Introduction
Hepatocellular carcinoma (HCC) is one of the most
prevalent neoplasms and the second leading cause of cancer-related
mortality worldwide (1). Despite
the application of surgical resection, ablation, embolization and
other therapeutic methods, the overall prognosis of patients with
HCC remains poor and the incidence of HCC is increasing annually
(2,3). Therefore, the underlying mechanisms
that promote the pathogenesis of this deadly disease must be
further investigated.
Metabolic reprogramming has long been linked to
cancer. One of the classical theories of metabolic abnormality in
tumors is the Warburg effect, which describes increased glycolysis
even under normal oxygen conditions (4). The shift of the metabolic pathway from
oxidative phosphorylation to glycolysis is more rapid to meet the
demands necessary for the process of tumor progression. However, it
is inefficient to generate ATP by consuming units of glucose under
glycolysis conditions (5).
Therefore, transformed cells need to uptake more glucose than
normal cells. The glucose transporter 1 (GLUT1) isoform is a key
rate-limiting protein in the transport of glucose and is
overexpressed in HCC (6). However,
the mechanism underlying increased glycolysis by targeting GLUT1 in
HCC is unknown.
The Forkhead box M1 (FOXM1) transcriptional factor
is a member of the Forkhead family that is involved in cell
proliferation and cell cycle progression. The expression of FOXM1
is frequently upregulated in many solid tumors (7). The overexpression of FOXM1 promotes
proliferation, invasion and epithelial-mesenchymal transition, and
is associated with a poor prognosis in HCC (8–10).
Recent studies also revealed that FOXM1 promotes glucose
consumption, lactate production and increased lactate dehydrogenase
A (LDHA) activity in different cells, suggesting a pivotal role for
FOXM1 in the regulation of cell aerobic glycolysis (11,12).
However, a mechanistic understanding of the involvement of FOXM1 in
HCC metabolism is lacking.
In the present study, we demonstrated that high
expression of FOXM1 promotes the reprogramming of glucose
metabolism in human HCC cells through the transactivation of GLUT1
expression, which results in the malignant progression of liver
cancer.
Materials and methods
Cell culture
All human HCC cell lines (HepG2, Hep3B, MHCC-97H and
SMMC-7721) used in the present study were obtained from the
Shanghai Cellular Biology Center (Shanghai, China). HCC cells were
cultured in Dulbeccos modified Eagles medium (DMEM) culture-medium
supplemented with 10% fetal bovine serum (FBS; HyClone, Logan, UT,
USA), penicillin (100 U/ml) and streptomycin (100 µg/ml) at 37̊C in
an atmosphere of humidified air containing 5% CO2.
Patient samples
In the present study, 100 samples were obtained at
the time of surgery at the Department of Hepatobiliary Surgery,
Xijing Hospital between 2005 and 2013. Clinicopathological
information (age, gender, tumor size, histological grade, liver
cirrhosis, hepatitis B virus infection, microvascular invasion and
TNM stage) for each patient was collected from medical records. Two
pathologists confirmed the pathologic diagnoses of all slides. The
protocol of the present study was approved by the Ethics Committee
of Xijing Hospital. The informed written consent to participate in
the present study was obtained from each patient.
FOXM1 and GLUT1 detection by
immunohistochemistry (IHC)
Paraffin-embedded HCC samples and their surrounding
liver tissues were used to construct the tissue microarrays (in
collaboration with Xi'an Dong-Ao Biosciences Co., Ltd. (Xi'an,
China). The TMA was dyed to detect FOXM1 and GLUT1 expression by
IHC. In brief, the slides were deparaffinized and rehydrated in
xylene and alcohol solutions and heated in boiling sodium citrate
buffer (10 mM, pH 6.0). After being maintained at a sub-boiling
temperature for 20 min, the slides were incubated in 3% hydrogen
peroxide for 15 min for endogenous peroxidase inactivation. Then,
the sections were blocked with 5% normal goat serum at room
temperature for 1 h. Subsequently, the slides were incubated with
rabbit anti-FOXM1 (ProteinTech Group, Wuhan, China) or with GLUT1
(Thermo Fisher Scientific, Inc., Waltham, MA, USA) polyclonal
antibody overnight at 4̊C. After a phosphate-buffered saline (PBS)
rinse, the sections were incubated with horseradish peroxidase
(HRP)-conjugated goat anti-rabbit antibody (Zhongshan Goldenbridge
Biotechnology Co., Ltd., Beijing, China) according to the
manufacturer's instructions. Then, the slides were incubated with
fresh 0.05% 3,3′-diaminobenzidine (DAB) for several minutes and
counterstained with hematoxylin. Two pathologists independently
examined the IHC staining. The scores were estimated according to
both the staining intensity and the number of stained cells, as
previously described (13).
Briefly, the staining intensity was scored as: 0, negative; 1,
weak; and 2, strong. The staining extent was determined according
to the percentage of positive staining cells: 0, negative; 1,
1–25%; 2, 26–50%; 3, 51–75%; and 4, 76–100%. Therefore, positive
expression was defined as a total score of >3, while negative
expression was defined as a total score of ≤3.
RNA extraction, cDNA synthesis and
quantitative real-time PCR
TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was
used to extracted the total RNA of the HCC cells according to the
procedure recommended by the manufacturer. Reverse transcription
was conducted with PrimeScript™ Master Mix (Takara Bio, Dalian,
China). Then, quantitative real-time PCR was performed with SYBR
Premix Ex Taq™ II (Takara) and detected on the Bio-Rad IQ5 (Bio-Rad
Laboratories, Hercules, CA, USA) system. β-actin was used as the
reference, and the ∆∆Ct method was used to analyze the gene
expression using iQ5 Optical System Software (Bio-Rad
Laboratories). The sequences of RT-qPCR primers used for mRNA
analysis in the present study are listed in Table I.
 | Table I.Primers for the RT-PCR. |
Table I.
