Introduction
Lung cancer is the most common cancer type, which is
responsible for around 1.4 million deaths worldwide each year
(1). NSCLC represents more than 80%
of lung cancer cases (2). Although
there have been advances in surgical techniques, chemotherapy and
radiotherapy for treating lung cancer, the overall survival of
patients remains low (3,4). Therefore, it is important to
investigate a more complete system for prognostic evaluation. It is
necessary to identify a more useful prognostic and predictive
markers for early molecular diagnosis or potential targets for more
precise risk analysis and development of new therapies.
The daily cycle of light and dark is a driving force
for various living organisms. Therefore, circadian rhythms are
basic regulators of an organism's biological activities. Circadian
rhythms are daily oscillations that are regulated by endogenous
clock genes. They make it easier for organisms to adapt to daily
environmental changes, including temperature, pressure and light.
They are also able to synchronize multiple molecular, biochemical,
physiological and behavioural processes (5,6).
Common core clock genes include CLOCK, BMAL1, Period1 (Per1),
Period2 (Per2), Period3 (Per3), cryptochrome1 (Cry1),
cryptochrome2, casein kinase1 epsilon and timeless (7). A number of epidemiological studies
have suggested that disruption of the normal circadian rhythm may
increase the risk of developing various types of cancer, including
breast (7), prostate (8), colorectal, liver and endometrial
cancers (9–11). Among all known clock genes, only the
members of the Per subfamily function as tumour suppressors in mice
(8). The neoplastic growth of
cancer cells may be restrained by overexpression of Per1 or Per2,
and their apoptotic rate may also be increased (8). Importantly, the involvement of Per1
and Per2 in ataxia telangiectasia mutated checkpoint kinase DNA
damage response pathways implicates the circadian system in tumour
suppression (9).
It has been reported that Per1 is downregulated in
NSCLC cell lines and inhibits cancer cell proliferation (7). In addition, Couto et al
(10) reported that Per3 was
downregulated in NSCLC, and polymorphisms in Per3 genes may be a
factor underlying NSCLC in Brazilian patients. According to these
reports, the members of the Per subfamily appear to be responsible
for tumour progression in NSCLC. However, the biological function
and clinicopathological significance of Per2 in NSCLC remains
unclear. In the present study, the role of Per2 and its relative
clinical significance in NSCLC were explored. It was identified
that loss of Per2 was associated with NSCLC progression, and
recovery of Per2 expression could suppress A549 cell growth and
migration in vitro and in vivo, suggesting that Per2
may serve as a potential molecular target for NSCLC.
Materials and methods
Clinical samples
A total of 31 pairs of NSCLC and matched
para-carcinoma tissues were collected from the Department of
Thoracic Surgery, Sichuan Cancer Hospital and Institute (Sichuan,
China) between March 2015 and May 2016. All patients were between
30 and 78 years old and among them 20 were male and 31 were female.
All patients provided written informed consent, and the study was
approved by the Ethical Committee of Sichuan Cancer Hospital &
Institute. Lung cancer tissues and matched para-carcinoma tissues
were immediately frozen in liquid nitrogen following surgical
resection and then stored at −80°C until use. The differentiation
and metastasis states were identified by experienced
pathologists.
Cell culture and lentivirus
transduction
A549 cells were purchased from the Institute of
Biochemistry and Cell Biology of Chinese Academy of Science
(Shanghai, China) and cultured in RPMI-1640 (Hyclone; GE Healthcare
Life Sciences, Logan, UT, USA) with 15% foetal bovine serum (FBS;
Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) at 37°C in
a humidified incubator containing 5% CO2. Overexpression
lentivirus of Per2 was purchased from Genechem (Shanghai GeneChem
Co., Ltd., Shanghai, China). A549 cells were infected with virus at
a multiplicity of infection of 60, and the expression rate of green
fluorescence protein was observed with a fluorescence microscope
(IX70; Olympus Corporation, Tokyo, Japan) on day 3. Finally, the
infected efficiency of A549 following 72 h of lentiviral
transduction was confirmed by reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) and western blot analysis.
Wound healing assay
A549 cells at a density of 3×105
cells/well were seeded in 6-well culture plates and cultured
overnight at 37°C. Then, a sterile, plastic 200-µl micropipette tip
was used to make a straight scratch on the confluent cell
monolayer. The well was washed with medium to remove detached
cells, then cells were cultured for a further 48 h at 37°C.
Finally, images were obtained under a light microscope and the
width of the wound was measured with the microscope and ImageJ
software v1.37 (National Institutes of Health, Bethesda, MD
USA).
