Introduction
Lung cancer is one of the most common cancers and is
a leading cause of mortality worldwide both in males and females.
The number of cancer-related deaths expected to occur in 2016 are
estimated based on the results obtained from 1998 through 2012 as
reported to the NCHS at state and national levels. Among males and
females, it is the second most commonly diagnosed cancer and the
leading cause of cancer-related deaths. In males, lung cancer is
expected to account for 14% (117,920) of the total cases and 27%
(5,920) of all cancer-related deaths in 2016. In females, lung
cancer is expected to account for 13% (106,470) of the total cases
and 26% (72,160) of all cancer-related deaths in 2016 (1). Approximately 80–85% of the newly
diagnosed cases of lung cancer are non-small cell lung cancer
(NSCLC). To date, the first-line therapy for advanced-stage NSCLC
is chemotherapy. The widespread use of early detection methods and
improvements in treatment, such as EGFR-TKIs (erlotinib) (2), gefitinib (3), afatinib (4), anaplastic lymphoma kinase inhibitor
(crizotinib) (5), ceritinib
(6), alectinib (7), anti-PD-1/PDL-1 immune checkpoint
inhibitor (MK-3475) (8), BMS-936558
(9), MPDL3280A (10) have led to a reduction in mortality
from lung cancer. Yet, it continues to be the leading cause of
cancer-related deaths. Better understanding of the molecules and
signaling pathways leading to lung cancer would facilitate the
development of more effective treatment strategies, with potential
improvements in the quality of life of patients. Thus,
identification of novel molecular mechanisms that lead to NSCLC
development and progression is still urgently needed.
The Testin gene was previously identified in a
common fragile site on chromosome 7q31.2 designated FRA7G. It is a
gene encoding a 421 amino-acid protein with three LIM domains.
There are three isoforms of human Testin, which differ in the size
of the 3-UTR encoded by exon 7 (11–13).
Testin mRNA is expressed in all normal human tissues, while low or
lack of Testin expression has been found in prostate cancer,
glioblastoma, endometrial carcinoma, ovarian, breast, uterine,
colon cancer, esophageal and gastric cancer, acute myelogenous and
acute lymphoblastic leukemia (ALL) and nasopharyngeal carcinoma
(14–30). Tatarelli et al observed lack
of expression in 22% of cancer cell lines and in 44% of the cell
lines derived from hematological malignancies. In most of these
cases the inactivation of Testin expression was due to methylation
of a CpG island. Analysis of the Testin coding region in 26 tumor
cell lines revealed three missense mutations (11). Other researchers also reported that
Testin expression is decreased or silenced partially by
hypermethylation and/or loss of heterozygosity in various human
cancers (16,18,26–29).
Testin is a novel focal adhesion protein with a role in cell
spreading. It interacts with a variety of cytoskeletal proteins,
including zyxin, mena, VASP, talin, and actin (31–33).
Yet, the potential role of Testin in the proliferation, invasion
and metastasis of NSCLC is still unknown. The aim of the present
study was to examine the relationship between the expression levels
of Testin and the proliferation and invasion of NSCLC cells in
vitro and tumor growth in NSCLC xenograft models in
vivo, in order to conduct a preliminary investigation into
whether Testin expression may be a suitable prognostic marker for
NSCLC in humans.
Materials and methods
Cell lines
The NSCLC cell lines LTEP-a-2, A549 and NCI-H1650
were obtained from Anhui Provincial Key Laboratory of Clinical
Basic Research on Respiratory Disease, The First Affiliated
Hospital of Bengbu Medical College. Human bronchial epithelial
cells (16HBE) were purchased from the Type Culture Collection of
the Chinese Academy of Sciences, Shanghai, China. The cell lines
were cultured in RPMI-1640 supplemented with 10% fetal bovine serum
(FBS) and 0.03% antibiotic-antimycotic (all from Gibco, Grand
Island, NY, USA) at 37°C in a 5% CO2 humidified
chamber.
Lentivirus transfection
Cancer cells (A549 and NCI-H1650 cells) were seeded
in a 96-well plate at a density of 3×104 cells/well. The
Testin overexpression vector and the empty vector (Cyagen
Biosciences Inc., Guangzhou, China) were used to transfect cells
using lentivirus transfection technique according to the
manufacturers protocol to establish Testin overexpression cell
lines and control cell lines. The total virus titer is
1×108 TU/ml. The virus titer was diluted with HBSS or
RPMI-1640 according to the cell number and the multiplicity of
infection. The appropriate multiplicity of infection was 40.
Furthermore, Polybrene (Sigma-Aldrich, St. Louis, MO, USA) was used
to significantly increase the viral transfection efficiency, which
plays an important role in the lentivirus transfection.
Semi-quantitative PCR analysis
Total RNA was extracted with TRIzol reagent
(Invitrogen Life Technologies, Carlsbad, CA, USA) according to the
manufacturers instructions. Total RNA was reverse transcribed to
cDNA in a 20 µl volume using a reverse transcription kit
(Invitrogen Life Technologies). Isoform 2 was used to design the
PCR primers. Primers designed and utilized for Testin were:
forward, 5-ACTGTGGCAGACATTACTGTGACA-3 and reverse,
5-GATAGCTATGGCTCGATACTTCTGGGTGC-3. The length of the Testin primer
was 440 bp. β-actin was used as an endogenous control for
quantitative DNA-PCR. Primers designed and utilized for β-actin
were listed as follows: forward, 5-TCACCAACTGGGACGACAT-3 and
reverse, 5-GCACAGCCTGGATAGCAAC-3. The length of the β-actin primer
was 192 bp. Annealing was performed at 72°C for Testin. All PCR
product electrophoreses were performed on a 2% agarose gel with
ethidium bromide and visualized using the Gel Imager system (Asia
Xingtai Mechanical and Electrical Equipment Co., Beijing, China).
The relative expression value of Testin mRNA is expressed as the
ratio between the target mRNA gray scale value and the β-actin gray
scale value. The experiments were repeated in triplicate to confirm
the findings.
Western blot analysis
Proteins were extracted using RIPA lysis buffer
(Beyotime, China) containing 0.1% phosphatase inhibitor cocktail
and protease inhibitor. The protein concentrations were determined
using the BCA protein assay kit (Beyotime, China). Equal amounts of
protein were separated by SDS-PAGE (Amresco, LLC, Solon, OH, USA),
electrotransferred to PVDF membranes (Biosharp, China) and blocked
in 5% non-fat dry milk.
