer is associated with tumor cell
proliferation and poor prognosis. J Mol Histol. 46:325–335. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen J, Gu J, Feng J, Liu Y, Xue Q, Ni T,
Wang Z, Jia L, Mao G and Ji L: TAB3 overexpression promotes cell
proliferation in non-small cell lung cancer and mediates
chemoresistance to CDDP in A549 cells via the NF-kappaB pathway.
Tumour Biol. 37:3851–3861. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xue Q, Lv L, Wan C, Chen B, Li M, Ni T,
Liu Y, Liu Y, Cong X, Zhou Y, et al: Expression and clinical role
of small glutamine-rich tetratricopeptide repeat (TPR)-containing
protein alpha (SGTA) as a novel cell cycle protein in NSCLC. J
Cancer Res Clin Oncol. 139:1539–1549. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Weng MT, Tsao PN, Lin HL, Tung CC, Change
MC, Chang YT, Wong JM and Wei SC: Hes1 increases the invasion
ability of colorectal cancer cells via the STAT3-MMP14 pathway.
PLoS One. 10:e01443222015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu J, Lu WG, Ye F, Cheng XD, Hong D, Hu
Y, Chen HZ and Xie X: Hes1/Hes5 gene inhibits differentiation via
down-regulating Hash1 and promotes proliferation in cervical
carcinoma cells. Int J Gynecol Cancer. 20:1109–1116. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gao F, Huang W, Zhang Y, Tang S, Zheng L,
Ma F, Wang Y, Tang H and Li X: Hes1 promotes cell proliferation and
migration by activating Bmi-1 and PTEN/Akt/GSK3β pathway in human
colon cancer. Oncotarget. 6:38667–38680. 2015.PubMed/NCBI
|
|
22
|
Ito T, Udaka N, Yazawa T, Okudela K,
Hayashi H, Sudo T, Guillemot F, Kageyama R and Kitamura H: Basic
helix-loop-helix transcription factors regulate the neuroendocrine
differentiation of fetal mouse pulmonary epithelium. Development.
127:3913–3921. 2000.PubMed/NCBI
|
|
23
|
Kitamoto M, Nakanishi T, Kira S, Kawaguchi
M, Nakashio R, Suemori S, Kajiyama G, Asahara T and Dohi K: The
assessment of proliferating cell nuclear antigen
immunohistochemical staining in small hepatocellular carcinoma and
its relationship to histologic characteristics and prognosis.
Cancer. 72:1859–1865. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Del Gobbo A, Pellegrinelli A, Gaudioso G,
Castellani M, Marino F Zito, Franco R, Palleschi A, Nosotti M,
Bosari S, Vaira V, et al: Analysis of NSCLC tumour heterogeneity,
proliferative and 18F-FDG PET indices reveals Ki-67 prognostic role
in adenocarcinomas. Histopathology. 68:746–751. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kamakura S, Oishi K, Yoshimatsu T,
Nakafuku M, Masuyama N and Gotoh Y: Hes binding to STAT3 mediates
crosstalk between Notch and JAK-STAT signalling. Nat Cell Biol.
6:547–554. 2004. View
Article : Google Scholar : PubMed/NCBI
|