Cell proliferation and colony
formation assays
A549 cells with or without lentiviral infection were
seeded at a density of 1.5×103 cells/per well into
96-well plates. The cell growth of A549 cell lines was determined
by Cell Counting Kit-8 (CCK-8) assay (Dojindo Molecular
Technologies, Inc., Kumamoto, Japan) once a day for 1 week. Assays
of each group were performed in quintuplicate wells.
For colony formation assays, A549 cells with or
without lentiviral transduction were seeded into 6-well plates at a
low density (200 cells/per well) and cultured for 2 weeks. Then,
cells were fixed with 4% paraformaldehyde at 37°C for 15 min and
stained with 1% crystal violet for 30 min at room temperature.
Finally, the number of colonies of substantial size (>50 cells
per colony) were imaged by light microscope, with three fields of
view per well. The experiments were repeated in triplicate.
Cell cycle analysis by DNA
content
Briefly, A549 cells with or without lentiviral
transduction for 48 h were harvested and washed by centrifugation
at 200 × g for 5 min at 25°C in 1X PBS buffer. Next, cells were
fixed in 70% ethanol at 4°C overnight and washed twice with cold 1X
PBS buffer. Then, cells were resuspended in 100 µl RNase A (10
µg/ml), incubated at 37°C for 30 min and immediately stained with
400 µl propidium iodide (500 µg/ml; Nanjing KeyGen Biotech Co.,
Ltd., Nanjing, China) at 4°C for 30 min in the dark. Finally, data
were acquired on a flow cytometer in triplicate (Coulter Epics XL;
Beckman Coulter, Inc., Brea, CA, USA) and analysed by Flow Jo7.6
software (FlowJo LLC, Ashland, OR, USA).
In vitro migration and invasion
assays
Migration assays and invasion assays were performed
using 24-well Transwell filters with 8 µm pore size polycarbonate
membrane (Corning, Inc., Corning, NY, USA) and an invasion assay
kit that included Matrigel (BD Biosciences, San Jose, CA, USA).
Briefly, 48 h following transfection, 2×104 (for the
migration assay) or 1×105 cells (for the invasion assay)
were resuspended in 200 µl serum-free RPMI-1640 and plated into the
upper chamber. The bottom of the lower chambers in the 24-well
plates was filled with 0.6 ml medium containing 10% FBS, which was
used as a chemoattractant. Following incubation at 37°C for 48 h,
the Transwell inserts were removed from the plate, and a
cotton-tipped applicator was used to carefully remove the media and
remaining cells that had not migrated or invaded from the top of
the membrane without damaging it. Then, cells were fixed with 600
µl of 4% paraformaldehyde, stained with 0.5% crystal violet and
incubated at room temperature for 5 min at 37°C. Then, the
Transwell membranes were allowed to dry. The lower chamber was
photographed by a light microscope and cells were counted in at
least five different fields of view.
Hematoxylin and eosin (H&E)
staining
Xenograft tissues were immersed in 4%
paraformaldehyde at 37°C for 4 h, then transferred to 70% ethanol.
Individual lobes of xenograft tissue biopsy material were placed in
processing cassettes, dehydrated through a serial alcohol gradient,
and embedded in paraffin wax blocks. Prior to staining, 5-µm-thick
lung tissue sections were dewaxed in xylene, rehydrated through
decreasing concentrations of ethanol, and washed in PBS. Tissues
were then stained at room temperature with H&E. Then, sections
were dehydrated through increasing concentrations of ethanol and
xylene. A light microscope was used to examine the tissues.
Immunohistochemistry (IHC)
analysis
The tumor xenograft were immersed in 4%
paraformaldehyde at 37°C for 4 h, and 5 µm thick slices were used
for IHC staining by the streptavidin-biotin-peroxidase complex
method. The antigen retrieval procedure was performed by heating
the samples in antigen retrieval solution containing 10 mM sodium
citrate buffer with a pressure cooker at 100°C for 150 sec. When
the slices were cooled, normal goat serum blocking solution
(Beyotime Institute of Biotechnology, Shanghai, China) was used for
blocking at 37°C for 2 h. Rabbit anti human Per2 (1:200; ab179813;
Abcam, Cambridge, MA, USA), Ki-67 (1:500; ab8191; Abcam),
E-Cadherin (1:100; ab133597; Abcam) and NM23 (1:100; ab154529;
Abcam) were incubated at 4°C overnight. Then, the Dako REAL™
EnVision™ kit (K500711; Dako; Agilent Technologies, Inc., Santa
Clara, CA, USA) was used to incubate the slices at 37°C for 30 min.
