Introduction
Brain-derived neurotrophic factor (BDNF), a member
of the neurotrophin family, plays an essential role in promoting
the growth, survival and differentiation of developing neurons in
the central and peripheral nervous systems (1). In addition, BDNF is also involved in
peripheral sensory and motor neuron regeneration at the site of
nerve injury (2). Apart from its
well-known effects on neurons, recent studies have demonstrated
that BDNF and its high-affinity tyrosine kinase (Trk) receptor TrkB
are constitutively expressed in a number of neural and non-neuronal
tumors, including neuroblastoma (3), hepatocellular carcinoma (4), multiple meloma (5), prostatic carcinoma (6), pancreatic ductal adenocarcinoma
(6), adenoid cystic carcinoma
(7) and retinoblastoma (8). Moreover, it has been reported that
BDNF and TrkB are preferentially expressed in more aggressive
neuroblastoma with N-myc amplification (9). BDNF is expressed in 33 and 100% of
typical (TC) and atypical (AC) pulmonary carcinoids, respectively,
indicating the unfavorable prognosis of patients (10). In addition to malignant cells, BDNF
can be produced by osteoblasts, and bone marrow (BM) endothelial
cells in BM stroma (5), implying
that the paracrine and autocrine functions of BDNF play critical
roles in interaction of MM plasma cells with BM microenvironment as
interleukin-6 (IL-6) (11).
Exogenous BDNF participates in the promotion of the growth and
survival of tumor cells, protection of tumor cells from
chemotherapy-induced apoptosis (12), enhancing invasive and migratory
capabilities of tumor cells in a dose-dependent manner (13), cooperation with TrkB in transforming
rat intestinal epithelia cells to malignant cells and suppression
of anoikis (apoptosis resulting from loss of cell-matrix
interactions) (14), induction of
the tube formation of human umbilical vein endothelial cells
(HUVEC) in vitro and modulation of angiogenesis in tumors
(15). These studies suggest the
potential roles of BDNF in tumorigenicity and the progression of
cancers; however, the mechanisms have not yet been completely
clarified.
RNA interference (RNAi) is a process of
post-transcriptional gene silencing in which double-stranded RNA
(dsRNA) inhibits gene expression in a sequence-dependent manner via
degradation of the corresponding mRNA (16–18).
RNAi is initiated by an event whereby dsRNA is recognized by Dicer
(19). The Dicer enzyme cleaves
dsRNA into 19–22 nucleotide short interfering RNA (siRNA). These
siRNA duplexes are incorporated into a protein complex called the
RNAi-induced silencing complex (RISC), where they are cleaved into
2 single strands (guide and passenger strand). The latter is
degraded, while the former, antisense siRNA, pairs with the
complementary sequence in mRNA to induce its cleavage (20). A previous study investigated the
RNAi-mediated inhibition of BDNF in rat myoblasts. The reduction in
the level of BDNF promoted myoblasts to exit the cell cycle and
initiate the myogenic differentiation program (21). RNAi-mediated, sequence-specific gene
silencing revealed that inhibition of BDNF expression enhanced
cocaine cytotoxicity in neuroblastoma SK-N-AS cells and primary rat
hippocampal neurons (22).
In the present study, small interfering RNA (siRNA)
was used as a tool for suppressing the expression of the endogenous
BDNF gene in HeLa, a cervical carcinoma cell line with high
expression of BDNF, in order to investigate the effects of RNA
interference on the proliferation, apoptosis, cell cycle
distribution and the invasive and migratory capabilities of HeLa
cells.
Materials and methods
Construction of the recombinant
eukaryotic human-BDNF siRNA expression vector
BDNF siRNA was designed according to the siRNA
design guidelines of Ambion Inc. (Carlsbad, CA, USA). General
design guidelines are as follows. The selected 19–22 base siRNA
sequences were designed with 30–50% guanine cytosine content to
avoid inverted repeats (23). Four
siRNAs were chosen based on the sequence of the human BDNF gene.
They covered different regions of the BDNF sequence and showed no
homology with other human genes. A scrambled siRNA was used as a
negative control. The target sequences and corresponding hairpin
siRNA sequences for the 5 siRNAs, designated siRNA1, siRNA2,
siRNA3, siRNA4 and scramble siRNA, are shown in Fig. 1. The structure of hairpin siRNA was
BamHI + sense + loop + antisense + termination signal +
SalI + HindIII. pGenesil-1 [enhanced green
fluorescent protein (EGFP) purchased from Genesil (Wuhan, China)]
was introduced for the construction of recombinant eukaryotic human
BDNF siRNA expression vectors. Five pairs of hairpin siRNA sense
and antisense sequences were synthesized, annealed and cloned into
the BamHI/HindIII cloning site of pGenesil-1,
respectively. The products were transformed into DH5α-competent
cells. Kanamycin-resistant colonies were chosen, identified by
restriction digestion, and further confirmed by DNA sequencing. The
synthesis of all DNA chains and DNA sequencing was performed by
Genesil.
Cell culture and transfection
The cervical carcinoma cell line HeLa was cultured
in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS),
100 U/ml penicillin, 100 U/ml streptomycin, and was routinely
maintained at 37̊C in 5% CO2. Medium was changed every 3
days.
HeLa cells were seeded in 6-well plates
(5×105/ml). Transient transfection was performed using
Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the
manufacturer's instructions. For stable transfection, HeLa cells
were cultured in the standard culture medium for 48 h after
transfection. G418 selection at 800 µg/ml was applied and continued
for 4 weeks until single colonies were formed. In parallel,
non-transfected cells were also placed in standard culture medium
to ensure the potency and selectivity of G418. Positive clones were
maintained in the culture medium with G418 at 300 µg/ml. To
determine the transfection efficiencies and expression effects of
BDNF siRNA in HeLa cells, EGFP expression was examined by
microscopy (magnification, ×400) at 24 h after transfection and
after the positive clones were established.
RNA extraction and
reverse-transcriptase polymerase chain reaction (RT-PCR)
Total RNA from HeLa cells was isolated using TRIzol
extraction according to the manufacturer's protocol (Invitrogen).
Complementary DNA (cDNA) was subsequently synthesized from total
RNA (5 µg) using RevertAid First-Strand cDNA Synthesis kit
(Fermentas, Burlington, Ontario, Canada). Then, 1 µg of cDNA was
subjected to PCR for the selected genes. Primers used were as
follows: 5′-GCAGCCTTCTTTTGTGTAACC-3′ and
5′-AGAGTGATGACCATCCTTTTC-3′ for BDNF (594 bp); as well as
5′-GAAGGTGAAGGTCGGAGTC-3′ and 5′-GAAGATGGTGATGGGATTC-3′ for
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (226 bp) as
control; 5′-GTTTCATAAGATCCCACTGGATGG-3′ and
5′-TGCTGCTTAGCTGCCTGAGAGTTA-3′ for TrkB (260 bp); as well as
5′-TGAGACCTTCAACACCCCAG-3′, and 5′-GCCATCTCTTGCTCGAAGTC-3′ for
β-actin (312 bp) as control. PCR profiles consisted of denaturation
at 94̊C for 1 min, annealing at 55̊C for 30 sec, and extension at
72̊C for 1 min. The samples were amplified for 32 cycles. PCR
products were separated by electrophoresis on 1.5% agarose gels,
stained with ethidium bromide and photographed.
Western blot analysis
Cells were washed twice with ice-cold
phosphate-buffered saline (PBS), and then harvested on ice with
NP40 lysis buffer, containing 50 mmol/l Tris-HCl, pH 7.4; 150
mmol/l NaCl; 1% NP40; 5 mmol/l EDTA; 5 mmol/l NaF; 2 mmol/l sodium
vanadate; 1 mmol/l phenylmethylsulfonyl fluoride; 5 mg/ml
leupeptin; and 5 mg/ml aprotinin. The protein content was
quantitated by Bio-Rad protein assay (Bio-Rad, Hercules, CA, USA).
The lysate was then boiled for 5 min for protein denaturation.