The membranes were incubated overnight at 4°C with
the following primary antibodies: polyclonal goat anti-Testin
(sc-34737; 1:50; Santa Cruz Biotechnology, Inc., Santa Cruz, CA,
USA) and monoclonal mouse anti-β-actin (ab8226; 1:2,000; Abcam,
Cambridge, MA, USA) as control. Then, the membranes were washed
with Tris-buffered saline and Tween-20 (TBST) for three times, and
incubated with a 1:2,000 dilution of HRP-conjugated secondary
antibodies: rabbit anti-goat IgG (ab6741; Abcam) and goat
anti-mouse IgG (ab97023; Abcam) at room temperature for 1 h. The
immunoblots were visualized using a chemiluminescence detection kit
(Pierce Chemical Co., Rockford, IL, USA).
Flow cytometric analysis
Cells were seeded in a 6-well plate
(1×104 cells/well) and incubated in 3 ml RPMI-1640
medium supplemented with 10% FBS in 5% CO2 at 37°C. As
the cells grew to the logarithmic phase of growth, the cells in the
6-well plates were collected by digestion with 1 ml 0.25% trypsin.
After being washed with pre-cooled phosphate-buffered saline (PBS)
twice, the cells were re-suspended in 300 µl binding buffer and
mixed with 5 µl Annexin V-PE/7-AAD and 5 µl propidium iodide
successively, followed by incubation at room temperature in the
dark for 15 min. The apoptosis rate of the cells was detected on a
flow cytometer within 1 h according to the manufacturer's
instructions.
Cell proliferation assay
The MTT assay was used to assess cellular
proliferation. Cells were seeded in a 96-well plate at a density of
2×104 cells/well. Then the cells were incubated in 5%
CO2 at 37°C for 72 h. As the cells grew to 80%
confluence, the freshly prepared MTT solution (5 mg/ml) was added
to each well (20 µl/well), and then incubated for an additional 2
h. Subsequently, the supernatant was removed from the well and 150
µl DMSO was added. After shaking, the asorbance of each well at 490
nm was measured using a microplate reader.
Clonogenic assay
The survival and proliferation potential of the
cells were assessed using clonogenic assays. The cells were
trypsinized, counted, and seeded in a 6-cm plate at 500 cells/well.
After incubation for 2 weeks, the colonies were fixed with
paraformaldehyde and stained with Giemsa staining solution, and the
number of colonies containing more than 50 cells was scored.
Invasion assay
Six hundred microliters of balanced mixture of the
conditional medium from Matrigel fibroblasts and the complete
medium was added to the lower compartment as the chemotactic
factor. Serum-free RPMI-1640 with 1×105 cells was added
to the upper compartment of the chamber. At the indicated time, the
non-invasive cells in the upper compartment were removed with a
cotton swab. The cells in the lower compartment of the chamber were
counted under a light microscope for a minimum of 10 random visual
fields.
In vivo tumor xenograft models
Four-week old female BALB/c athymic nude mice
(Comparative Medicine Centre of Yangzhou University, China) were
housed in an environmentally controlled room (22±2°C, 40–60%
humidity and a 12-h light cycle). Cancer cells (1×106)
were subcutaneously inoculated into the fossa axillaris of mice at
5 weeks of age. The injection was made through the subcutaneous
layer of the cervicodorsal part of the animals. The growth of
primary tumors was monitored by measuring the tumor diameters for 5
weeks. Tumor length (L) and width (W) were measured twice a week
using a caliper, and tumor volume (V) was calculated by the
equation: V = (W2xL)/2. After 5 weeks, the mice were
scarificed under anesthesia, the tumor masses were removed, weighed
and fixed in 10% neutral buffered formaldehyde solution and
paraffin-embedded for histological analysis or preserved at
−80°C.
Immunohistochemistry
Tumor sections (3-µm) were cut from formalin-fixed
paraffin-embedded blocks and mounted on positive-charged slides.
The primary antibody was goat anti-Testin polyclonal antibody
(Santa Cruz Biotechnology). The paraffin sections were placed in a
xylene bath for 10 min to remove paraffin, and repeated again and
then placed in an ethanol gradient for rehydration. Antigen
retrieval was performed with EDTA (pH 8.0) repair solution in a
microwave, cooled to room temperature, treated with 3%
H2O2 for 10 min for inactivation of
endogenous peroxidase, rinsed with 1X PBST (0.1% Tween), incubated
with 5% rabbit serum at room temperature for 15 min, and then
incubated with primary antibody (1:100) at 4°C overnight. The
sections were then rinsed and incubated with biotin-labeled
secondary antibody (SP KIT; Beijing Zhongshan Golden Bridge
Biotechnology Co., Ltd., Beijing, China) at 37°C for 15 min, rinsed
in 1X PBST (0.1% Tween) and then incubated with horseradish
peroxidase (SP KIT; Beijing Zhongshan Golden Bridge Biotechnology
Co., Ltd.) at 37°C for 15 min. The sections were then treated with
DAB for 10 min and the reaction was terminated. H&E staining
was performed. The sections were fixed with hydrochloride ethanol
and then mounted for analysis. All sections were observed in at
least five areas at a magnification of ×400 by at least two
investigators in a blinded manner. Cytoplasm and nuclei were
counterstained with hematoxylin solution. The total number of cells
and positive cells were counted and the staining was scored as the
percentages of positive cells: 0 (no staining) for specimens with
positive cells ≤5%; 1 (weak staining) for specimens with positive
cells >5% and ≤25%; 2 (moderate staining) for specimens with
positive cells >25% and ≤50%; 3 (strong staining) for specimens
with positive cells >50%. Specimens with scores of ≤1 were
regarded as negative; specimens with scores of >1 were regarded
as positive.
Statistical analysis
Data are expressed as the mean and standard
deviation (SD), and statistical analysis was performed using
software SPSS version 18.0. The differences among groups were
analyzed by one-way ANOVA followed by Bonferronis multiple
comparison test. Differences were considered significant at
P<0.05.