IHC staining intensity was independently scored under a light
microscope by two anatomical pathologists. The staining intensity
(negative=0, weak=1, moderate=2 or strong=3) and the proportion of
positively stained cells (<25%=1, 25–50%=2, >50 and
<75%=3, ≥75%=4) were scored. The immunostaining was
semi-quantitatively categorized by multiplying the intensity and
the quantity scores to yield a staining index (values from 0 to
12). A staining index of 3–12 was regarded as high expression,
while a staining index of 0–2 was regarded as low expression.
Animal studies
This experiment was conducted in accordance with the
Care and Use of Laboratory Animals (12) and was approved by the Ethical
Committee of Sichuan Cancer Hospital & Institute. Sixteen
4-week-old male BALB/c-nude mice weighing 12–15 g were purchased
from the Laboratory Animal Centre of Sichuan University (Chengdu,
China) and reared in specific-pathogen-free conditions. The mice
were randomly and equally divided into experimental and control
groups, in which the experimental group mice underwent tumour cell
transplantation with Per2 overexpression and the control group mice
underwent tumour cell transplantation without Per2 overexpression.
For the subcutaneous transplantation tumour model, 2×106
A549 cells that did or did not overexpress Per2 were resuspended in
0.1 ml PBS and then injected subcutaneously into the backs of the
mice. Tumour growth was measured with Vernier calipers over the
course of 30 days following transplantation of tumour cells, and
tumour volume was calculated according to the formula: Tumour
volume = 0.5× length × width2. For the lung metastasis
model, 1×105 A549 cells diluted in 100 µl PBS was
intravenously injected via the mouse tail vein. The metastasis
score was determined by the pathological condition of the
metastatic mice following treatment for 4 weeks.
RNA extraction and RT-qPCR
Total RNA was extracted from cells using a Total RNA
kit (Omega Bio-Tek, Inc., Norcross, GA, USA) according to the
manufacturer's protocol. Reverse transcription to cDNA was
perfroemd using Prime Script™ RT Master mix (Takara Biotechnology
Co., Ltd., Dalian, China). The cDNA was used for qPCR using the
CFX96 Real-Time Quantitative PCR system (Bio-Rad Laboratories,
Inc.) with SYBR® Premix ExTaq™ (Takara Biotechnology
Co., Ltd.) according to the manufacturer's protocol. Relative mRNA
expression levels were determined by the cycle threshold (Ct)
normalized against GAPDH using the 2−ΔΔCt formula
(13). Experiments were performed
in triplicate. The primers used in this study were as follows
(5′-3′): Per2-forward (F), CAGGTGAAAGCCAATGAAGAG; Per2-reverse (R),
GGGAGGTGAAACTGTGGAAC; Bax-F, CCCGAGAGGTCTTTTTCCGAG; Bax-R,
CCAGCCCATGATGGTTCTGAT; P53-F, ACTTGTCGCTCTTGAAGCTAC; P53-R,
GATGCGGAGAATCTTTGGAACA; P21-F, CCAACAAACTTAACGTGCCAC; P21-R,
AGGCTCAACAGTAACTGCATC; Nm23-F, CATTGCGATCAAACCAGATG; Nm23-R:
CAGAAGTCTCCACGGATGGT; E-Cadherin-F, CGAGAGCTACACGTTCACGG;
E-Cadherin-R, GGGTGTCGAGGGAAAAATAGG; vascular endothelial growth
factor (VEGF)-F, GGAGGAGGAAGAAGAGAAGGAA; VEGF-R,
GTGGAGGTAGAGCAGCAAGG; CD44-F, CAGCACCATTTCAACCACAC; CD44-R,
GCCAAACCACTGTTCCTTCT; cMyc-F, CCGAGGAGAATGTCAAGAGG; cMyc-R,
ACGCACAAGAGTTCCGTAGC; GAPDH-F, GAAGGTGAAGGTCGGAGTC; and GAPDH-R,
GAAGATGGTGATGGGATTTC.