Protein samples containing an equal amount (60 µg) of protein were
separated by electrophoresis on 10% polyacrylamide-SDS gels and
transferred onto nitrocellulose membranes. Non-specific binding of
antibodies was blocked by incubation in Tris-buffered saline (TBS)
containing 0.1% Tween-20 (TBS-T) and 5% non-fat milk for 1 h,
followed by overnight incubation with rabbit anti-human BDNF
(SC-546) (1:500) or rabbit anti-human TrkB (SC-12) (both from Santa
Cruz Biotechnology, Santa Cruz, CA, USA) (1:500) or rabbit
polyclonal IgG to GAPDH (1:1,000) at 4̊C. Rabbit anti-pAKT (1:500)
and anti-AKT (1:1,000) were obtained from Cell Signaling Technology
(Beverly, MA, USA). After washing, the membranes were incubated
with horseradish peroxidase (HRP)-conjugated goat anti-rabbit
secondary antibodies (1:5,000) at room temperature for 1 h.
Immunoreactive bands were visualized using the ECL kit.
MTT assay
Cell proliferation was assessed using the
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide
(MTT; Sigma Chemical Co., St. Louis, MO, USA) assay. Cells were
seeded in a 96-well plate at a density of 1.5×104
cells/well and allowed to adhere overnight. Transient transfection
was performed using Lipofectamine 2000 (Invitrogen), according to
the manufacturer's instructions. After 12, 24, 48, 72 and 120 h of
transfection, 20 µl of 5 mg/µl MTT solution was added into each
well of the plate and cells were incubated at 37̊C for an
additional 4 h. The culture supernatant was removed, 150 µl of
dimethyl sulfoxide (DMSO; Sigma) was added to dissolve the
crystals. Spectrophotometric absorbance of the samples was measured
by a microtiter plate reader at 570 nm. Considering that the
initial cell concentrations of each group may be not identical,
data at 12 h was set as the control. The cell proliferation rate
was calculated as follows: Proliferation rate (%) = (sample
absorbance/control absorbance) × 100. Each value represents 6
replicates, and each experiment was repeated 3 times.
Flow cytometry
HeLa cells were divided into non-transfected
(Pnon), pGenesil-1-tranfected (P0) and
positive experimental groups (PBDNF1). Cells in the
Pnon, P0 and PBDNF1 groups were
cultured in 6-well plates. After 48 h of incubation, a total of
1×106 cells were harvested, washed twice with PBS and
fixed with cold 70% ethanol overnight at −20̊C. Fixed cells were
centrifuged at 1,200 rpm and washed with PBS. Cells were stained in
the dark with propidium iodide (PI; 50 µg/ml; Sigma) and 0.1%
RNaseA (Invitrogen) at 4̊C for 1 h. Measurement of nuclear DNA was
performed using flow cytometry (FCM). The results obtained
reflected the percentage of cells in each phase of the cell
cycle.
Hoechst 33258 staining
After 24 h of incubation in 6-well plates, the
supernatant were removed. Then, the cells were fixed with cold
methanol (0̊C) for 10 min and washed twice with PBS. Hoechst 33258
(1 mg/ml) was added and incubation for 30 min at 4̊C was carried
out in a dark place. Cells were then washed with PBS and the
apoptotic cells were observed using an Olympus BH-2 fluorescence
microscope (magnification, ×200; Olympus, Tokyo, Japan). Cells
stained bright blue were considered as apoptotic cells. Random
cells (500) were counted and the apoptotic rate was calculated as
follows: Hoechst 33258-stained cells/500 cells×100%.
Migration and invasion assays
Cell migration was quantified by the number of cells
that directionally migrated through a 8-µm-pore polycarbonate
filter (porosity, 8 µm; Costar, Appleton Woods, Birmingham, UK) in
Boyden chambers. Briefly, the lower surface of the filter was
coated with 10 mg of gelatin. HeLa cells were serum-starved
overnight and resuspended in serum-free medium, and then 200 µl of
the cell suspension was replaced onto the upper chamber of each
well at a final concentration of 1×106 cells/ml. FBS
(10%)-containing medium was added to the bottom chamber and cells
were allowed to migrate for 6 h at 37̊C. Non-migrated cells on the
upper membrane surface were removed with a cotton swab. The
migratory cells attached to the lower membrane surfaces were fixed
with 4% paraformaldehyde in PBS and stained with Wright staining.
Cells were counted at a magnification of ×400 using standard
microscopy, and the mean number of cells/field in 5 random fields
was recorded. Triplicate filters were used and the experiments were
repeated 3 times. The invasion assay was performed as above except
that the upper surface of the filters were coated with 25 µg
Matrigel, the cell concentration was adjusted to 2×105
cells/ml and the time was prolonged to 24 h.
Statistical analysis
SPSS for Windows (SPSS 13.0; SPSS, Inc., Chicago,
IL, USA) was used to analyze the data. Data are expressed as the
mean ± standard deviation (SD) from at least 3 separate
experiments. Student's t-test was used to determine the significant
difference between 2 groups. For comparison between 3 or more
groups, one-way ANOVA test was used to determine statistical
significance. The level of significance was set at p<0.05.
Results
High expression levels of BDNF and
TrkB in HeLa cells
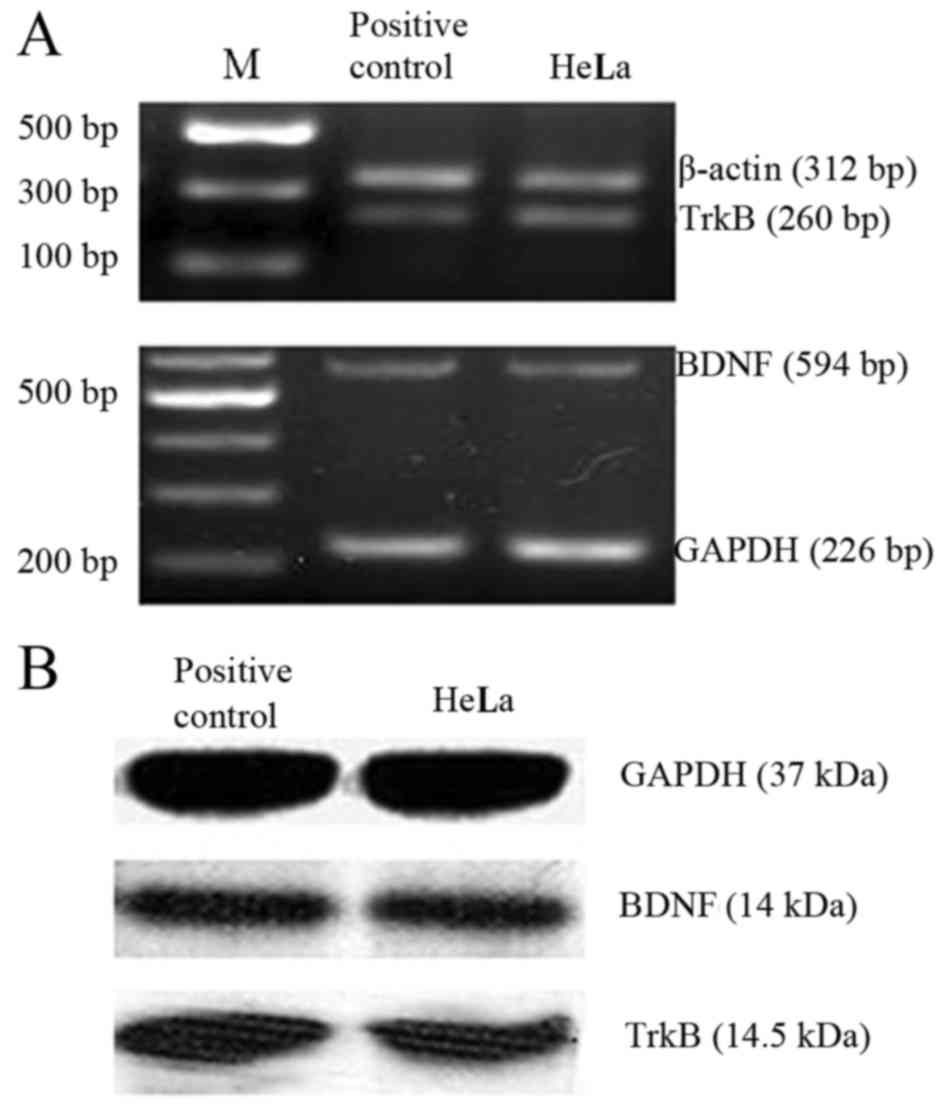
As shown in Fig. 2,
RT-PCR and western blot analysis revealed that BDNF and TrkB were
highly expressed in the HeLa cells at both the mRNA (Fig. 2A) and protein level (Fig. 2B). BDNF may provide autocrine
support for TrkB-expressing HeLa cells as reported in the nervous
system (24).