Primers for the RT-PCR.
| Gene | Primers (5′-3′) |
|---|
| FOXM1 | F
5′-GGGCGCACGGCGGAAGATGAA3′ |
|
| R
5′-CCACTCTTCCAAGGGAGGGCTC3′ |
| ALDO | F
5′-ATGCCACTCTCAACCTCAATGCTATC-3′ |
|
| R
5′-TTATTTTCTTGGGTGGGTATTCTGG-3′ |
| ENDOLASE | F
5′-CTGATGCTGGAGTTGGATGG-3′ |
|
| R
5′-CCATTGATCACGTTGAAGGC-3′ |
| GLUT1 | F
5′-GGCCAAGAGTGTGCTAAAGAA-3′ |
|
| R
5′-ACAGCGTTGATGCCAGACAG-3′ |
| GLUT4 | F
5′-CTTCATCATTGGCATGGGTTT-3′ |
|
| R
5′-CGGGTTTCAGGCACTTTTAGG-3′ |
| GAPDH | F
5′-CAGCCTCAAGATCATCAGCA-3′ |
|
| R
5′-TGTGGTCATGAGTCCTTCCA-3′ |
| G6PI | F
5′-AGGCTGCTGCCACATAAGGT-3′ |
|
| R
5′-AGCGTCGTGAGAGGTCACTTG-3′ |
| HK2 | F
5′-GATTTCACCAAGCGTGGACT-3′ |
|
| R
5′-CCACACCCACTGTCACTTTG-3′ |
| LDHA | F
5′-CAGCTTGGAGTTTGCAGTTAC-3′ |
|
| R
5′-TGATGGATCTCCAACATGG-3′ |
| LDHB | F
5′-CCTAGAGCTCACTAGTCACAG-3′ |
|
| R
5′-CTCCTGTGCAAAATGGCAAC-3′ |
| PFK-L | F
5′-GGACAGGAAAGAGGAAGTGAC-3′ |
|
| R
5′-CGTAGATGAGGAAGACTTTGGC-3′ |
| PFK-M | F
5′-ATTCGGGCTGTGTTCTGG-3′ |
|
| R
5′-TGGCTAGGATTTTGAGGATGG-3′ |
| PFK-P | F
5′-CATCGACAATGATTTCTGCGG-3′ |
|
| R
5′-CCATCACCTCCAGAACGAAG-3′ |
| PGAM1 | F
5′-CCTGGAGAACCGCTTC-3′ |
|
| R
5′-CATGGGCTGCAATCAGTACAC-3′ |
| PGK1 | F
5′-CGGTAGTCCTTATGAGCC-3′ |
|
| R
5′-CATGAAAGCGGAGGTTCT-3′ |
| PKM1 | F
5′-CTATCCTCTGGAGGCTGTGC-3′ |
|
| R
5′-CCATGAGGTCTGTGGAGTGA-3′ |
| PKM2 | F
5′-GGGTTCGGAGGTTTGATG-3′ |
|
| R
5′-ACGGCGGTGGCTTCTGT-3′ |
| β-actin | F
5′-TAGTTGCGTTACACCCTTTCTTG-3′ |
|
| R
5′-TCACCTTCACCGTTCCAGTTT-3′ |
shRNAs and plasmid transfection
The shRNA of FOXM1 and the shRNA-NC not targeting
any known mammalian gene were synthesized by Shanghai GenePharma
Co. (Shanghai, China). The plasmid pcDNA3.1-FOXM1 and control
vector pcDNA3.1 were previously described (14). Transfection of shRNAs and plasmids
was performed with Lipofectamine 2000 (Invitrogen) according to the
manufacturer's instructions.
Western blotting
Ice-cold PBS was used to wash the cells 3 times.
Then, protein was extracted using strong RIPA buffer (Beyotime,
Shanghai, China) with protease inhibitor (Thermo Fisher Scientific,
Waltham, MA, USA). The lysates were separated by 8% SDS-PAGE and
then transferred to nitrocellulose membranes. After blocking with
5% milk for 1 h, the membranes were incubated with primary
antibodies diluted in 5% milk overnight at 4̊C. HRP-conjugated
secondary antibody (Abcam, Cambridge, MA, USA) was incubated with
the membranes for 1 h at room temperature. After washing with
Tris-buffered saline with Tween-20 (TBST), the blots were
visualized with the ChemiDoc™ XRS+ using Image Lab™ software
(Bio-Rad Laboratories).
Glucose uptake and lactate production
assays
The glucose assay kit (BioVision, Milpitas, CA, USA)
was used to detect relative glucose uptake of cells before and
after transfection in accordance with the manufacturer's
instructions. The relative lactate production among different
groups was measured via using the lactate assay kit (BioVision)
according to the protocol.
Luciferase reporter assay
Cells were incubated at 37̊C overnight in 6-well
plates. Each group was co-transfected with GLUT1 promoter
reporters, shFOXM1 or pcDNA3.1-FOXM1 plasmid. Twenty-four hours
later, whole-cell lysates were prepared with passive lysis buffer.
Then, the collections were used for reporter detection using the
Dual-Luciferase Reporter System (Promega, Madison, WI, USA) and the
reactions were measured.
Statistical analysis
All statistical analyses were performed using SPSS
17.0 statistical software (SPSS, Inc., Chicago, IL, USA). Students
t-test was performed for two group comparisons. One-way ANOVA and
Dunnett's multiple comparison test were used for multiple group
comparisons. The expression of FOXM1 and GLUT1 in HCC samples and
their relationship with clinicopathological characteristics were
analyzed using χ2 tests. A P<0.05 was considered to
indicate a statistically significant result.