Western blot analysis
Antibodies for Per2 (ab179813), p53 (#48818), BAX
(#2774), P21 (#2947), E-Cadherin (#14472), NM23 (#3338), VEGF
(#2463), CD44 (#3570), c-Myc (#5605) and β-actin (#3700) were
purchased from Abcam or Cell Signaling Technology, Inc. (Danvers,
MA, USA) and all the antibodies were rabbit anti-human. Cells were
harvested, lysed with RIPA buffer (Sigma-Aldrich; Merck KgaA,
Darmstadt, Germany) supplemented with 1 mmol/l PMSF (Boster
Biological Technology, Pleasonton, CA, USA) and then centrifuged at
14,000 × g at 4°C for 10 min. Protein concentrations of the
extracts were measured using a bicinchoninic acid protein assay kit
(Nanjing KeyGen Biotech Co., Ltd.). Proteins (20 µg) were separated
through 10% SDS-PAGE gels, and then transferred to polyvinylidene
difluoride membranes. Following blocking in Tris-buffered saline
with Tween-20 containing 5% non-fat milk for 60 min, the membranes
were incubated with primary antibodies (P53, Bax, c-Myc, Per2,
NM23, CD44, E-cadherin and VEGF primary antibody: 1:1,000 dilution;
P21 primary antibody: 1:500 dilution; β-actin as a loading control:
1:2,500 dilution) overnight at 4°C. The membranes were then
incubated with the secondary antibodies(mouse anti-rabbit and
horseradish peroxidase-linked antibody, 1:5,000 dilution; Cell
Signaling Technology, Inc.). Following incubation in enhanced
chemiluminescence solution, the proteins on the membranes were
detected using Bio-Rad Universal Hood III and analysed using Image
Lab™ software 2.0 (Bio-Rad Laboratories, Inc.).
Statistical analysis
Data are presented as the mean ± standard deviation,
with the exception of tissue expression data, which are presented
as the median ± range. The differences in Per2 expression between
NSCLC tissues and matched para-carcinoma adjacent normal lung
tissues were compared by a two-tailed independent samples Student's
t-test. All analyses were performed using GraphPad Prism 6.0
software (GraphPad Software, Inc., La Jolla, CA, USA). P<0.05
was considered to indicate a statistically significant
difference.
Results
Per2 is downregulated in NSCLC tissues
and associated with clinicopathological characteristics
Per2 gene exhibits low expression in certain cancer
tissues. In the present study, the expression of Per2 in lung
cancer was evaluated by performing RT-qPCR in different
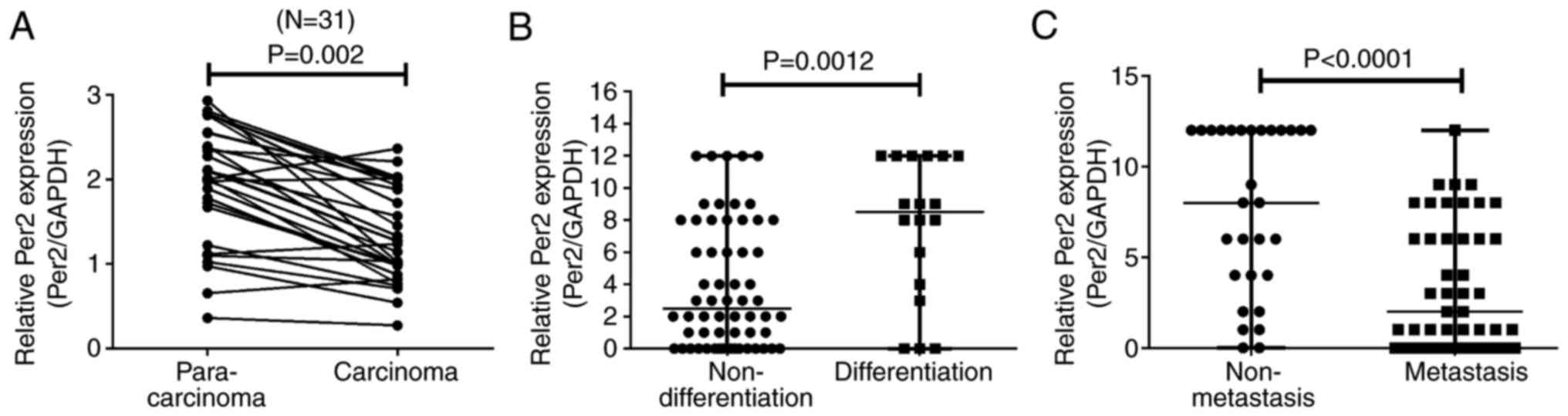
histological subtypes of NSCLC tissue. As presented in Fig. 1A, compared with matched controls,
the Per2 mRNA level was strongly downregulated in 83.87% (26 of 31)
of NSCLC samples (P<0.01 vs. control). As indicated in Fig. 1B and C, Per2 expression was
significantly associated with pathological differentiation
(P<0.01) and lymph node metastasis (P<0.0001). Increased Per2
expression indicated increased differentiation and reduced lymph
node metastasis. These findings suggested that Per2 may be involved
in NSCLC development.