Results
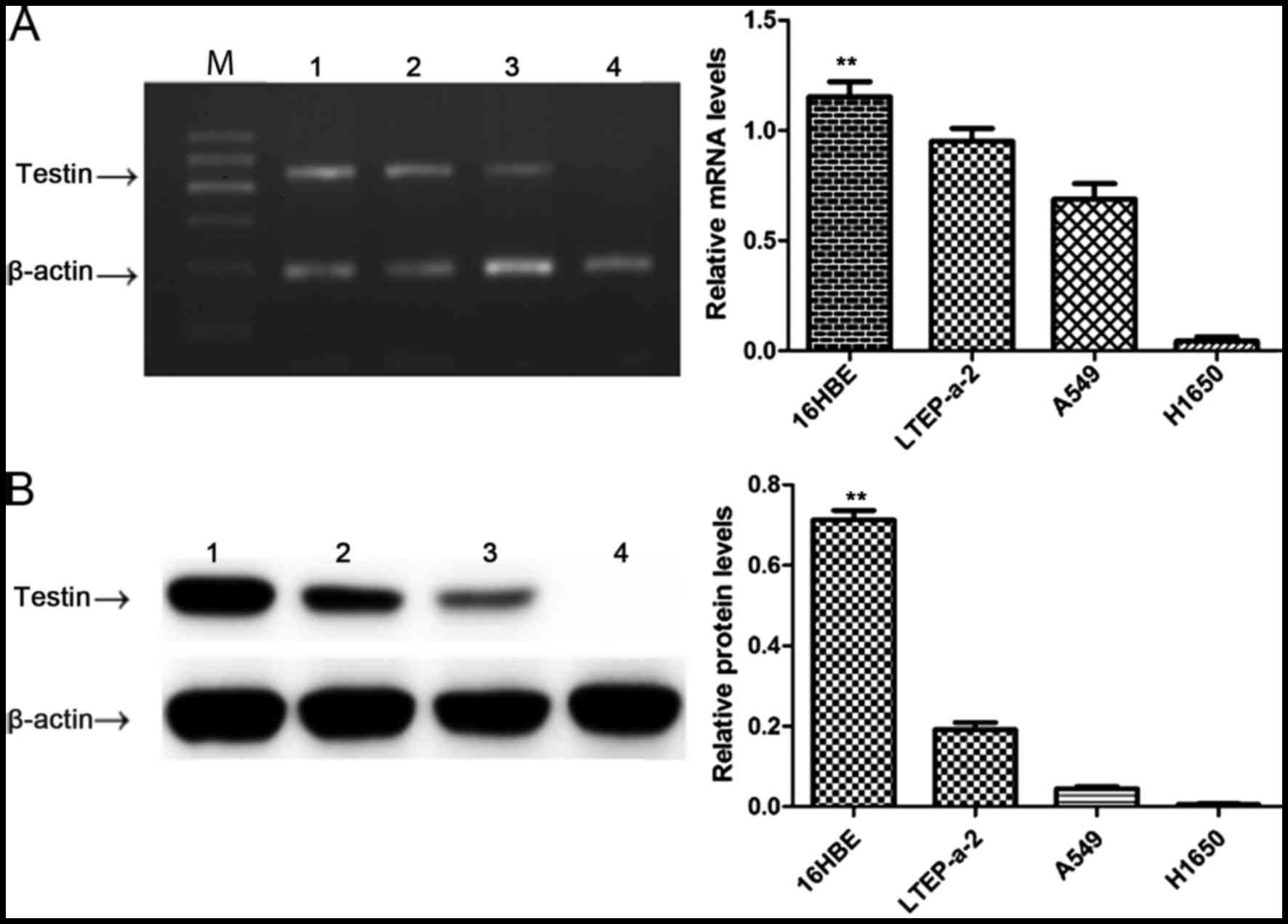
Testin mRNA and protein are reduced in
the NSCLC cell lines
In order to clarify the relationship between Testin
and NSCLC, we first compared the expression level of Testin in
NSCLC cell lines LTEP-a-2, A549 and NCI-H1650 and 16HBE cells by
semi-quantitative PCR and western blot analysis. The Testin mRNA
(Fig. 1A) and protein levels
(Fig. 1B) were significantly
reduced in the LTEP-a-2, A549 and NCI-H1650 cells compared with
these levels in the 16HBE cells (P<0.01), suggesting an
association between decreased expression of Testin mRNA and protein
levels and the carcinogenesis of NSCLC.
Testin gene inhibits proliferation,