Results
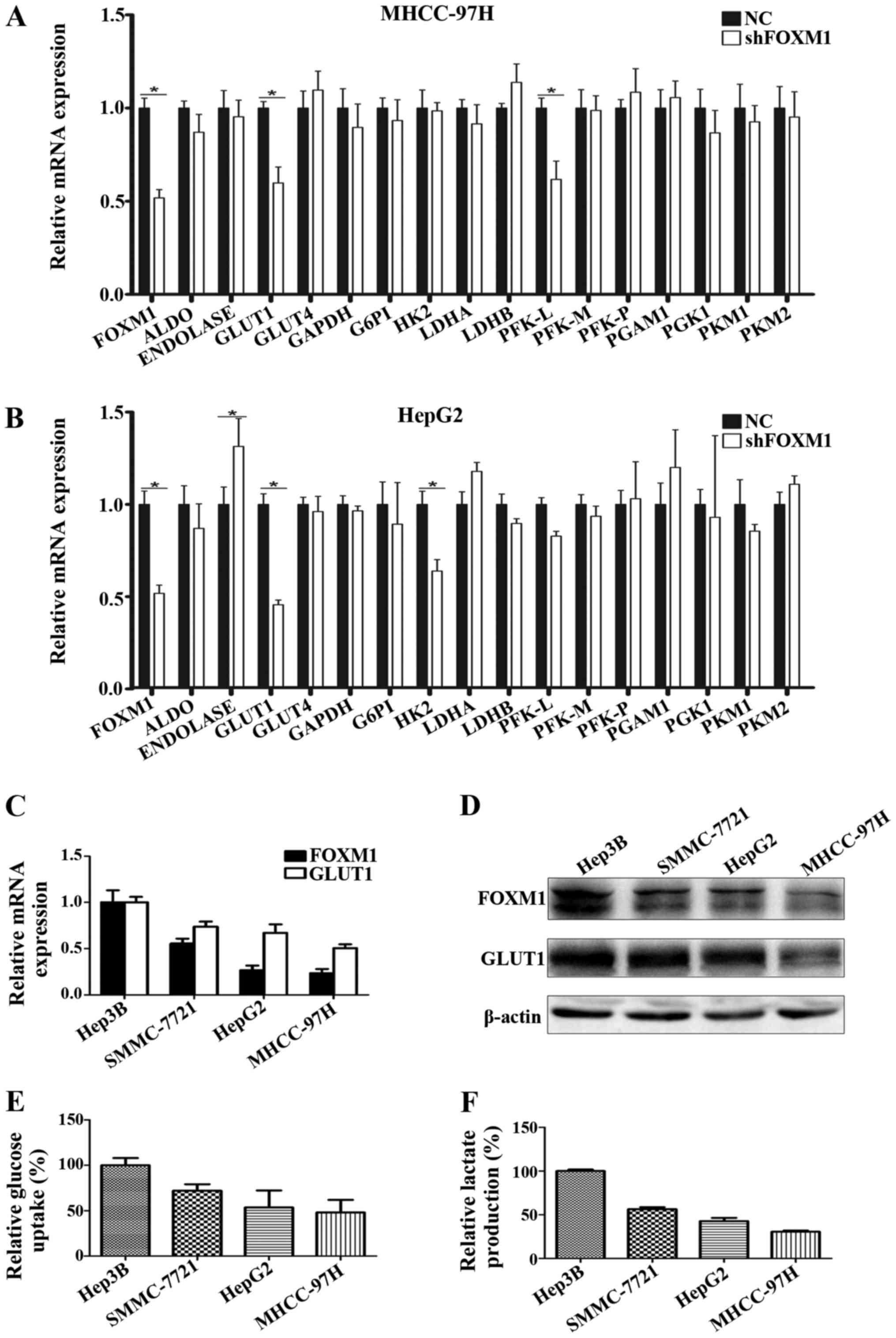
FOXM1 promotes glycolysis in HCC
cells
As previously reported, numerous oncogenes and
tumor-suppressor genes participate in cancer metabolism by
regulating the expression of key metabolism-related molecules. To
investigate the role of FOXM1 in HCC glycolysis, we evaluated the
effect of FOXM1 on the expression of glycolysis-related molecules.
Among these key glycolysis-related molecules, we found that only
GLUT1 mRNA expression was significantly downregulated in both the
HepG2 and MHCC-97H HCC cell lines when FOXM1 was knocked down by
shRNA (Fig. 1A and B). To further
confirm the effect of FOXM1 on GLUT1 expression and HCC glycolysis,
we assessed the correlation of FOXM1 expression with GLUT1
expression, glucose uptake and lactate production in 4 different
HCC cell lines. As shown in Fig.
1C-F, expression of FOXM1 was tightly associated with the
expression of GLUT1, glucose uptake and lactate production.
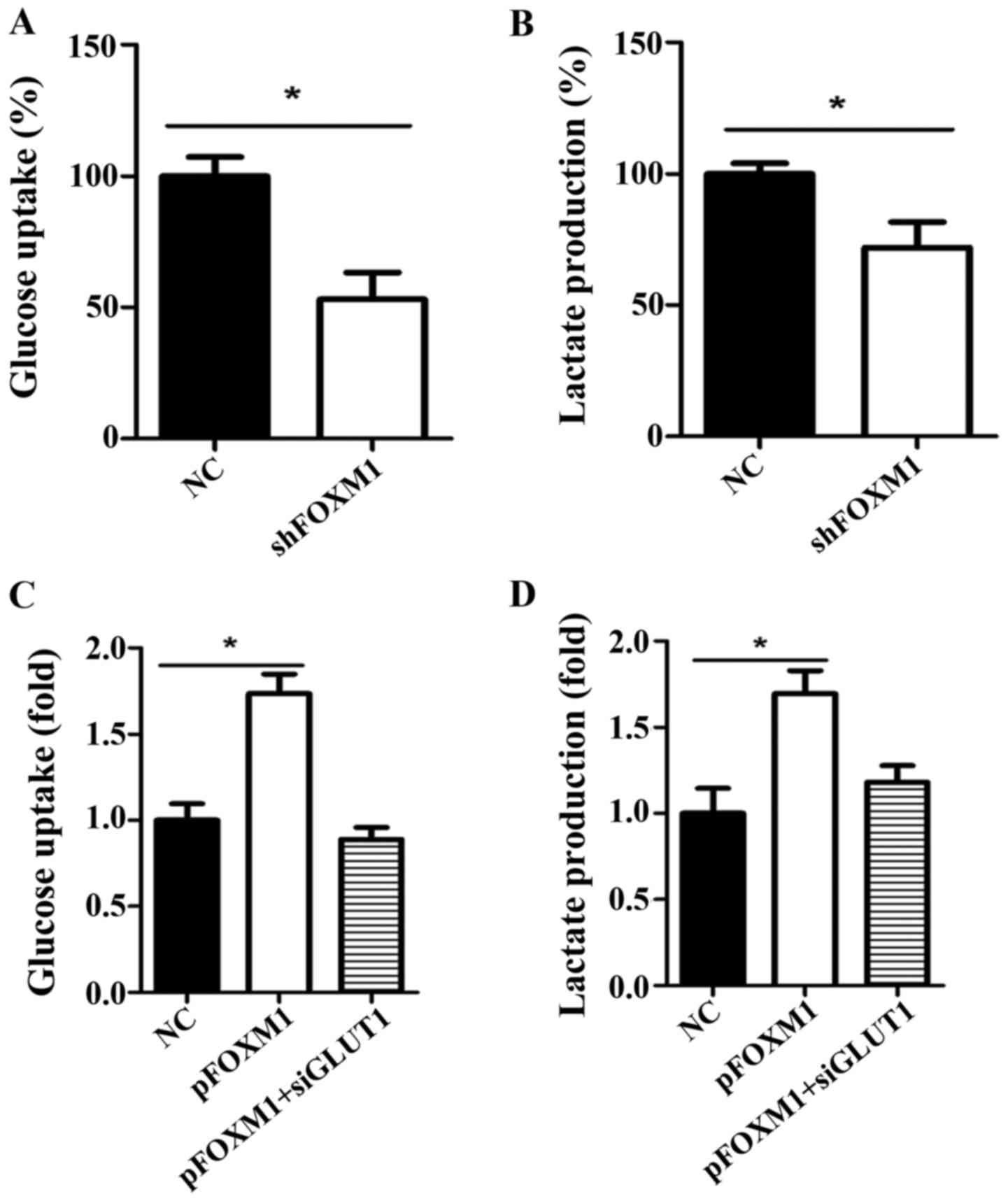
FOXM1 positively regulates the Warburg
effect
The Warburg effect is characterized by consuming
more glucose and producing more lactate than normal cells when the
glycolysis pathway is activated in cancer cells. To investigate
whether FOXM1 impacts the Warburg effect in HCC, we detected the
level of glucose uptake and lactate production when the expression
of FOXM1 was altered using shRNA or an overexpression plasmid. We
knocked down FOXM1 expression in Hep3B cells, which have a
relatively high expression of FOXM1 among the 4 HCC cell lines used
in the present study. We found that the decreased expression of
FOXM1 significantly reduced glucose uptake and lactate production
in the Hep3B cells (Fig. 2A and B).