Overexpression of Per2 decreases A549
cell growth by blocking the cell cycle at G0/G1 phase
Based on the finding that Per2 expression was
decreased in NSCLC samples, a Per2-expressing lentivirus was
constructed and used to infect A549 cells, in order to determine
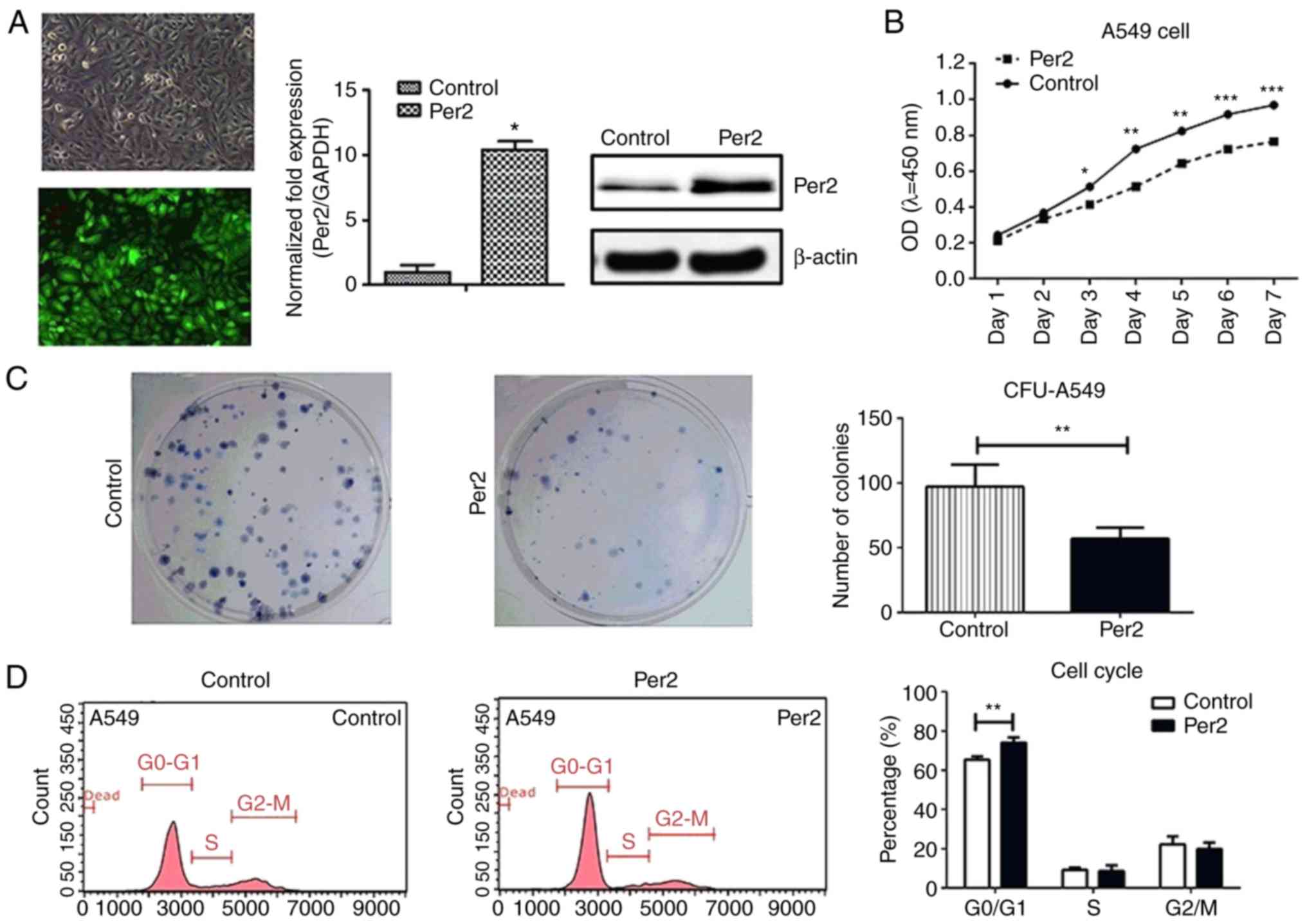
the potential role of Per2 in NSCLC. As presented in Fig. 2A, the mRNA expression of Per2
increased 11-fold and Per2 protein expression also increased in
A549 cells following infection with Per2-expressing lentivirus.
Next, a CCK-8 assay, colony formation assay and cell cycle analysis
were performed to detect the effect of overexpression of Per2 on
A549 cell growth. The results of the CCK-8 and colony formation
assays demonstrated that overexpression of Per2 suppressed A549
cell proliferation and colony formation compared with the control
group (P<0.05; Fig. 2B and C).