The recombinant eukaryotic BDNF siRNA
expression vectors were successfully constructed and transfected
into HeLa cells stably with high efficiencies
As shown in Fig.
3A-C, 4 experimental groups of plasmids pBDNF1, pBDNF2, pBDNF3
and pBDNF4, and the negative control group plasmid pScr were
successfully generated. The multiple clone sites of pGenesil-1 are:
HindIII, shRNA, BamHI, U6 promotor, EcoRI,
SalI and XbaI. A specific SalI site was
designed and synthesized into the shRNA sequences. Recombinant
vectors were digested to a 400 bp fragment by SalI with the
correct insertion. Most of the HeLa cells were effectively
transfected with the recombinant BDNF siRNA expression vectors. The
transient transfection efficiencies were ~60–80%. EGFP expression
was not significantly reduced after the positive clones were
generated (Fig. 3D-F).
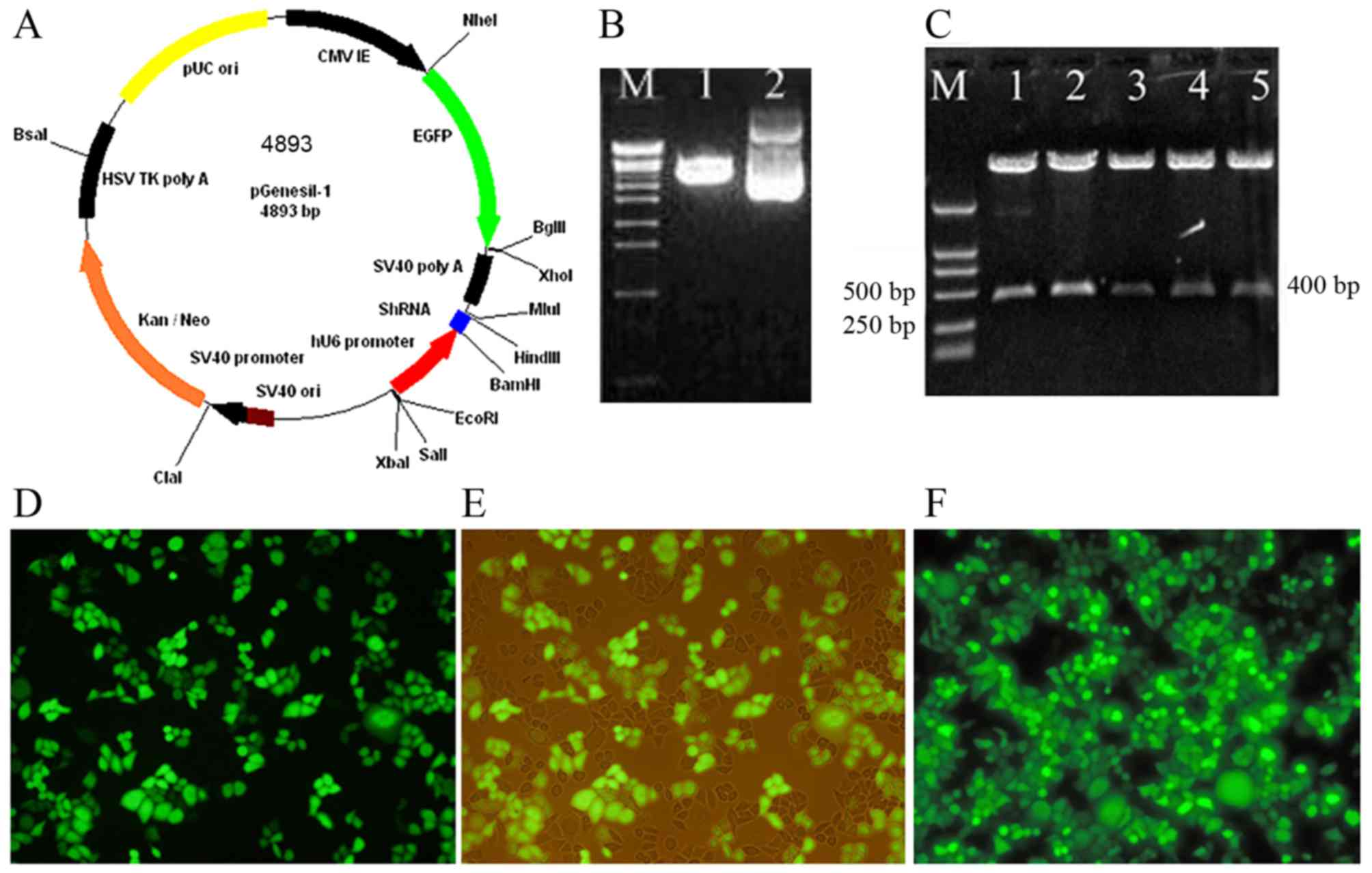 | Figure 3.Recombinant eukaryotic BDNF siRNA
expression vectors were successfully constructed and stably
transfected into HeLa cells with high efficiency. (A) Schematic
drawing of the pGenesil-1 vector. The hU6-RNA promoter was cloned
in front of the gene-specific targeting sequence. (B) Restrictive
enzyme digestion analysis of pGenesil-1 by agarose gel
electrophoresis. Lane M, EcoT-14 maker (19, 329, 7,743, 6,223,
4,254, 3,472, 2,690, 1882, 1,489, 925, 421 and 74 bp); lane 1,
pGenesil-1 digested by BamHI and HindIII; lane 2,
undigested pGenesil-1. (C) A specific SalI site was designed
and synthesized into the sequences of shRNA. Recombinant vectors
were digested to a 400 bp fragment by SalI with the correct
insertion. Lane M, DL2000 maker (2,000, 1,000, 750, 500, 250 and
100 bp); lanes 1–5 represent pBDNF1, pBDNF2, pBDNF3, pBDNF4 and
pScr digested by SalI, respectively. (D and E) The