In contrast, overexpression of FOXM1 in MHCC-97H cells, which have
the lowest FOXM1 expression of the HCC cells, significantly
increased glucose uptake and lactate production (Fig. 2C and D). Moreover, the increased
glucose uptake and lactate production caused by FOXM1 expression
was decreased when GLUT1 was knocked down using a specific siRNA.
These results suggest that FOXM1 positively regulates the Warburg
effect in HCC cells, which is dependent on the expression of
GLUT1.
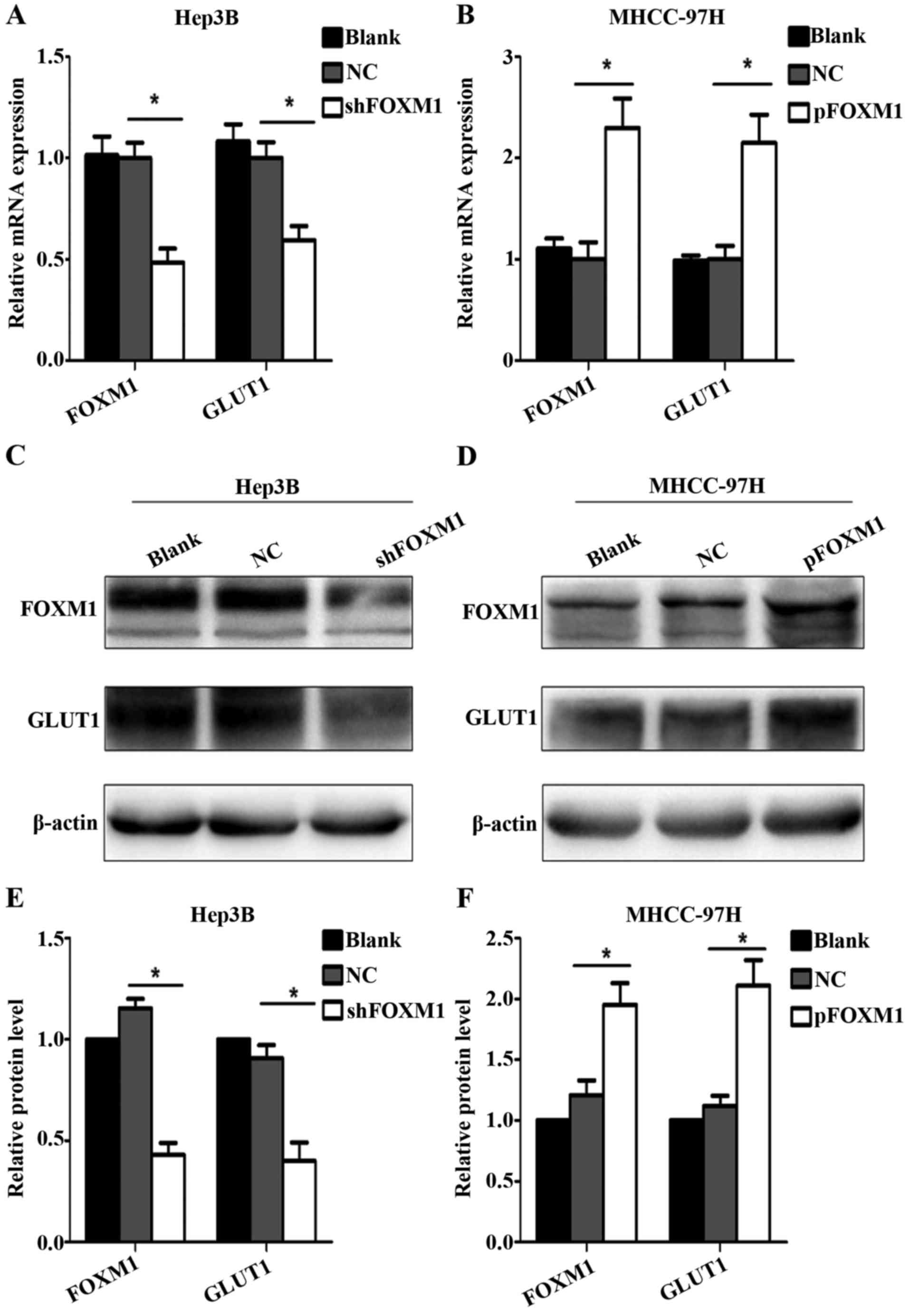
FOXM1 promotes GLUT1 expression
To further determine whether FOXM1 regulates the
expression of GLUT1, RT-qPCR and western blot assay were used. Upon
downregulation of FOXM1, GLUT1 expression was decreased at both the
mRNA and protein levels in the Hep3B cells compared with the blank
and negative control groups (Fig. 3A, C
and E). By comparison, upregulation of the FOXM1 in MHCC-97H
cells promoted the expression of GLUT1 (Fig. 3B, D and F).