invasion and colony formation of NSCLC cells and induces cancer
cell apoptosis
In order to explore additional functions of Testin
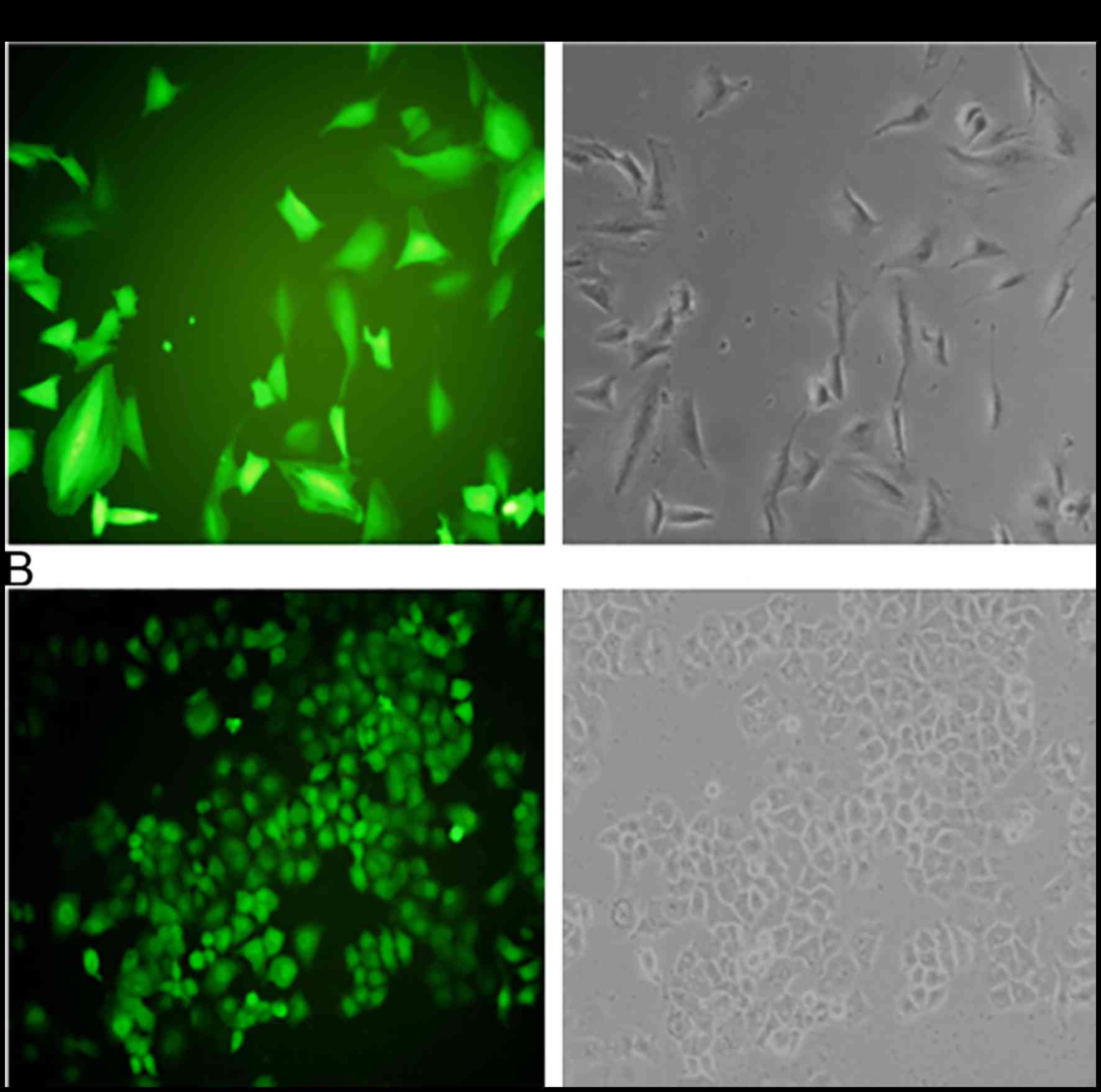
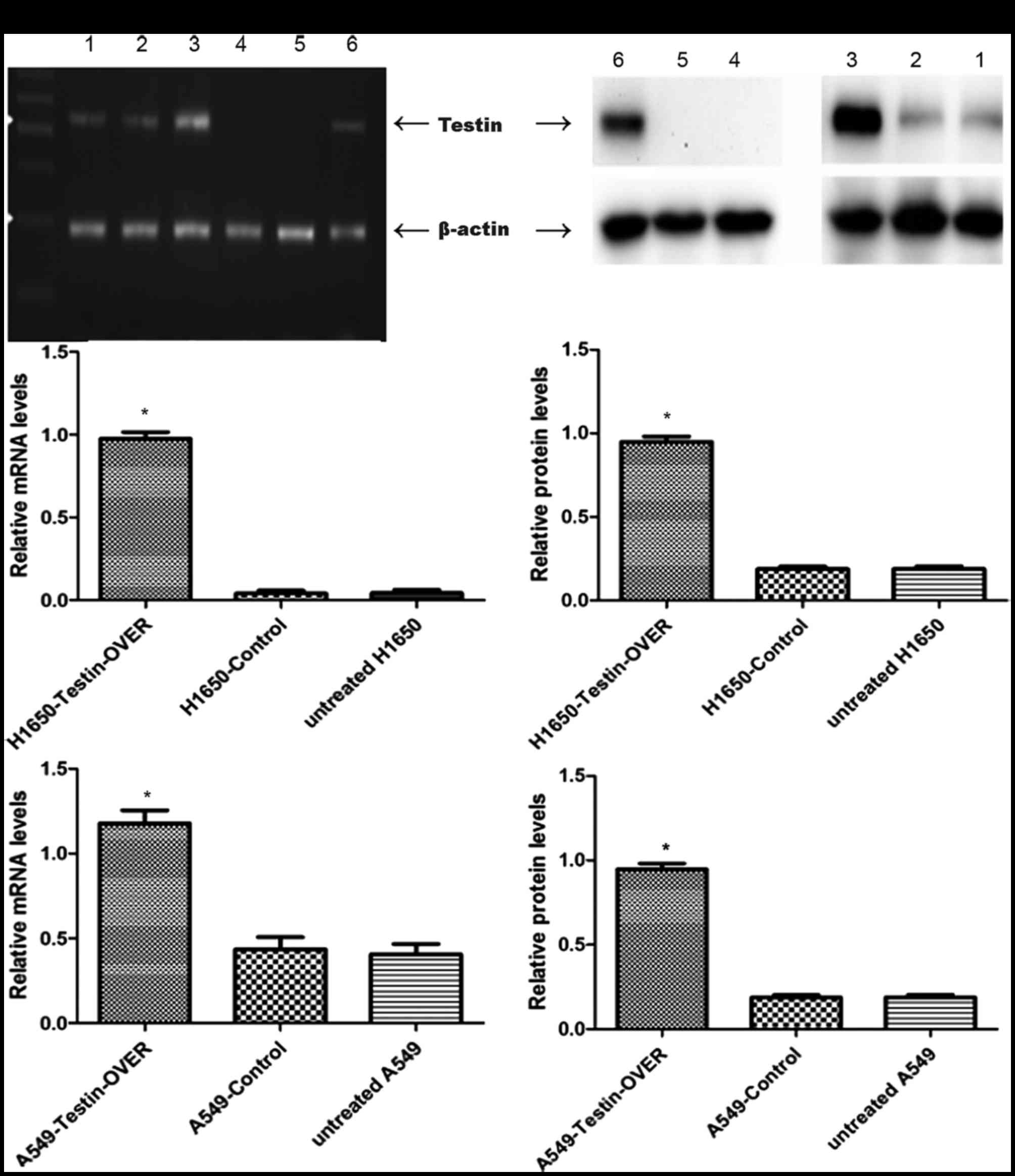
in NSCLC, we used the NSCLC cell lines A549 and NCI-H1650 to
establish stable cells that constitutively overexpressed the Testin
mRNA and protein (Fig. 2). The
transfection efficiency was confirmed using semi-quantitative PCR
(Fig. 3A)and western blot analysis
(Fig. 3B). A549 and NCI-H1650 cells
transfected with the Testin overexpression vector showed
significantly increased Testin mRNA levels and protein expression
compared with the control cells.