transfection efficiencies and expression effects of BDNF siRNA were
estimated at 24 h after transfection and (F) after positive clones
were established. The EGFP expression was observed by fluorescence
microscopy; (E) is the same image as (D) except for the cells were
observed by light and fluorescence mixture. |
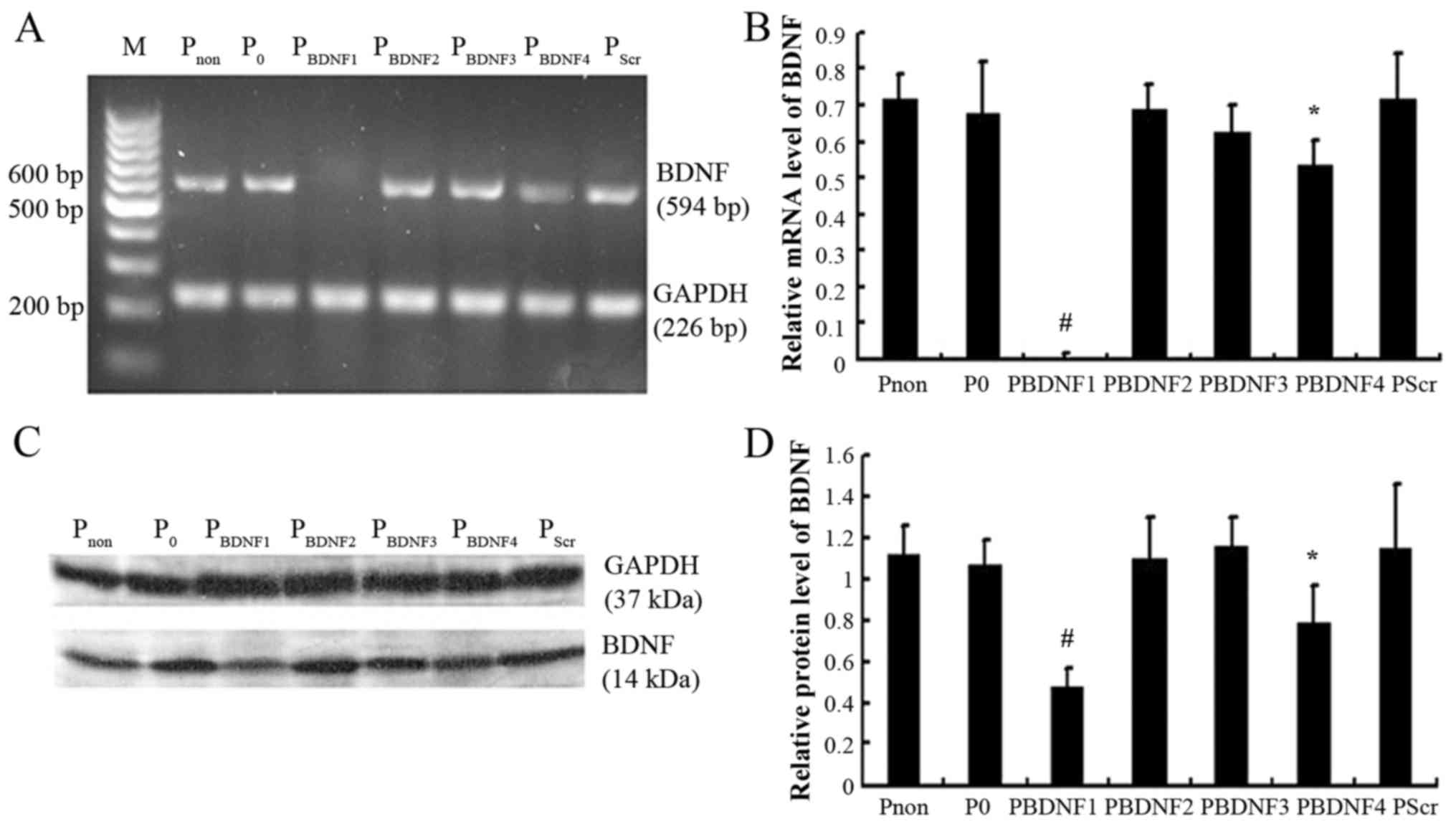
siRNA1 is the most efficient sequence
among the siRNAs examined
HeLa cells were divided into non-transfected
(Pnon), pGenesil-1-tranfected (P0), pScr
(PScr) and positive experimental groups
(PBDNF1, PBDNF2, PBDNF3 and
PBDNF4, respectively). As shown in Fig. 4, semi-quantitative RT-PCR and
western blot analysis were performed when the positive clones had
been established. The results revealed that siRNA1 (p<0.01) and
siRNA4 (p<0.05) both led to significant reductions in BDNF
expression without marked changes in GAPDH (GAPDH, data not shown),
while pGenesil-1, siRNA2, siRNA3 and scramble siRNA barely altered
the expression of BDNF (p>0.05). Compared with the level of BDNF
in the Pnon group, siRNA1 led to nearly 99% reduction in
the relative mRNA level, while a 58% decrease in the relative
protein level was noted. siRNA1 was selected as the most efficient
siRNA for use in the next experiments. Stable HeLa cell clones in
the Pnon, P0 and PBDNF1 groups
were used as experimental objects.
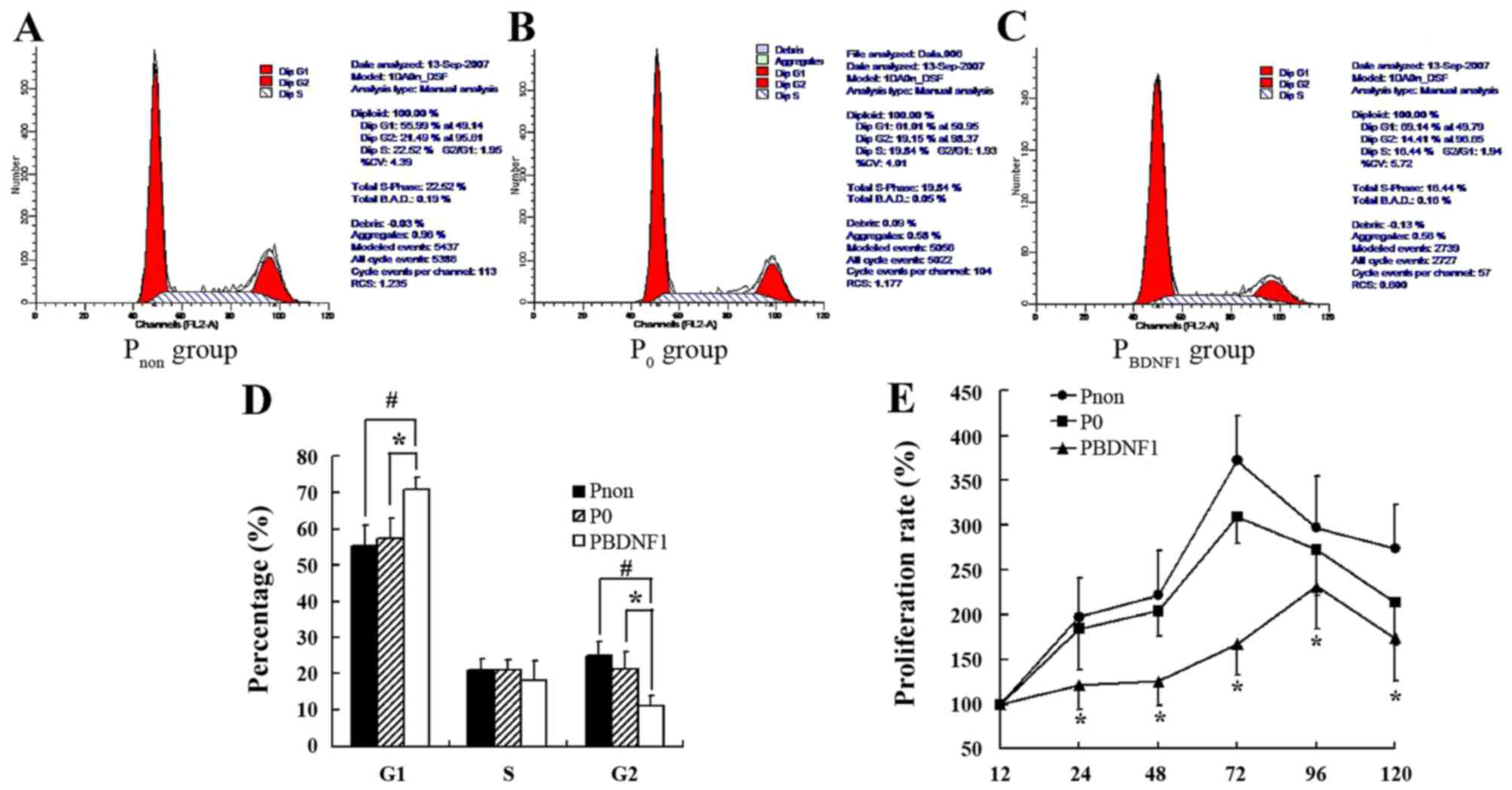
Downregulation of BDNF expression
suppresses the proliferation of HeLa cells and induced cell cycle
arrest in the G1 phase
FCM was performed to evaluate the cell cycle profile
in the BDNF-siRNA-transfected HeLa cells. As shown in Fig. 5A-D, the percentage of cells in the
G1 phase in the PBDNF1 group (70.73±4.15%) was much
higher than that observed in the Pnon (55.33±5.64%)
(p<0.01) and P0 group (57.47±2.98%) (p<0.05).