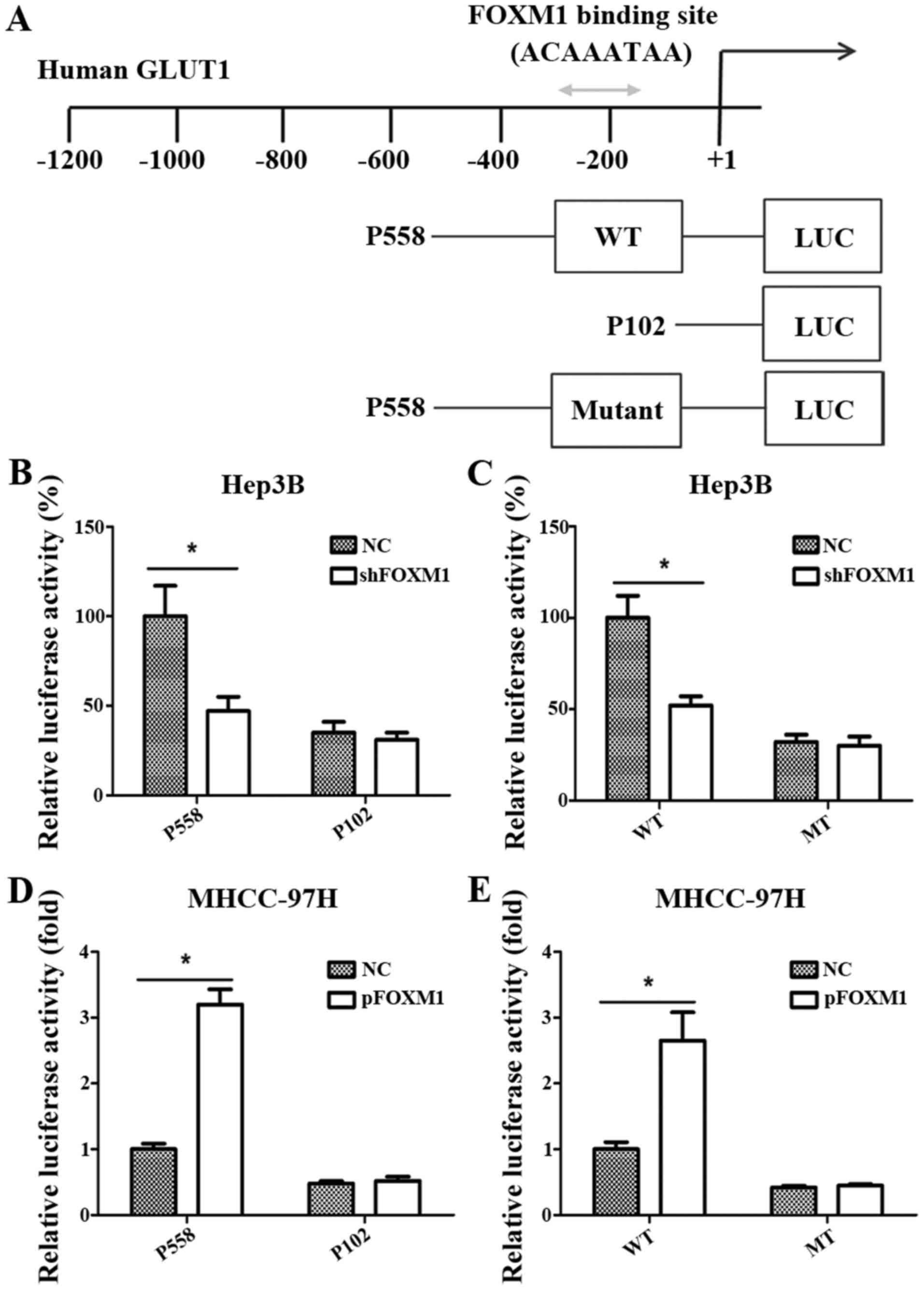
GLUT1 is a direct target of FOXM1
To further verify whether GLUT1 is a direct target
gene of FOXM1 transcription activation, we analyzed the sequences
of the GLUT1 promoter and identified a putative FOXM1-binding site
in its promoter region (Fig. 4A)
(12). Then, 3 reporter gene
constructs were produced based on the potential binding site. These
reporter constructs were co-transfected with pcDNA3.1-FOXM1,
shFOXM1 or control vector into HCC cells, and the promoter activity
was then measured. As shown in Fig. 4B
and D, the altered expression of FOXM1 significantly changed
the activity of the GLUT1 promoter in the P558 construct, but not
in the P102 construct, which did not contain the putative FOXM1
binding site. As shown in Fig. 4C and
E, FOXM1 knockdown or overexpression significantly reduced or
increased respectively the activity of the wild-type pLuc-GLUT1
construct in the Hep3B and MHCC-97H cells. However, the altered
expression of FOXM1 did not change the activity of the mutant
pLuc-GLUT1 construct.
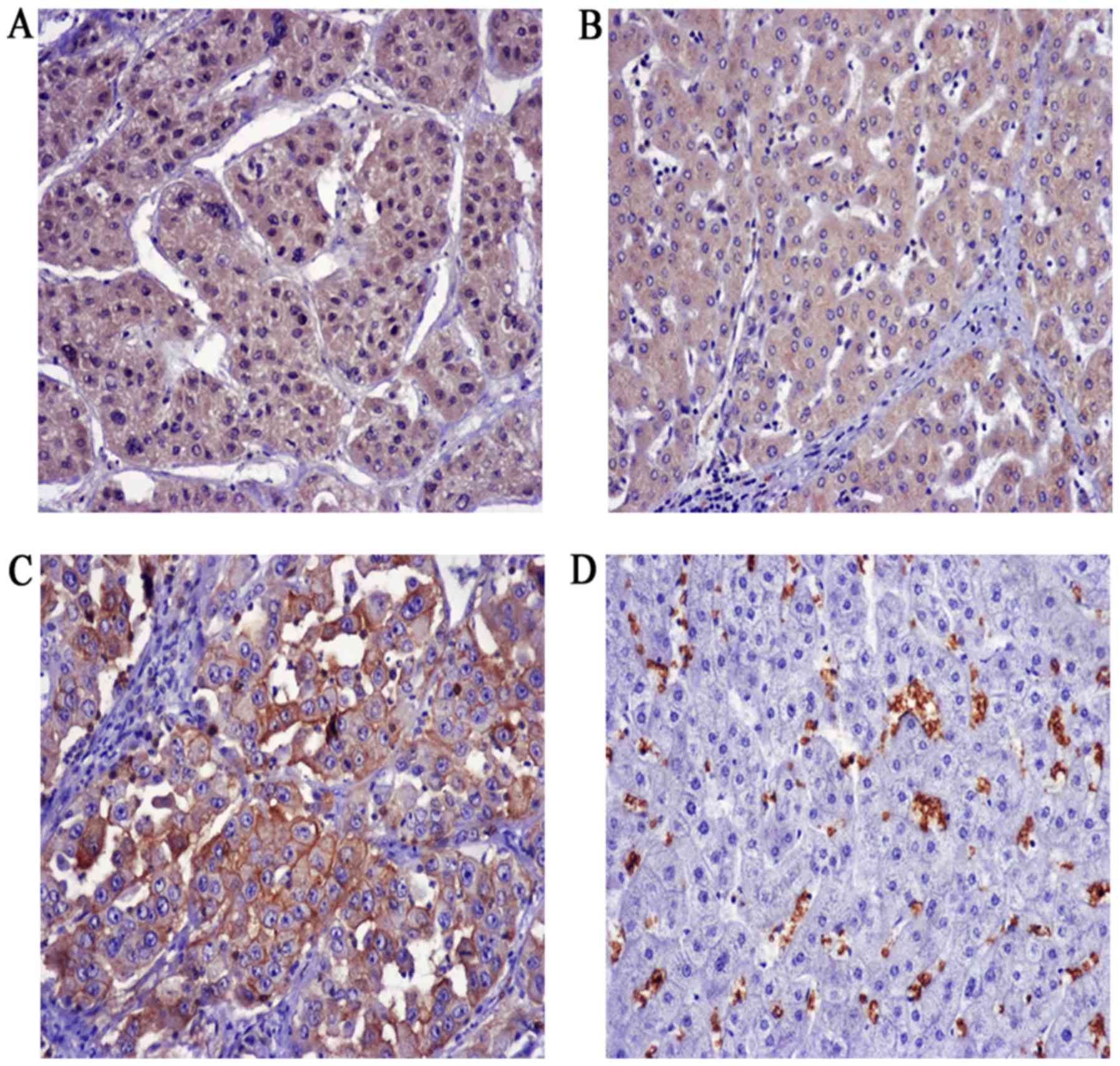
Expression of FOXM1 and GLUT1 in HCC
and adjacent liver tissues
To explore the expression of FOXM1 and GLUT1 in HCC,
100 HCC patients were enrolled in the present study. We found that
FOXM1 was mainly expressed in the cytoplasm as well as the nucleus,
whereas GLUT1 was exclusively expressed in plasma membranes in the
HCC patients (Fig. 5). Positive
staining for FOXM1 in the HCC and adjacent normal tissues was
64/100 and 35/100, respectively, and 24/100 and 0/100 for GLUT1,
respectively.