Furthermore, flow cytometry was performed to analyse the influence
of Per2 on the cell cycle and it was identified that the percentage
of A549 cells that remained in G1 phase was significantly increased
in the Per2 overexpression group compared with the control group
(76.67±10.22 vs. 66.80±11.38%; P<0.01), while fewer cells stayed
in S phase compared with the control group (6.01±2.74 vs.
10.03±3.01%) after 48 h of culture (Fig. 2D). These results indicated that
increased Per2 expression in A459 cells inhibits tumour cell
proliferation by inducing cell cycle arrest at G0/G1 phase.
Overexpression of Per2 suppresses A549
cell migration and invasion ability
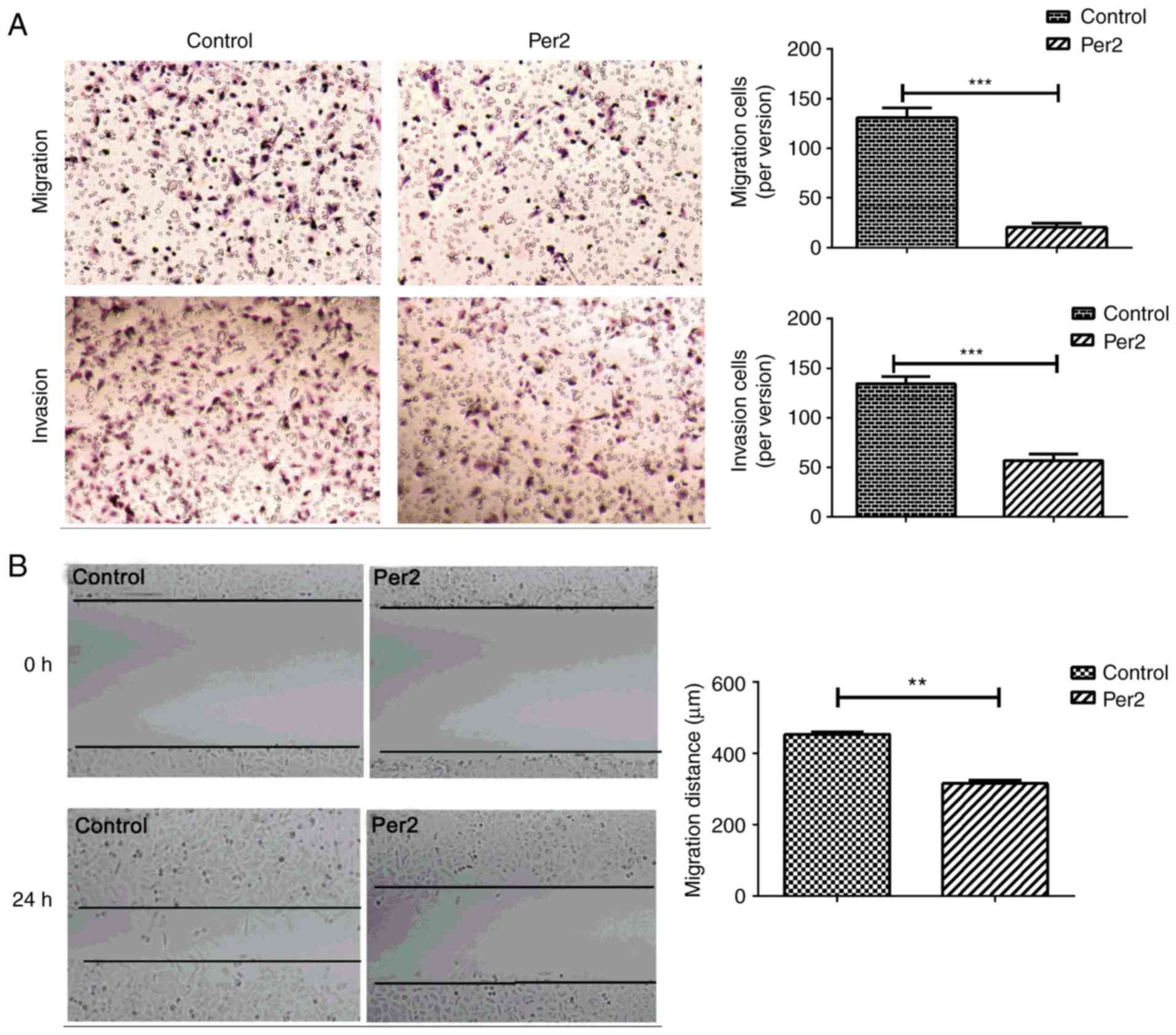
Metastasis begins when the tumour invades into the
vascular stroma and migrates to the blood (14). Therefore, the influence of Per2 on
tumour cell migration and invasion ability was evaluated with in
vitro Transwell assays. Overexpression of Per2 significantly
inhibited the number of migrating and invading A549 cells compared
with the negative control group (migration: 130.80±9.987 vs.