Meanwhile, the percentage of cells in the G2 phase correspondingly
decreased in the PBDNF1 group (11.15±2.88%) compared
with the Pnon (24.83±3.67%) (p<0.01) and
P0 group (21.28±5.38%) (p<0.05). The percentage of
cells in the S phase had no significant change in the 3 groups
(p>0.05) (Fig. 5A-D). Together,
the results showed that BDNF-siRNA induced cell cycle arrest at the
G1 phase and decreased the distribution of cells in the G2 phase.
The growth curves of cells in the 3 groups as determined by the MTT
assay showed significant growth inhibition of the
BDNF-siRNA-transfected HeLa cells (Fig.
5E).
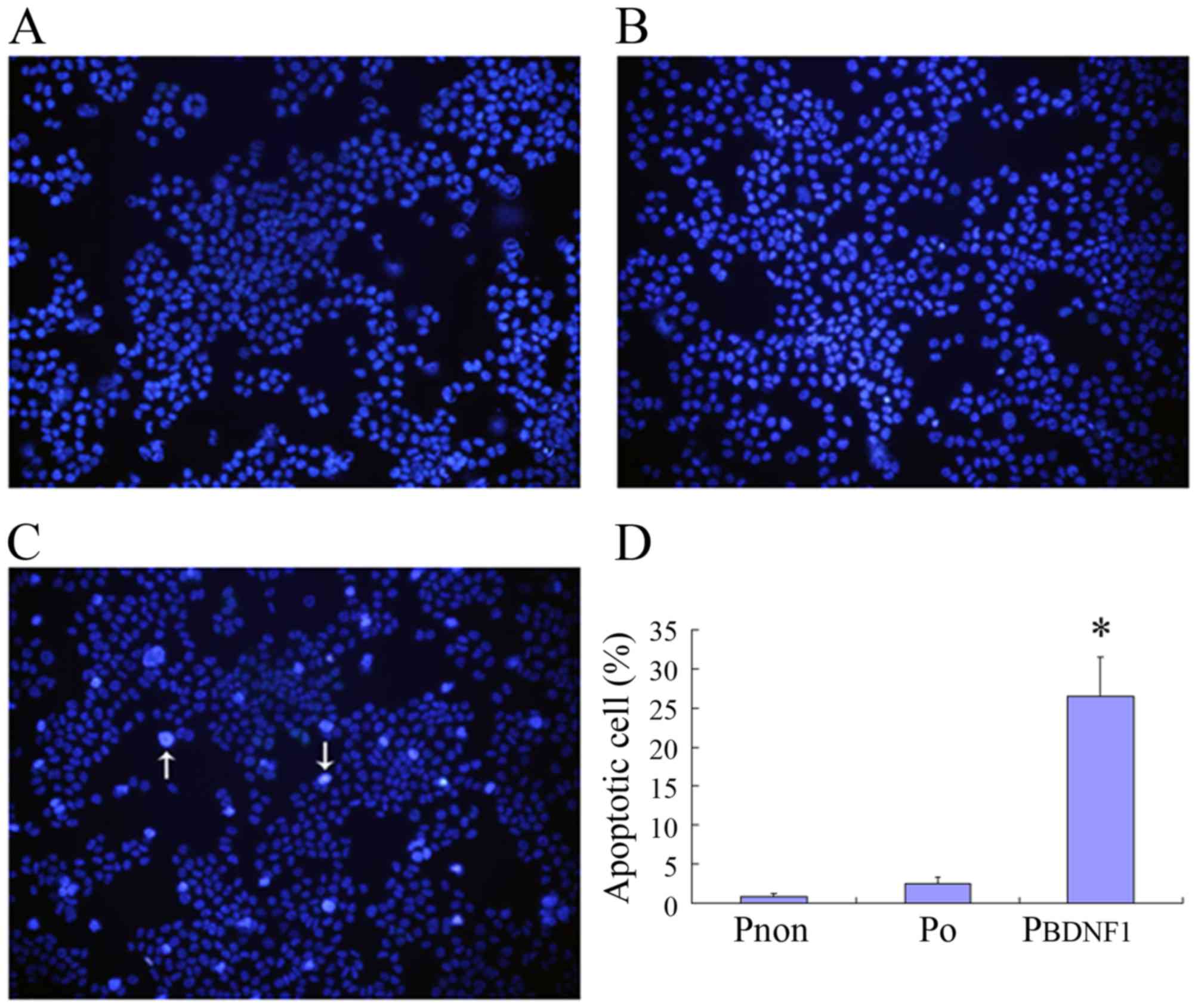
Downregulation of BDNF expression
induces the apoptosis of HeLa cells
To analyze the involvement of BDNF in cell
apoptosis, Hoechst 33258 staining was performed to investigate the
apoptosis-related changes in cell morphology and to further
evaluate the apoptotic rates in the Pnon, P0
and PBDNF1 groups. Observation with fluorescence
microscopy (magnification, ×200) revealed a significant increase in
the number of cells in the PBDNF1 group showing nuclear
condensation and fragmentation which was not observed in the
Pnon and P0 groups (Fig. 6A-C). As shown in Fig. 6D, the percentage of
apoptotic/necrosis cells in the PBDNF1 group
(27.14±4.57%) was much higher than those observed in the
Pnon (0.84±0.39%) and the P0 group
(2.68±1.02%) (p<0.01).
Downregulation of BDNF expression
suppresses the migratory and invasive capabilities of HeLa
cells
To evaluate the role of BDNF in cell migration and
invasion, Transwell assay was used as a tool to determine the
migratory and invasive capabilities of HeLa cells in the
Pnon, P0 and PBDNF1 groups. As
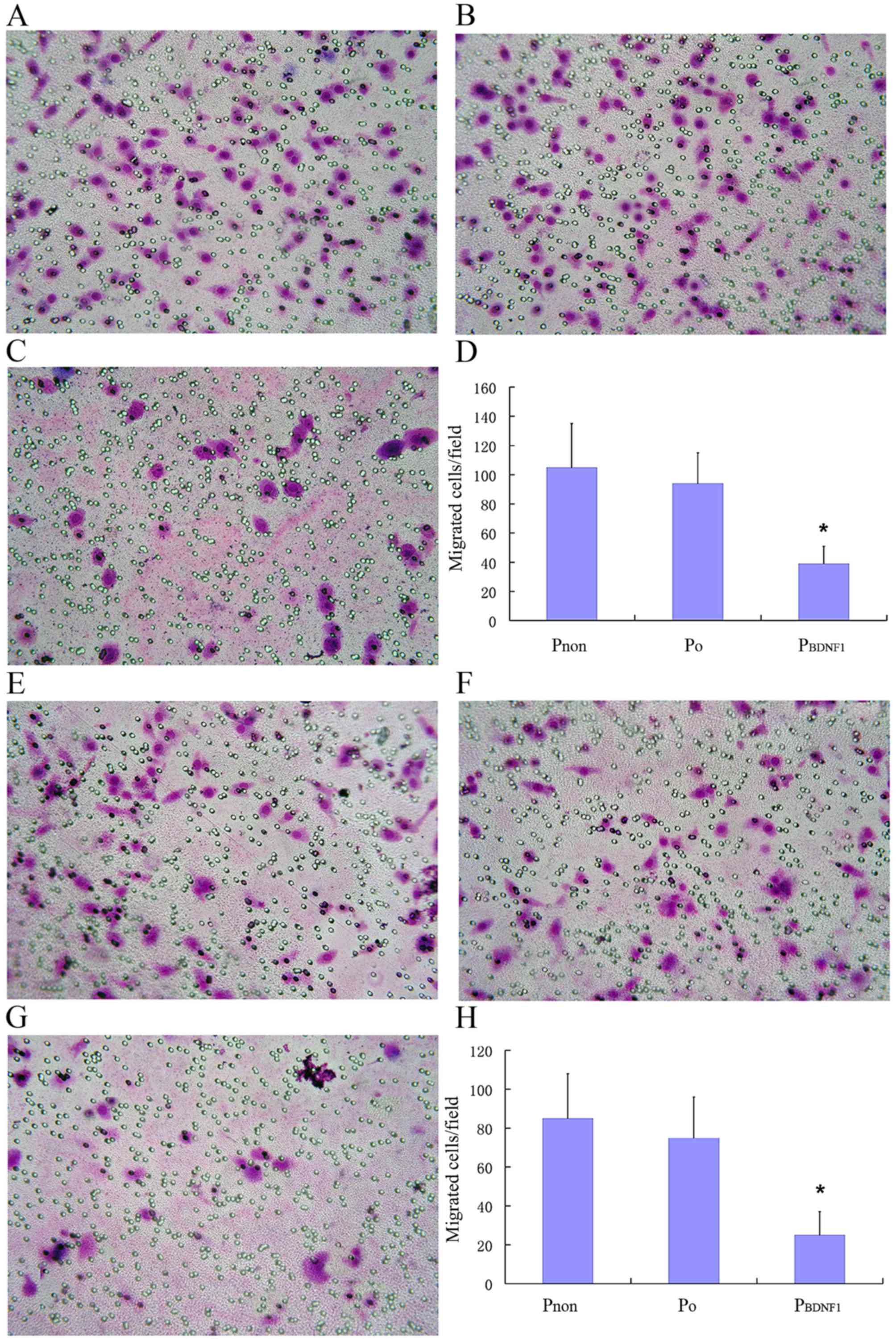
shown in the Fig. 7A-D, the number
of cells in the P0 group that had migrated to the
underside of the filters was similar to that of the cells in the
Pnon group, whereas cells in the PBDNF1 group
showed a significantly reduced migratory capability compared with
the other 2 groups (p<0.01). Migrated cells/field in the
PBDNF1 group (37±17), were significantly less than those
in the Pnon (105±31) and P0 group (92±28).