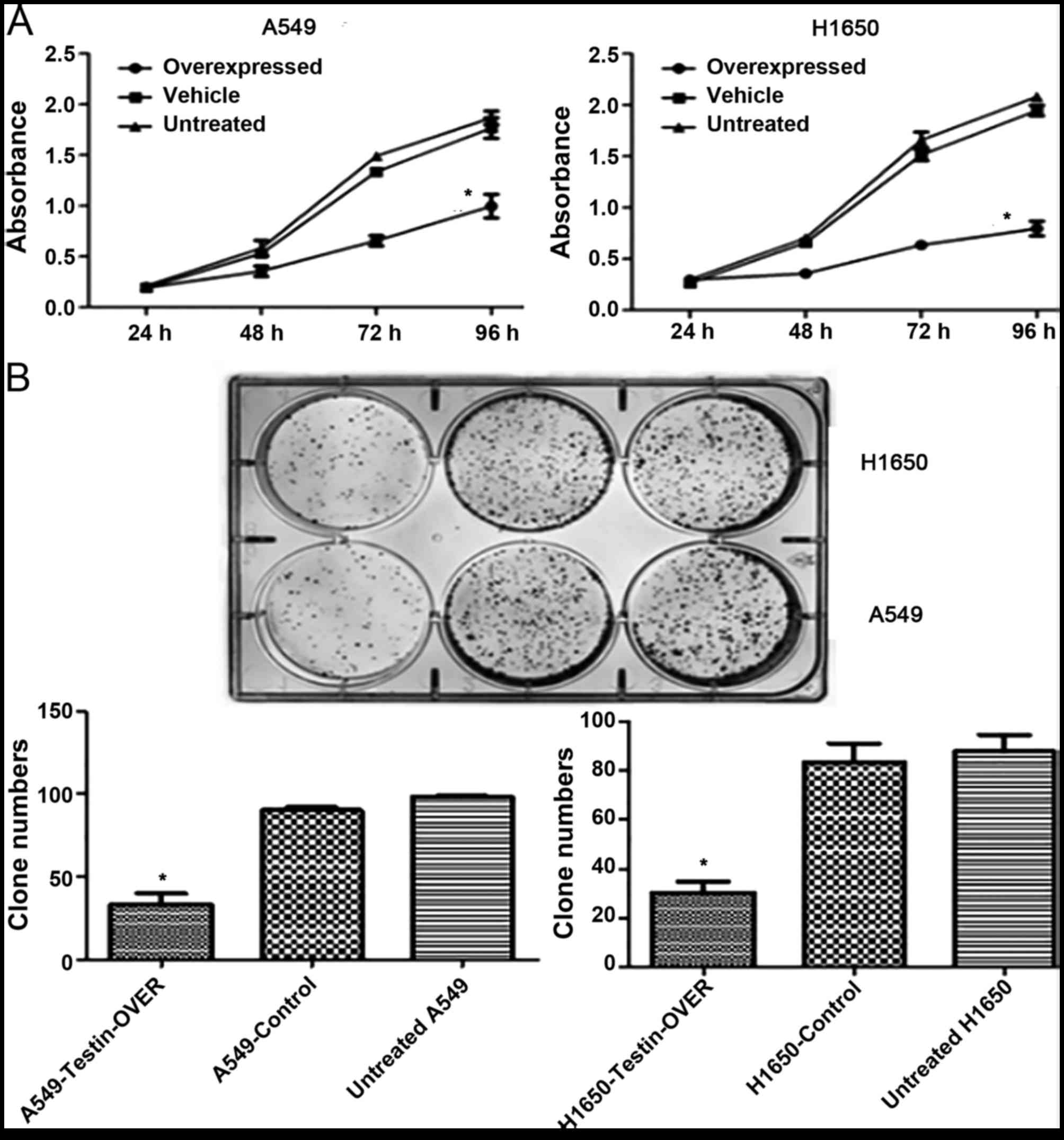
We next investigated the effect of Testin
overexpression on cell proliferation. The MTT assay showed that
overexpression of Testin significantly inhibited proliferation of
the A549 and NCI-H1650 cells compared with control cells
(P<0.05) (Fig. 4A). Clonogenic
assay showed that overexpression of Testin in the A549 and
NCI-H1650 cells markedly reduced colony formation efficiency
compared with the control cells (P<0.05) (Fig. 4B). Invasion assay showed that
overexpression of Testin significantly inhibited invasion of the
A549 and NCI-H1650 cells compared with the control cells
(P<0.05) (Fig. 4C). Flow
cytometric analysis showed that overexpression of the Testin gene
in the A549 and NCI-H1650 cells significantly induced cancer cell
apoptosis compared with the control cells (P<0.05) (Fig. 4D). These results suggest that Testin
plays a significant role in inhibiting the proliferation, invasion
and colony formation of NSCLC cells.
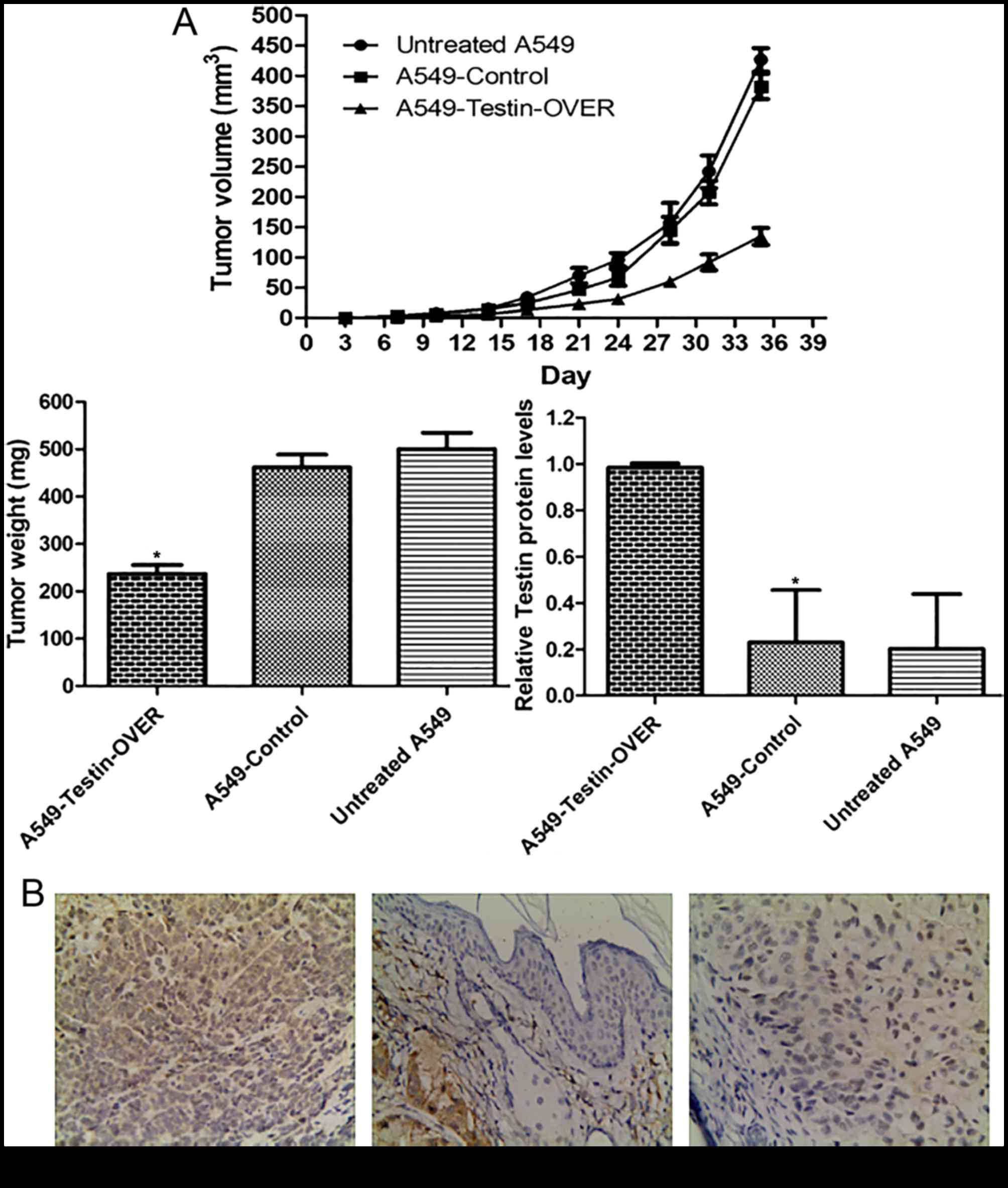
Testin gene inhibits NSCLC cell
xenograft formation and growth in vivo
In order to investigate the tumor-suppressing
function of Testin in vivo, the Testin-overexpressing A549
cells (A549-Testin-OVER), A549-Control cells and untreated A549
cells (1×106) were subcutaneously inoculated into the
fossa axillaris of 5-week-old female BALB/c athymic nude mice.
During the process, we found that the mice injected with the
A549-Testin-OVER cells formed tumors later than those in the
control groups. Whereas large tumors were formed in the mice
injected with the control cells within 5 weeks, tumor growth was
greatly reduced in the mice injected with the A549-Testin-OVER
cells (Fig. 5A).
After sacrifice at 5 weeks, the tumor masses were
removed, weighed and fixed in 10% neutral buffered formaldehyde
solution. Tumors were stained for Testin and representative images
are shown in Fig. 5B. The
A549-Testin-OVER group (98.32±1.76%) had higher Testin expression
while the A549-control (22.92±21.46%) and untreated A549 group
(20.14±22.5%) had lower Testin expression (P<0.05). These data
showed that Testin plays a critical role in the inhibition of NSCLC
cell xenograft formation and growth in vivo.
Discussion
It has been established that Testin is a candidate
human tumor-suppressor gene in several types of cancers, including
prostate, ovarian, breast and gastric cancer. But its role in NSCLC
remains unknown. Our study is the first attempt to elucidate the
tumor-suppressor role of Testin in the proliferation, invasion and
colony formation of NSCLC cells in in vitro models and in
the inhibition of NSCLC cell xenograft formation and growth in
in vivo models. Testin encodes a protein containing a PET
domain at the NH2-terminus, which is involved in protein-protein
interactions, and three LIM domains at the COOH-terminus. The LIM
domain is also a common protein-protein interaction motif that was
originally discovered in the products of the lin-11, Isl-1, and
mec-3 genes (34–37). One LIM domain consists of a loosely
conserved cysteine-rich consensus sequence including two separate