20.20±4.164; invasion: 134.20±7.165 vs. 56.80±6.763; both
P<0.0001; Fig. 3A). Similarly,
results of the wound healing assay revealed that wound gap closure
and migration was significantly decreased for A549 cells
overexpressing Per2 (P<0.01; Fig.
3B). These results demonstrated that Per2 may act as an
anti-oncogene in suppressing A549 cell growth and metastasis.
Overexpression of Per2 decreases
apoptosis- and metastasis-associated gene and protein expression in
A549 cells
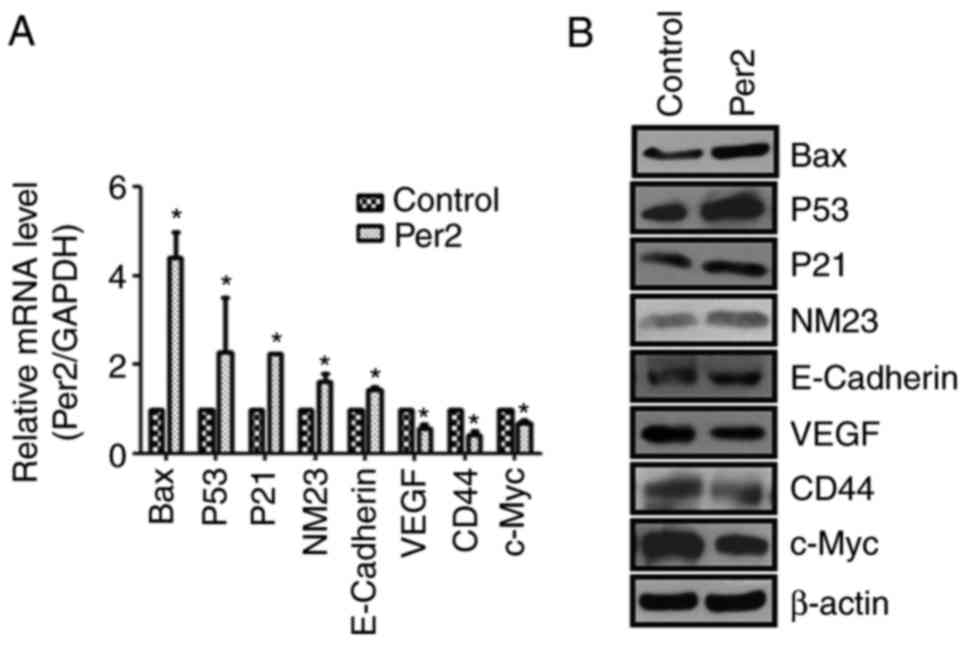
To further elucidate the molecular mechanisms
underlying Per2 effects on cell growth and metastasis,
apoptosis-associated genes Bax, P53, P21 and NM23,
metastasis-associated genes E-Cadherin, VEGF and CD44, and c-Myc
oncogene were evaluated. RT-qPCR and western blotting results are
presented in Fig. 4. Overexpression
of Per2 markedly increased the expression of Bax, P53, P21 and NM23
and suppressed VEGF, CD44 and c-Myc expression. These findings
indicated that the antitumour effect of Per2 is associated with
these apoptosis- and metastasis-associated genes.