Migratory capability was impaired in the BDNF-knockdown cells
compared with that noted in the non-transfected cells within at
least a 2-fold reduction (p<0.01). As shown in Fig. 7E-H, similar to the migration assay,
the number of invaded cells/field in the PBDNF1 group
(24±12), was significantly less than those in the Pnon
(85±26) and P0 groups (75±20). Invasive capability was
significantly impaired in the BDNF-knockdown cells compared with
non-transfected cells with at least a 3-fold reduction (p<0.01).
Using stable cell lines expressing shRNA against BDNF, these
results indicate that endogenous BDNF is essential for the ability
of HeLa cells to migrate and invade normally.
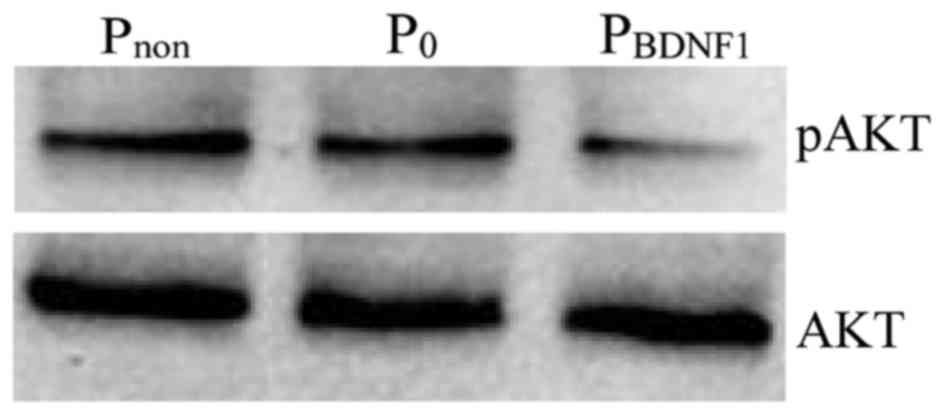
Downregulation of BDNF expression
inhibits the activation states of PI3K signaling
We next sought to study the effect of BDNF on the
activation states of PI3K, a member of the signaling pathways known
to be involved in mediating tumor cell proliferation and migration.
AKT is a downstream target of PI3K-generated signals and becomes
activated after phosphorylation of Ser473. The results showed that
the level of phosphorylated AKT was significantly impaired in the
BDNF-knockdown HeLa cells compared with the level noted in the
non-transfected cells (Fig. 8).
Discussion
The present study was designed to investigate the
biological role of BDNF in cervical carcinoma cell line HeLa. We
showed that both BDNF and also its high-affinity receptor TrkB are
expressed by HeLa cells. It has been documented that an autocrine
loop exists between BDNF and TrkB in malignant tumors such as
neuroblastoma (25), multiple
myeloma (5) and ovarian cancer
(26). In the autocrine loop, high
expression of endogenous BDNF induced expression of TrkB. Hence, we
postulated that BDNF may provide autocrine support for
TrkB-expressing HeLa cells. Cervical carcinoma is the second
leading cause of cancer morbidity and mortality among women
worldwide, particularly in developing countries. It is well known
that infection with high-risk human papilloma virus (HPV) is the
predominant risk factor for cervical carcinoma. Although estrogens
are a human carcinogen for a variety of cancers (27–29),
its effect on cervical carcinoma has not received much attention.
However, it has been revealed that estrogens (E) and the estrogen
receptor (ER) are overexpressed in cervical carcinoma tissue
samples. The E/ER signaling pathway is essential for stimulating
the expression of HPV E6 and E7 mRNA and cervical carcinoma cell
proliferation, anchorage-independent growth and resistance to
drug-induced apoptosis (30). The
BDNF gene contains a sequence with close homology to the estrogen
response element (ERE) and estrogen-ligand complexes are capable of
binding this sequence and protecting it from DNase degradation
(31). Ovariectomy in female rats
was found to reduce BDNF expression and exogenous estrogen
replacement restores it (32).
Gonadectomized male rats show an increase in BDNF mRNA after
estrogen treatment (33). Recent
studies have also found that estrogen regulates BDNF expression via
non-receptor-dependent mechanisms, involving disinhibition of
GABA-ergic neurons (34). It
remains unknown whether BDNF/TrkB is expressed in cervical
carcinoma tissue samples, whether the expression level is
correlated with the clinical stage, and whether BDNF-estrogen
interaction is involved in the pathological mechanism of cervical
carcinoma. We are currently conducting studies to address these
issues.
In the present study, recombinant eukaryotic BDNF
siRNA expression vectors were successfully constructed and
transfected into HeLa cells. siRNA1 was selected as the most
efficient siRNA used in the present study. siRNA-induced silencing
of endogenous BDNF expression suppressed the proliferation of HeLa
cells and induced cell cycle arrest in the G1 phase. These
investigations are consistent with previous findings which
demonstrated that BDNF promoted multiple myeloma (35), pancreatic cancer (36) and hepatocellular carcinoma cell
(4) proliferation in a
dose-dependent manner. Administration of BDNF to hepatocellular
carcinoma cell lines induced significantly increased expression of
cyclin D1 (4). Cyclin D1 is a key
modulator in the G1 phase which controls the cell cycle switch to S
phase. Upregulation of cyclin D1 by exogenous BDNF accelerated the
G1 phase process and promoted tumor cell proliferation. Hence, we
postulated that interference of endogenous BDNF expression in HeLa
cells may alter the cell cycle profile in the G1 phase resulting in
downregulation of the proliferation rate. However, BDNF/TrkB signal
transduction in neuroblastoma cell lines SMS-KCN and SH-SY5Y, and
retinoblastoma cell line RBL-15 are distinct from those observed in
the present study. BDNF was barely able to alter cell proliferation
or change cell cycle distribution (3,25). The
data indicate that the role of BDNF in promoting proliferation is
still controversial; much research must be carried out to fully
elucidate the issue.
Hoechst 33258 staining assay revealed that
interference of BDNF expression also increased cell apoptosis.