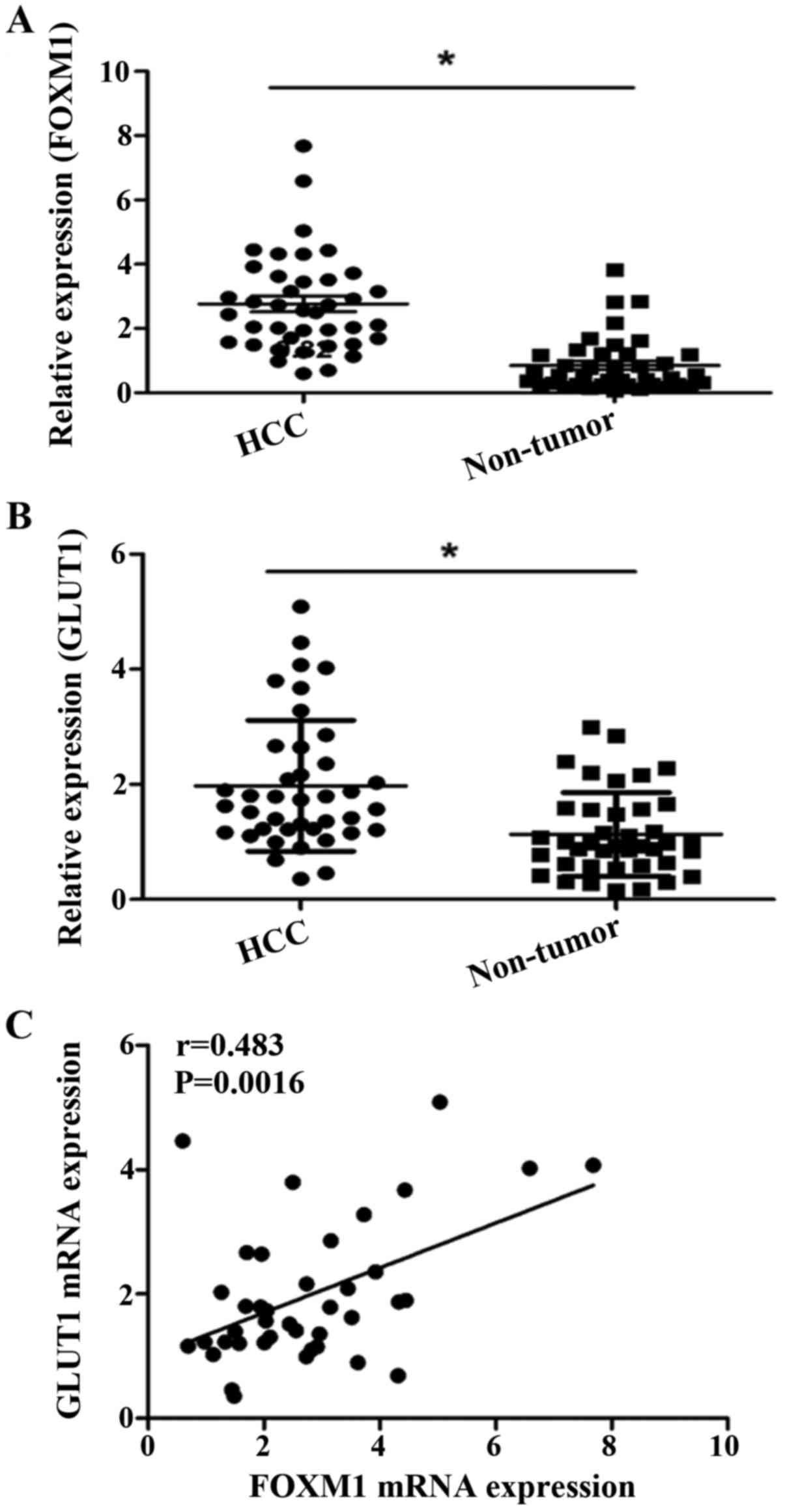
We further showed that the FOXM1 and GLUT1 genes
were also increased at the transcriptional level. The real-time
quantitative RT-PCR assay demonstrated that the mRNA expression
level of FOXM1 and GLUT1 were significantly increased in the HCC
tissues compared to the levels noted in the adjacent non-cancerous
tissues (Fig. 6A and B).
Correlation between FOXM1 and GLUT1 in
HCC
We evaluated the correlation between FOXM1 and GLUT1
expression at both the mRNA and protein levels. Our data showed a
positive correlation between the expression of FOXM1 and GLUT1
(r=0.483, P=0.0016) at the mRNA level (Fig. 6C). We also analyzed the results of
the HCC staining and also found a positive correlation between
FOXM1 and GLUT1 expression at the protein level (Table II). Notably, further statistical
analysis demonstrated that the nuclear localization of FOXM1 was
more closely associated with GLUT1 expression, which suggests a
crucial role of FOXM1 in GLUT1 transactivation (Table III).
 | Table II.Correlation analysis of FOXM1 and
GLUT1 expression in HCC cases. |
Table II.
Correlation analysis of FOXM1 and
GLUT1 expression in HCC cases.
|
|
| GLUT1 expression |
|
|
|---|
|
|
|
|
|
|
|---|
|
| No. of patients | Negative | Positive | r | P-value |
|---|
| FOXM1 expression |
|
|
|
|
|
|
Negative | 36 | 36 | 0 | 0.421 |
<0.001a |
|
Positive | 64 | 40 | 24 |
|
|
 | Table III.Correlation analysis of GLUT1 and
FOXM1 nuclear expression in FOXM1-positive staining samples. |
Table III.