zinc fingers. Testin protein can also be a component of focal
adhesions and cell junctions, which can interact with other
cytoskeleton-associated proteins, such as talin, mena,
vasodilator-stimulated phosphoprotein, and actin. Previous studies
demonstrated that Testin inhibited the growth of breast and uterine
as well as ovarian cancer cell proliferation through
caspase-dependent and caspase-independent apoptosis (18,19).
Downregulation of Testin has been reported to have a significant
association with highly aggressive breast tumor subtypes, such as
triple-negative and luminal B tumors, along with the prognostic
relevance of nuclear expression of survivin (30). Weeks et al discovered that
100% of the ALL samples (n=20) were methylated at the Testin
promoter, whereas the matched remission (n=5), normal bone marrow
(n=6) and normal PBL (n=5) samples were unmethylated. Expression of
Testin in hyperdiploid, TEL-AML+, BCR-ABL+,
and E2A-PBX+ subtypes of B lineage ALL was markedly
reduced compared to that in normal bone marrow progenitor cells and
in B cells. In addition, Testin methylation and silencing was
demonstrated in 9 out of 10 independent B ALL propagated as
xenografts in NOD/SCID mice. Thus, Testin is the most frequently
methylated and silenced gene yet reported in ALL (25). Zhu et al further confirmed
that the Testin gene inhibited invasion, metastasis, and
angiogenesis through miR-29b-mediated MMP-2 inhibition in breast
cancer (22).
In the present study, firstly we identified that
Testin expression was reduced in NSCLC cell lines LTEP-a-2, A549
and NCI-H1650 compared with that in 16HBE cells, which implied that
Testin is a candidate tumor-suppressor gene in NSCLC. Secondly, to
further explore the detailed tumor-suppressor function of Testin,
we used the NSCLC cell lines A549 and NCI-H1650 to establish stable
cells that constitutively overexpressed the Testin mRNA and
protein. The transfection efficiency was confirmed using
semi-quantitative PCR and western blot analysis. We found that
overexpression of Testin significantly inhibited proliferation,
invasion, and colony formation in NSCLC cell lines. In tumor
xenograft models, Testin also markedly inhibited lung cancer cell
xenograft formation and growth. These data further support the
tumor suppressor role of Testin in NSCLC. These findings imply that
Testin is possibly an individual therapeutic target for NSCLC
patients. These results require further validation in larger
cohorts. Our present results further confirmed that Testin could
significantly inhibit cancer cell proliferation and invasion, but
the possible suppressing mechanism of Testin gene and its role in
angiogenesis of lung cancer are still unknown. Angiogenesis has
been reported to be essential for tumor metastasis (38), thus in subsequent studies we will
examine the effect of Testin on angiogenesis, which partly
contributes to the metastasis of NSCLC. Furthermore, Ki-67 is a
nuclear located protein that is closely linked to cell
proliferation. It is present in all active phases of the cell
cycle, but absent from resting cells, thus, indicating the
proliferating cell fraction (39).