Kurokawa et al (37)
constructed a rat retinal ischemia-reperfusion injury model and
found that exogenous BDNF intravitreally injected immediately after
reperfusion decreased the number of caspase-2-positive cells in the
retinal ganglion cell layer. Administration of K252α (a type of
TrkB inhibitor) was able to activate caspase-3 and furthermore
induce apoptosis in lung adenocarcinoma cell line A549 (38). BDNF may reduce neuron apoptosis by
increasing the expression of the Bcl-2 anti-apoptosis protein and
by inhibiting intracellular calcium overload (39). Anoikis is defined as apoptosis
caused by lack of cell-matrix interactions (40), which has been suggested to act as a
barrier to metastasis. Transgenic co-expression of BDNF/TrkB in rat
intestinal epithelial cells resulted in complete transformation of
the cells from normal cells to malignant cells. Transformed cells
showed the capability of anti-apoptosis in systemic circulation and
seeded to a distant place forming secondary tumors (14). A similar phenomenon was observed in
ovarian cancer (41), BDNF and TrkB
were found to be overexpressed in epithelial ovarian cancers and
the BDNF/TrkB/PI3K-AKT signaling pathway may mediate anoikis
suppression. Suppression of anoikis by BDNF may increase the
survival of grafted Schwann cells in the case of therapy for spinal
cord injury (42). In the present
study, it was found that interference of BDNF expression
significantly enhanced cell apoptosis and PI3K/AKT was involved in
BDNF signal transduction in the cervical carcinoma cells. This
evidence suggests that BDNF/TrkB functions as an anti-apoptosis
signal to modulate tumor cell survival.
Metastasis is a major factor in the malignancy of
cancers, and is often responsible for the failure of cancer
treatment. Yu et al (41)
clarified that the overexpression of BDNF/TrkB was significantly
higher in greater omentum metastatic lesions and multicellular
spheroids in ascites than in the corresponding primary lesions. In
the present study, the migratory capability was impaired in the
BDNF-knockdown cells compared with the non-transfected cells with
at least a 2-fold reduction while the invasive capability was
significantly attenuated with at least a 3-fold reduction. BDNF is
a novel cytokine which induces the metastasis of HeLa cells as well
as in multiple myeloma (35), and
lung adenocarcinoma (43). Invasion
of tumor cells into neighboring tissues requires degradation of the
extracellular matrix by proteases. A recent study demonstrated that
at least 2 discrete domains within the tissue-type plasminogen
activator (tPA) gene promoter contribute to the BDNF response
(44). Enhanced TrkB expression in
neuroblastoma cells was associated with a significant increase in
the secretion of subsets of matrix metalloproteinases (MMPs)
(MMP-1, MMP-2 and MMP-9) and the urokinase-type plasminogen
activator (uPA) and tPA, which resulted in an increase in their
invasive capability via increased activity of proteolytic networks
(45). These findings explain why
BDNF/TrkB expression contributes to the migration and invasion of
tumor cells.
In conclusion, we report the expression of BDNF/TrkB
in human cervical carcinoma cell line HeLa. siRNA targeting the
BDNF gene was used to prove that BDNF promotes HeLa cell
proliferation and suppresses apoptosis, migration and invasion. In
addition to the biological roles observed in the present study,
BDNF was also found to act as a pro-angiogenic factor essential for
the formation of tumor blood vessels (46). A new hypothesis suggests that tumors
initiate their own innervations by the release of neurotrophic
factors including nerve growth factor (NGF) and BDNF. By this
process, which is termed neoneurogenesis, tumor cells come into
close contact to nerve cells, forming a neuro-neoplastic synapse.
Through these synapses, neurotransmitters are directly supplied to
the tumor, which has impact on tumor growth and metastasis
formation (47,48). Such findings provide hints to the
possible mechanisms of the BDNF/TrkB signaling pathway in
tumorigenesis which warrants further investigation for the
possibility of alternative therapeutic targets.
Acknowledgements
The present study was supported by grants from the
National Natural Sciences Foundation of China (no. 81272625 for
C.-Y.S.; no. 81302042 to Z.-B.C.) and the Important New Drug
Discovery (no. 2011ZX09302-002 to Y.H.).
Glossary
Abbreviations
Abbreviations:
|
BDNF
|
brain-derived neurotrophic factor
|
|
siRNA
|
small interfering RNA
|
|
Trk
|
tyrosine kinase
|
|
IL-6
|
interleukin-6
|
|
RNAi
|
RNA interference
|
|
dsRNA
|
double-stranded RNA
|
|
RISC
|
RNAi-induced silencing complex
|
|
FBS
|
fetal bovine serum
|
|
cDNA
|
complementary DNA
|
|
PBS
|
phosphate-buffered saline
|
|
TBS
|
Tris-buffered saline
|
|
MTT
|
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium
bromide
|
|
DMSO
|
dimethyl sulfoxide
|
|
RT-PCR
|
reverse-transcriptase polymerase chain
reaction
|
|
EGFP
|
enhanced green fluorescent protein
|
|
HPV
|
human papilloma virus
|
|
ER
|
estrogen receptor
|
|
MMPs
|
matrix metalloproteinases
|
|
uPA
|
urokinase-type plasminogen
activator
|
|
tPA
|
tissue-type plasminogen activator
|
|
NGF
|
nerve growth factor
|
References
|
1
|
Lewin GR and Barde YA: Physiology of the
neurotrophins. Annu Rev Neurosci. 19:289–317. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Novikov L, Novikova L and Kellerth JO:
Brain-derived neurotrophic factor promotes axonal regeneration and
long-term survival of adult rat spinal motoneurons in vivo.
Neuroscience. 79:765–774. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lattanzio F, Carboni L, Carretta D,
Candeletti S and Romualdi P: Treatment with the neurotoxic Aβ
(25–35) peptide modulates the expression of neuroprotective factors
Pin1, Sirtuin 1, and brain-derived neurotrophic factor in SH-SY5Y
human neuroblastoma cells. Exp Toxicol Pathol. 68:271–276. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Guo D, Hou X, Zhang H, Sun W, Zhu L, Liang
J and Jiang X: More expressions of BDNF and TrkB in multiple
hepatocellular carcinoma and anti-BDNF or K252a induced apoptosis,
supressed invasion of HepG2 and HCCLM3 cells. J Exp Clin Cancer
Res. 30:972011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pearse RN, Swendeman SL, Li Y, Rafii D and
Hempstead BL: A neurotrophin axis in myeloma: TrkB and BDNF promote
tumor-cell survival. Blood. 105:4429–4436. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Miknyoczki SJ, Wan W, Chang H, Dobrzanski
P, Ruggeri BA, Dionne CA and Buchkovich K: The neurotrophin-trk
receptor axes are critical for the growth and progression of human
prostatic carcinoma and pancreatic ductal adenocarcinoma xenografts
in nude mice. Clin Cancer Res. 8:1924–1931. 2002.PubMed/NCBI
|
|
7
|
Jia S, Wang W, Hu Z, Shan C, Wang L, Wu B,
Yang Z, Yang X and Lei D: BDNF mediated TrkB activation contributes
to the EMT progression and the poor prognosis in human salivary
adenoid cystic carcinoma. Oral Oncol. 51:64–70. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Stephan H, Zakrzewski JL, Bölöni R,
Grasemann C, Lohmann DR and Eggert A: Neurotrophin receptor
expression in human primary retinoblastomas and retinoblastoma cell
lines. Pediatr Blood Cancer. 50:218–222. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Thiele CJ, Li Z and McKee AE: On
Trk - the TrkB signal transduction pathway is an
increasingly important target in cancer biology. Clin Cancer Res.
15:5962–5967. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ricci A, Graziano P, Mariotta S, Cardillo
G, Sposato B, Terzano C and Bronzetti E: Neurotrophin system
expression in human pulmonary carcinoid tumors. Growth Factors.
23:303–312. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rosean TR, Tompkins VS, Tricot G, Holman
CJ, Olivier AK, Zhan F and Janz S: Preclinical validation of
interleukin 6 as a therapeutic target in multiple myeloma. Immunol
Res. 59:188–202. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li Z, Zhang Y, Tong Y, Tong J and Thiele
CJ: Trk inhibitor attenuates the BDNF/TrkB-induced protection of
neuroblastoma cells from etoposide in vitro and in vivo. Cancer
Biol Ther. 16:477–483. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kupferman ME, Jiffar T, El-Naggar A,
Yilmaz T, Zhou G, Xie T, Feng L, Wang J, Holsinger FC, Yu D, et al:
TrkB induces EMT and has a key role in invasion of head and neck
squamous cell carcinoma. Oncogene. 29:2047–2059. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Douma S, Van Laar T, Zevenhoven J,
Meuwissen R, Van Garderen E and Peeper DS: Suppression of anoikis
and induction of metastasis by the neurotrophic receptor TrkB.