Correlation analysis of GLUT1 and
FOXM1 nuclear expression in FOXM1-positive staining samples.
|
|
| GLUT1
expression |
|
|
|---|
|
|
|
|
|
|
|---|
|
| No. of
patients | Negative | Positive | r | P-value |
|---|
| FOXM1 nuclear
expression |
|
|
|
|
|
|
Negative | 25 | 22 | 3 | 0.422 |
<0.001a |
|
Positive | 39 | 18 | 21 |
|
|
Correlation between FOXM1 and GLUT1
expression with the clinicopathological features
As shown in Table
IV, the expression of FOXM1 and GLUT1 was significantly
associated with tumor histological grade and TNM stage in HCC. In
addition, GLUT1 expression was also related to microvascular
invasion.
 | Table IV.Correlation between FOXM1 and GLUT1
expression levels and clinicopathological features of the HCC
patients. |
Table IV.
Correlation between FOXM1 and GLUT1
expression levels and clinicopathological features of the HCC
patients.
|
|
| FOXM1
expression |
|
|
| GLUT1
expression |
|
|---|
|
|
|
|
|
|
|
|
|
|---|
|
Characteristics | No. of
patients | Negative | Positive | r | P-value | r | Negative | Positive | P-value |
|---|
| Gender |
|
|
|
|
|
|
|
|
|
|
Male | 74 | 29 | 45 | −0.112 | 0.344 | 60 | 14 | −0.201 | 0.062 |
|
Female | 26 | 7 | 19 |
|
| 16 | 10 |
|
|
| Age (years) |
|
|
|
|
|
|
|
|
|
|
<45 | 27 | 9 | 18 | 0.034 | 0.817 | 20 | 7 | 0.027 | 0.796 |
|
≥45 | 73 | 27 | 46 |
|
| 56 | 17 |
|
|
| Histological
grade |
|
|
|
|
|
|
|
|
|
| G1 | 15 | 11 | 4 | 0.275 | 0.004a | 15 | 0 | 0.326 | 0.005a |
| G2 | 69 | 21 | 48 |
|
| 53 | 16 |
|
|
| G3 | 16 | 4 | 12 |
|
| 8 | 8 |
|
|
| Tumor size
(cm) |
|
|
|
|
|
|
|
|
|
|
<5 | 35 | 17 | 18 | 0.192 | 0.080 | 28 | 7 | 0.069 | 0.625 |
| ≥5 | 65 | 19 | 46 |
|
| 48 | 17 |
|
|
| Liver
cirrhosis |
|
|
|
|
|
|
|
|
|
| No | 39 | 15 | 24 | 0.041 | 0.831 | 32 | 7 | 0.113 | 0.339 |
|
Yes | 61 | 21 | 40 |
|
| 44 | 17 |
|
|
| HBsAg status |
|
|
|
|
|
|
|
|
|
|
Negative | 75 | 27 | 48 | ≤0.001 | 1.000 | 56 | 19 | −0.054 | 0.788 |
|
Positive | 25 | 9 | 16 |
|
| 20 | 5 |
|
|
| Microvascular
invasion |
|
|
|
|
|
|
|
|
|
| No | 72 | 30 | 42 | 0.189 | 0.067 | 60 | 12 | 0.275 | 0.009a |
|
Yes | 28 | 6 | 22 |
|
| 16 | 12 |
|
|
| TNM stage |
|
|
|
|
|
|
|
|
|
|
I–II | 31 | 20 | 11 | 0.398 |
<0.001a | 29 | 2 | 0.275 | 0.005a |
|
III–IV | 69 | 16 | 53 |
|
| 47 | 22 |
|
|
Discussion
FOXM1 is a proliferation related transcription
factor, which plays crucial roles in normal cell proliferation,
organogenesis and cancer progression (15). The aberrant expression of FOXM1 is
involved in cancer proliferation (16,17),
epithelial-mesenchymal transition, metastasis (10) and stem cell features (18) in hepatocellular carcinoma (HCC).
However, the significance of FOXM1 in HCC glycolysis has not been
established. In the present study, we assessed the role of FOXM1 in
HCC glucose metabolism. The glucose uptake and lactate production
assays demonstrated that FOXM1 expression was consistent with
glycolysis levels in several HCC cell lines. Furthermore, we found
that knockdown or overexpression of FOXM1 significantly suppressed
or promoted glycolysis in HCC cells, respectively.
To determine whether FOXM1 promotes the Warburg
effect in HCC, we further explored the underlying molecular
mechanism of FOXM1 regulation of HCC glycolysis. A previous study
showed that oncogenes and tumor-suppressor genes participate in
cancer metabolism via the dysregulation of key metabolism-related
molecules, including glycolysis-related transporters (GLUTs and
MCTs) and glycolytic enzymes (G6PI and HK2). For example, HIF,
either alone or together with the oncogene MYC, was shown to
promote glycolysis by activating glucose transporters, glycolytic
enzymes and lactate dehydrogenase A (LDHA) (5,19–21).
Therefore, we detected the expression changes of all
glycolysis-related molecules in HepG2 and MHCC-97H cell lines when
FOXM1 was knocked down. We found that knockdown of FOXM1 expression
did not change the expression of most of the molecules, except for
GLUT1, in the different HCC cell lines. Further experiments
confirmed that FOXM1 promotes HCC glycolysis by regulation of the
expression of GLUT1. As previously published, FOXM1 is a typical
transcription factor that belongs to the Forkhead box family, which
is evolutionarily conserved and is defined by having a common
DNA-binding domain called Forkhead or winged-helix domain (22). We further assessed whether FOXM1
affects GLUT1 expression via transcriptional regulation. A series
of luciferase assays verified the direct transactivating role of
FOXM1 on GLUT1. Taken together, our data strongly support the
hypothesis that FOXM1 promotes glycolysis in HCC by transactivating
GLUT1 expression.
Tumor metabolism is an important link between the
tumor microenvironment and tumor progression (23). As described previously, cancer cells
use the glycolysis pathway to generate ATP as well as the byproduct
lactate. The accumulation of lactate in the extracellular matrix
(ECM) causes the acidification of the tumor microenvironment. The
acidic environment is favorable for the activation of MMPs, uPA and
other critical proteases to facilitate degradation of the ECM and,
subsequently, tumor invasion and metastasis. Additionally, the
aberrant expression of glycolytic transporters and enzymes has been
linked to tumor progression (24).
Therefore, we evaluated the association between FOXM1 and GLUT1
expression with several clinicopathological characteristics to
determine whether FOXM1 and GLUT1 expression contributes to HCC
progression. The statistical analysis showed a significant
association between the expression of FOXM1 and GLUT1 with the
tumor histological grade and TNM stage in HCC. In addition, GLUT1
expression was also related to microvascular invasion. These
results indicate that the overexpression of FOXM1 and GLUT1 may
play important roles in HCC progression.