Ki-67 is a prognostic biomarker in several tumor entities, for
example, breast cancer, lymphoma, neuroendocrine neoplasia and
NSCLC (40–45). The Ki-67 index could also be
employed to investigate the association between Testin expression
and NSCLC cell proliferation.
To the best of our knowledge, this is the first
study to indicate the potential role of Testin in the occurrence
and development of NSCLC. Expression of Testin was generally lower
in NSCLC cell lines compared with that noted in human bronchial
epithelial cells. Our findings showed that Testin plays a
significant role in the proliferation, invasion and colony
formation of NSCLC cells and in the inhibition of NSCLC cell
xenograft formation and growth. Testin is a potential therapeutic
target for NSCLC patients.
Acknowledgements
The authors thank the staff at the Department of
Respiration, The First Affiliated Hospital of Bengbu Medical
College and Anhui Provincial Key Laboratory of Clinical Basic
Research on Respiratory Disease, The First Affiliated Hospital of
Bengbu Medical College. This study was supported by grants from the
Natural Science Fund of the Education Department of Anhui Province
(no. KJ2014A158).
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
De Grève J, Van Meerbeeck J, Vansteenkiste
JF, Decoster L, Meert AP, Vuylsteke P, Focan C, Canon JL, Humblet
Y, Berchem G, et al: Prospective evaluation of first-line erlotinib
in advanced non-small cell lung cancer (NSCLC) carrying an
activating EGFR mutation: A multicenter academic phase II study in
Caucasian patients (FIELT). PLoS One. 11:e01475992016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kazandjian D, Blumenthal GM, Yuan W, He K,
Keegan P and Pazdur R: FDA Approval of gefitinib for the treatment
of patients with metastatic EGFR mutation-positive non-small cell
lung cancer. Clin Cancer Res. 22:1307–1312. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang JC, Hirsh V, Schuler M, Yamamoto N,
OByrne KJ, Mok TS, Zazulina V, Shahidi M, Lungershausen J, Massey
D, et al: Symptom control and quality of life in LUX-Lung 3: A
phase III study of afatinib or cisplatin/pemetrexed in patients
with advanced lung adenocarcinoma with EGFR mutations. J Clin
Oncol. 31:3342–3350. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shaw AT, Kim DW, Nakagawa K, Seto T, Crinó
L, Ahn MJ, De Pas T, Besse B, Solomon BJ, Blackhall F, et al:
Crizotinib versus chemotherapy in advanced ALK-positive lung
cancer. N Engl J Med. 368:2385–2394. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shaw AT, Kim DW, Mehra R, Tan DS, Felip E,
Chow LQ, Camidge DR, Vansteenkiste J, Sharma S, De Pas T, et al:
Ceritinib in ALK-rearranged non-small-cell lung cancer. N Engl J
Med. 370:1189–1197. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ou SH, Ahn JS, De Petris L, Govindan R,
Yang JC, Hughes B, Lena H, Moro-Sibilot D, Bearz A, Ramirez SV, et
al: Alectinib in crizotinib-refractory ALK-rearranged
non-small-cell lung cancer: A phase II global study. J Clin Oncol.
34:661–668. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rizvi NA, Mazières J, Planchard D,
Stinchcombe TE, Dy GK, Antonia SJ, Horn L, Lena H, Minenza E,
Mennecier B, et al: Activity and safety of nivolumab, an anti-PD-1
immune checkpoint inhibitor, for patients with advanced, refractory
squamous non-small-cell lung cancer (CheckMate 063): A phase 2,
single-arm trial. Lancet Oncol. 16:257–265. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Garon EB, Leighl NB, Rizvi NA,
Blumenschein GR, Balmanoukian AS, Eder JP, Goldman JW, Hui R, Soria
JC, Gangadhar TC, et al: Safety and clinical activity of MK-3475 in
previously treated patients with non-small cell lung cancer. J Clin
Oncol. 32:80202014.
|
|
10
|
Casaluce F, Sgambato A, Sacco PC,
Palazzolo G, Maione P, Rossi A, Ciardiello F and Gridelli C:
Emerging drugs targeting PD-1 and PD-L1: Reality or hope? Expert
Opin Emerg Drugs. 19:557–569. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tatarelli C, Linnenbach A, Mimori K and
Croce CM: Characterization of the human TESTIN gene localized in
the FRA7G region at 7q31.2. Genomics. 68:1–12. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tobias ES, Hurlstone AF, MacKenzie E,
McFarlane R and Black DM: The TES gene at 7q31.1 is methylated in
tumours and encodes a novel growth-suppressing LIM domain protein.
Oncogene. 20:2844–2853. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mruk DD and Cheng CY: Rat and mouse
testicular testin is different from the human tumor suppressor gene
TESTIN (Tes): Authors response to the letter of Dr. S. Kapoor.
Spermatogenesis. 2:3052012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Drusco A, Zanesi N, Roldo C, Trapasso F,
Farber JL, Fong LY and Croce CM: Knockout mice reveal a tumor
suppressor function for Testin. Proc Natl Acad Sci USA.
102:10947–10951. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chêne L, Giroud C, Desgrandchamps F,
Boccon-Gibod L, Cussenot O, Berthon P and Latil A: Extensive
analysis of the 7q31 region in human prostate tumors supports TES
as the best candidate tumor suppressor gene. Int J Cancer.
111:798–804. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mueller W, Nutt CL, Ehrich M,
Riemenschneider MJ, von Deimling A, van den Boom D and Louis DN:
Downregulation of RUNX3 and TES by hypermethylation in
glioblastoma. Oncogene. 26:583–593. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gu Z, Ding G, Liang K, Zhang H, Guo G,
Zhang L and Cui J: TESTIN suppresses tumor growth and invasion via
manipulating cell cycle progression in endometrial carcinoma. Med
Sci Monit. 20:980–987. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Qiu H, Zhu J, Yuan C, Yan S, Yang Q and
Kong B: Frequent hypermethylation and loss of heterozygosity of the
testis derived transcript gene in ovarian cancer. Cancer Sci.