Nature. 430:1034–1039. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sun CY, Hu Y, Wang HF, He WJ, Wang YD and
Wu T: Brain-derived neurotrophic factor inducing angiogenesis
through modulation of matrix-degrading proteases. Chin Med J.
119:589–595. 2006.PubMed/NCBI
|
|
16
|
Sharp PA: RNA interference - 2001. Genes
Dev. 15:485–490. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hutvágner G and Zamore PD: RNAi: Nature
abhors a double-strand. Curr Opin Genet Dev. 12:225–232. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hannon GJ and Rossi JJ: Unlocking the
potential of the human genome with RNA interference. Nature.
431:371–378. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bernstein E, Caudy AA, Hammond SM and
Hannon GJ: Role for a bidentate ribonuclease in the initiation step
of RNA interference. Nature. 409:363–366. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hammond SM, Boettcher S, Caudy AA,
Kobayashi R and Hannon GJ: Argonaute2, a link between genetic and
biochemical analyses of RNAi. Science. 293:1146–1150. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mousavi K and Jasmin BJ: BDNF is expressed
in skeletal muscle satellite cells and inhibits myogenic
differentiation. J Neurosci. 26:5739–5749. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yan QS, Feng MJ and Yan SE: RNA
interference-mediated inhibition of brain-derived neurotrophic
factor expression increases cocaine's cytotoxicity in cultured
cells. Neurosci Lett. 414:165–169. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Reynolds A, Leake D, Boese Q, Scaringe S,
Marshall WS and Khvorova A: Rational siRNA design for RNA
interference. Nat Biotechnol. 22:326–330. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Eaton MJ and Whittemore SR: Autocrine BDNF
secretion enhances the survival and serotonergic differentiation of
raphe neuronal precursor cells grafted into the adult rat CNS. Exp
Neurol. 140:105–114. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nakamura Y, Suganami A, Fukuda M, Hasan
MK, Yokochi T, Takatori A, Satoh S, Hoshino T, Tamura Y and
Nakagawara A: Identification of novel candidate compounds targeting
TrkB to induce apoptosis in neuroblastoma. Cancer Med. 3:25–35.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Streiter S, Fisch B, Sabbah B, Ao A and
Abir R: The importance of neuronal growth factors in the ovary. Mol
Hum Reprod. 22:3–17. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Clemons M and Goss P: Estrogen and the
risk of breast cancer. N Engl J Med. 344:276–285. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Burry K and Cain JM: Estrogen replacement
therapy and risk of ovarian cancer in postmenopausal women. JAMA.
288:2538author reply 2539. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen GG, Zeng Q and Tse GM: Estrogen and
its receptors in cancer. Med Res Rev. 28:954–974. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nair HB, Luthra R, Kirma N, Liu YG,
Flowers L, Evans D and Tekmal RR: Induction of aromatase expression
in cervical carcinomas: Effects of endogenous estrogen on cervical
cancer cell proliferation. Cancer Res. 65:11164–11173. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sohrabji F, Miranda RC and Toran-Allerand
CD: Identification of a putative estrogen response element in the
gene encoding brain-derived neurotrophic factor. Proc Natl Acad Sci
USA. 92:11110–11114. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Singh M, Meyer EM and Simpkins JW: The
effect of ovariectomy and estradiol replacement on brain-derived
neurotrophic factor messenger ribonucleic acid expression in
cortical and hippocampal brain regions of female Sprague-Dawley
rats. Endocrinology. 136:2320–2324. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Solum DT and Handa RJ: Estrogen regulates
the development of brain-derived neurotrophic factor mRNA and
protein in the rat hippocampus. J Neurosci. 22:2650–2659.
2002.PubMed/NCBI
|
|
34
|
Blurton-Jones M, Kuan PN and Tuszynski MH:
Anatomical evidence for transsynaptic influences of estrogen on
brain-derived neurotrophic factor expression. J Comp Neurol.
468:347–360. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hu Y, Sun CY, Wang HF, Guo T, Wei WN, Wang
YD, He WJ, Wu T, Tan H and Wu TC: Brain-derived neurotrophic factor
promotes growth and migration of multiple myeloma cells. Cancer
Genet Cytogenet. 169:12–20. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ketterer K, Rao S, Friess H, Weiss J,
Büchler MW and Korc M: Reverse transcription-PCR analysis of
laser-captured cells points to potential paracrine and autocrine
actions of neurotrophins in pancreatic cancer. Clin Cancer Res.
9:5127–5136. 2003.PubMed/NCBI
|
|
37
|
Kurokawa T, Katai N, Shibuki H, Kuroiwa S,
Kurimoto Y, Nakayama C and Yoshimura N: BDNF diminishes caspase-2
but not c-Jun immunoreactivity of neurons in retinal ganglion cell
layer after transient ischemia. Invest Ophthalmol Vis Sci.
40:3006–3011. 1999.PubMed/NCBI
|
|
38
|
Perez-Pinera P, Hernandez T, García-Suárez
O, de Carlos F, Germana A, Del Valle M, Astudillo A and Vega JA:
The Trk tyrosine kinase inhibitor K252a regulates growth of lung
adenocarcinomas. Mol Cell Biochem. 295:19–26. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu Z, Ma D, Feng G, Ma Y and Hu H:
Recombinant AAV-mediated expression of human BDNF protects neurons
against cell apoptosis in Abeta-induced neuronal damage model. J
Huazhong Univ Sci Technolog Med Sci. 27:233–236. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Frisch SM and Screaton RA: Anoikis
mechanisms. Curr Opin Cell Biol. 13:555–562. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yu X, Liu L, Cai B, He Y and Wan X:
Suppression of anoikis by the neurotrophic receptor TrkB in human
ovarian cancer. Cancer Sci. 99:543–552. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Koda M, Someya Y, Nishio Y, Kadota R,
Mannoji C, Miyashita T, Okawa A, Murata A and Yamazaki M:
Brain-derived neurotrophic factor suppresses anoikis-induced death
of Schwann cells. Neurosci Lett. 444:143–147. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sinkevicius KW, Kriegel C, Bellaria KJ,
Lee J, Lau AN, Leeman KT, Zhou P, Beede AM, Fillmore CM, Caswell D,
et al: Neurotrophin receptor TrkB promotes lung adenocarcinoma
metastasis. Proc Natl Acad Sci USA. 111:10299–10304. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Daniel PB, Lux W, Samson AL, Schleuning
WD, Niego B, Weiss TW, Tjärnlund-Wolf A and Medcalf RL: Two
conserved regions within the tissue-type plasminogen activator gene
promoter mediate regulation by brain-derived neurotrophic factor.
FEBS J. 274:2411–2423. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hecht M, Schulte JH, Eggert A, Wilting J
and Schweigerer L: The neurotrophin receptor TrkB cooperates with
c-Met in enhancing neuroblastoma invasiveness. Carcinogenesis.
26:2105–2115. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Usui T, Naruo A, Okada M, Hayabe Y and
Yamawaki H: Brain-derived neurotrophic factor promotes angiogenic
tube formation through generation of oxidative stress in human
vascular endothelial cells. Acta Physiol. 211:385–394. 2014.
View Article : Google Scholar
|
|
47
|
Mancino M, Ametller E, Gascón P and
Almendro V: The neuronal influence on tumor progression. Biochim
Biophys Acta. 1816:105–118. 2011.PubMed/NCBI
|
|
48
|
Entschladen F, Palm D, Niggemann B and
Zaenker KS: The cancer's nervous tooth: Considering the neuronal
crosstalk within tumors. Semin Cancer Biol. 18:171–175. 2008.
View Article : Google Scholar : PubMed/NCBI
|