Although, our experiments investigated the
regulatory role and mechanism of FOXM1 on glycolysis in HCC cells,
the present study has some limitations. Firstly, the number of
patients enrolled in the present study was relatively limited.
Therefore, large scale studies are needed to validate these
findings in the future. In addition, further experiments are
required to validate the role of the FOXM1-GLUT1 axis in HCC
glycolysis in vivo.
In summary, the present study showed that FOXM1
promotes glycolysis in HCC. We demonstrated that FOXM1 binds to the
GLUT1 promoter to regulate the expression of GLUT1 in HCC cells.
Moreover, overexpression of FOXM1 and GLUT1 plays an important role
in HCC progression. Collectively, these results suggest that the
FOXM1-GLUT1 axis is a potential therapeutic target in HCC.
Acknowledgements
We thank Fuqin Zhang for advice on performing the
experiments. The present study was supported by grants from the
National Nature Science Foundation of China (nos. 81270549 and
30872480).
References
|
1
|
David F and Jacques F: The global and
regional burden of cancer. World Cancer Report 2014. Bernard WS and
Christopher PD: International Agency for Research on Cancer; Paris:
pp. 16–53. 2014
|
|
2
|
Llovet JM: Updated treatment approach to
hepatocellular carcinoma. J Gastroenterol. 40:225–235. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Villanueva A: Rethinking future
development of molecular therapies in hepatocellular carcinoma: A
bottom-up approach. J Hepatol. 59:392–395. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Warburg O: On the origin of cancer cells.
Science. 123:309–314. 1956. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cairns RA, Harris IS and Mak TW:
Regulation of cancer cell metabolism. Nat Rev Cancer. 11:85–95.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Amann T, Maegdefrau U, Hartmann A, Agaimy
A, Marienhagen J, Weiss TS, Stoeltzing O, Warnecke C, Schölmerich
J, Oefner PJ, et al: GLUT1 expression is increased in
hepatocellular carcinoma and promotes tumorigenesis. Am J Pathol.
174:1544–1552. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zona S, Bella L, Burton MJ, de Nestal
Moraes G and Lam EW: FOXM1: An emerging master regulator of DNA
damage response and genotoxic agent resistance. Biochim Biophys
Acta. 1839:1316–1322. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu QF, Liu C, Tai MH, Liu D, Lei L, Wang
RT, Tian M and Lü Y: Knockdown of FoxM1 by siRNA interference
decreases cell proliferation, induces cell cycle arrest and
inhibits cell invasion in MHCC-97H cells in vitro. Acta Pharmacol
Sin. 31:361–366. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Park HJ, Gusarova G, Wang Z, Carr JR, Li
J, Kim KH, Qiu J, Park YD, Williamson PR, Hay N, et al:
Deregulation of FoxM1b leads to tumour metastasis. EMBO Mol Med.
3:21–34. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Meng FD, Wei JC, Qu K, Wang ZX, Wu QF, Tai
MH, Liu HC, Zhang RY and Liu C: FoxM1 overexpression promotes
epithelial-mesenchymal transition and metastasis of hepatocellular
carcinoma. World J Gastroenterol. 21:196–213. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Davis DB, Lavine JA, Suhonen JI,
Krautkramer KA, Rabaglia ME, Sperger JM, Fernandez LA, Yandell BS,
Keller MP, Wang IM, et al: FoxM1 is up-regulated by obesity and
stimulates beta-cell proliferation. Mol Endocrinol. 24:1822–1834.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cui J, Shi M, Xie D, Wei D, Jia Z, Zheng
S, Gao Y, Huang S and Xie K: FOXM1 promotes the warburg effect and
pancreatic cancer progression via transactivation of LDHA
expression. Clin Cancer Res. 20:2595–2606. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xia L, Mo P, Huang W, Zhang L, Wang Y, Zhu
H, Tian D, Liu J, Chen Z, Zhang Y, et al: The
TNF-α/ROS/HIF-1-induced upregulation of FoxMI expression promotes
HCC proliferation and resistance to apoptosis. Carcinogenesis.
33:2250–2259. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu M, Dai B, Kang SH, Ban K, Huang FJ,
Lang FF, Aldape KD, Xie TX, Pelloski CE, Xie K, et al: FoxM1B is
overexpressed in human glioblastomas and critically regulates the
tumorigenicity of glioma cells. Cancer Res. 66:3593–3602. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Laoukili J, Stahl M and Medema RH: FoxM1:
At the crossroads of ageing and cancer. Biochim Biophys Acta.
1775:92–102. 2007.PubMed/NCBI
|
|
16
|
Yu M, Tang Z, Meng F, Tai M, Zhang J, Wang
R, Liu C and Wu Q: Elevated expression of FoxM1 promotes the tumor
cell proliferation in hepatocellular carcinoma. Tumour Biol.
37:1289–1297. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hu C, Liu D, Zhang Y, Lou G, Huang G, Chen
B, Shen X, Gao M, Gong W, Zhou P, et al: LXRα-mediated
downregulation of FOXM1 suppresses the proliferation of
hepatocellular carcinoma cells. Oncogene. 33:2888–2897. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kopanja D, Pandey A, Kiefer M, Wang Z,
Chandan N, Carr JR, Franks R, Yu DY, Guzman G, Maker A, et al:
Essential roles of FoxM1 in Ras-induced liver cancer progression
and in cancer cells with stem cell features. J Hepatol. 63:429–436.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Goda N and Kanai M: Hypoxia-inducible
factors and their roles in energy metabolism. Int J Hematol.
95:457–463. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kim JW, Tchernyshyov I, Semenza GL and
Dang CV: HIF-1-mediated expression of pyruvate dehydrogenase
kinase: A metabolic switch required for cellular adaptation to
hypoxia. Cell Metab. 3:177–185. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Semenza GL, Roth PH, Fang HM and Wang GL:
Transcriptional regulation of genes encoding glycolytic enzymes by
hypoxia-inducible factor 1. J Biol Chem. 269:23757–23763.
1994.PubMed/NCBI
|
|
22
|
Wierstra I and Alves J: FOXM1, a typical
proliferation-associated transcription factor. Biol Chem.
388:1257–1274. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kroemer G and Pouyssegur J: Tumor cell
metabolism: Cancer's Achilles' heel. Cancer Cell. 13:472–482. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Han T, Kang D, Ji D, Wang X, Zhan W, Fu M,
Xin HB and Wang JB: How does cancer cell metabolism affect tumor
migration and invasion? Cell Adh Migr. 7:395–403. 2013. View Article : Google Scholar : PubMed/NCBI
|