101:1255–1260. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sarti M, Sevignani C, Calin GA, Aqeilan R,
Shimizu M, Pentimalli F, Picchio MC, Godwin A, Rosenberg A, Drusco
A, et al: Adenoviral transduction of Testin gene into breast and
uterine cancer cell lines promotes apoptosis and tumor reduction in
vivo. Clin Cancer. 11:806–813. 2005.
|
|
20
|
Ohkouchi S, Kawamoto N, Koga M, Sakanashi
F, Shichijo S, Saijo Y, Nukiwa T, Itoh K and Yamada A:
Identification of a CTL-directed epitope encoded by an intron of
the putative tumor suppressor gene Testin of the common fragile
site 7G region: A peptide vaccine candidate for HLA-B52+
and HLA-62+ cancer patients. Eur J Immunol.
33:2964–2973. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Griffith E: Using RNA interference to
knock down the adhesion protein TES. Methods Mol Biol. 370:97–108.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhu J, Li X, Kong X, Moran MS, Su P,
Haffty BG and Yang Q: Testin is a tumor suppressor and prognostic
marker in breast cancer. Cancer Sci. 103:2092–2101. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Long JG, Zhang CQ and Zhao L: Expression
of Testin in human colorectal carcinoma and its clinical
significance. Chin Oncol. 19:428–432. 2009.(In Chinese).
|
|
24
|
Yu H, Ling TS, Shu QW and Shi RH: The
association of TESTIN and Caspase-3 protein expressions with
clinicopathological features and prognosis of esophageal squamous
cell carcinoma. Chin J Dig. 30:47–52. 2010.
|
|
25
|
Weeks RJ, Ludgate JL, LeMée G and Morison
IM: TESTIN induces rapid death and suppresses proliferation in
childhood B acute lymphoblastic leukaemia cells. PLoS One.
11:e01513412016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dong R, Pu H, Wang Y, Yu J, Lian K and Mao
C: TESTIN was commonly hypermethylated and involved in the
epithelial-mesenchymal transition of endometrial cancer. APMIS.
123:394–400. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Huang W, Weng DS, Pan ZZ, Pan K, Ding PR,
Zhou J, Wang H, Zhang HK, Li JJ and Xia JC: Expression and clinical
significance of TESTIN in primary gastric cancer. Ai Zheng.
27:984–988. 2008.(In Chinese). PubMed/NCBI
|
|
28
|
Weeks RJ, Kees UR, Song S and Morison IM:
Silencing of TESTIN by dense biallelic promoter methylation is the
most common molecular event in childhood acute lymphoblastic
leukaemia. Mol Cancer. 9:1632010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhong Z, Zhang F and Yin SC: Effects of
TESTIN gene expression on proliferation and migration of the 5-8F
nasopharyngeal carcinoma cell line. Asian Pac J Cancer Prev.
16:2555–2559. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sarti M, Pinton S, Limoni C, Carbone GM,
Pagani O, Cavalli F and Catapano CV: Differential expression of
testin and survivin in breast cancer subtypes. Oncol Rep.
30:824–832. 2013.PubMed/NCBI
|
|
31
|
Garvalov BK, Higgins TE, Sutherland JD,
Zettl M, Scaplehorn N, Köcher T, Piddini E, Griffiths G and Way M:
The conformational state of Tes regulates its zyxin-dependent
recruitment to focal adhesions. J Cell Biol. 161:33–39. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Coutts AS, MacKenzie E, Griffith E and
Black DM: TES is a novel focal adhesion protein with a role in cell
spreading. J Cell Sci. 116:897–906. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Griffith E, Coutts AS and Black DM: RNAi
knockdown of the focal adhesion protein TES reveals its role in
actin stress fibre organisation. Cell Motil Cytoskeleton.
60:140–152. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Freyd G, Kim SK and Horvitz HR: Novel
cysteine-rich motif and homeodomain in the product of the
Caenorhabditis elegans cell lineage gene lin-11. Nature.
344:876–879. 1990. View
Article : Google Scholar : PubMed/NCBI
|
|
35
|
Karlsson O, Thor S, Norberg T, Ohlsson H
and Edlund T: Insulin gene enhancer binding protein Isl-1 is a
member of a novel class of proteins containing both a homeo- and a
Cys-His domain. Nature. 344:879–882. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Michelsen JW, Sewell AK, Louis HA, Olsen
JI, Davis DR, Winge DR and Beckerle MC: Mutational analysis of the
metal sites in an LIM domain. J Biol Chem. 269:11108–11113.
1994.PubMed/NCBI
|
|
37
|
Dawid IB, Toyama R and Taira M: LIM domain
proteins. C R Acad Sci III. 318:295–306. 1995.PubMed/NCBI
|
|
38
|
Weis SM and Cheresh DA: Tumor
angiogenesis: Molecular pathways and therapeutic targets. Nat Med.
17:1359–1370. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
39
|
Scholzen T and Gerdes J: The Ki-67
protein: From the known and the unknown. J Cell Physiol.
182:311–322. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Stuart-Harris R, Caldas C, Pinder SE and
Pharoah P: Proliferation markers and survival in early breast
cancer: A systematic review and meta-analysis of 85 studies in
32,825 patients. Breast. 17:323–334. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
He X, Chen Z, Fu T, Jin X, Yu T, Liang Y,
Zhao X and Huang L: Ki-67 is a valuable prognostic predictor of
lymphoma but its utility varies in lymphoma subtypes: Evidence from
a systematic meta-analysis. BMC Cancer. 14:1532014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
van Velthuysen ML, Groen EJ, van der Noort
V, van de Pol A, Tesselaar ME and Korse CM: Grading of
neuroendocrine neoplasms: Mitoses and Ki-67 are both essential.
Neuroendocrinology. 100:221–227. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Berghoff AS, Ilhan-Mutlu A, Wöhrer A,
Hackl M, Widhalm G, Hainfellner JA, Dieckmann K, Melchardt T, Dome
B, Heinzl H, et al: Prognostic significance of Ki67 proliferation
index, HIF1 alpha index and microvascular density in patients with
non-small cell lung cancer brain metastases. Strahlenther Onkol.
190:676–685. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Del Gobbo A, Pellegrinelli A, Gaudioso G,
Castellani M, Marino F Zito, Franco R, Palleschi A, Nosotti M,
Bosari S, Vaira V, et al: Analysis of NSCLC tumour heterogeneity,
proliferative and 18F-FDG PET indices reveals Ki67 prognostic role
in adenocarcinomas. Histopathology. 68:746–751. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ji Y, Zheng M, Ye S, Chen J and Chen Y:
PTEN and Ki67 expression is associated with clinicopathologic
features of non-small cell lung cancer. J Biomed Res. 28:462–467.
2014.PubMed/NCBI
|