Overexpression of Per2 reduces tumour
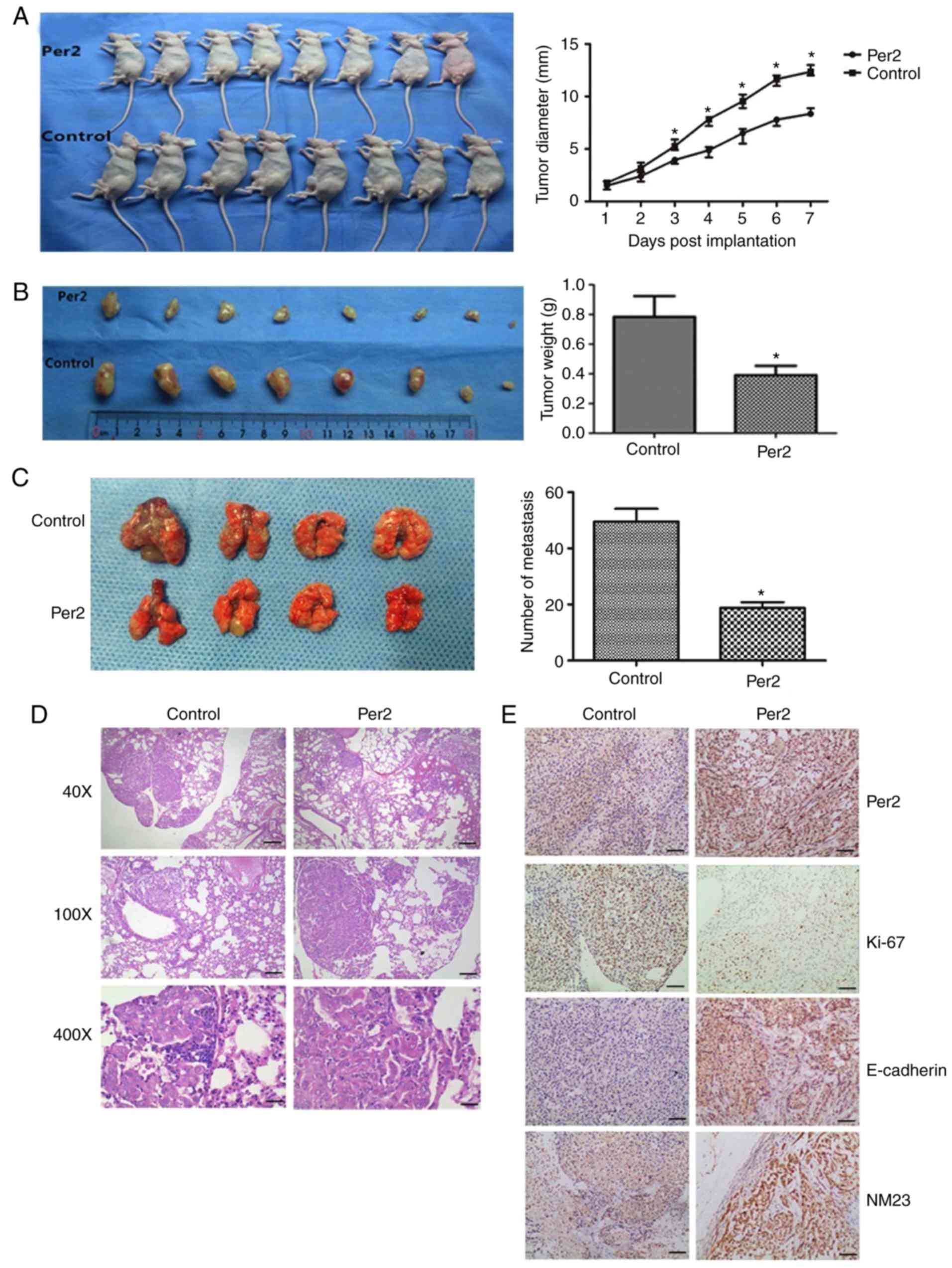
formation of A549 cells in a nude mouse xenograft model
To evaluate whether Per2 exhibited antitumour
effects in vivo, A549 cells that were or were not transduced
with Per2-overexpressing lentivirus were subcutaneously implanted
into the backs of BALB/c-nude mice. Consistent with the current
in vitro results, Per2-lentivirus A549 NSCLC cells exhibited
delayed tumour formation compared with negative controls
(P<0.05; Fig. 5A). At 30 days
after implanting tumour cells, the tumour weight of the
Per2-overexpressing group was 0.328±0.171 g, while that of the
negative control group was 0.785±0.249 g; therefore, the tumour
growth inhibition rate of the Per2-overexpressing group was 63.56%
(P<0.05; Fig. 5B). Furthermore,
the number of lung metastasis tumours in the Per2-overexpressing
group was significantly decreased when compared with those of the
control group, and the H&E staining identified less metastasis
in the Per2-overexpressing group (Fig.
5C and D). In addition, IHC analysis was performed on animal
tumour samples. As indicated in Fig.
5E, the score of Per2 expression in the Per2-overexpression
group (6.38±2.504) was increased compared with the control group
(3.12±2.232). For E-cadherin, NM23 and Ki-67 protein expression,
IHC indicated that overexpression of Per2 could increase expression
of the tumour growth suppressor E-cadherin and tumour metastasis
inhibitor NM23, and lower the expression of Ki-67. Collectively,
these results suggested that overexpression of Per2 reduces the
capacity of A549 cells to form tumours in vivo.
Discussion
Core circadian genes play an important role in
tissue homeostasis and tumourigenesis. Multiple cancer types are
commonly caused by the disruption of circadian rhythms (15). Thus, improved understanding of the
underlying biological role of circadian rhythms in NSCLC would help
to elucidate the occurrence and development of lung cancer and
bring about new ideas for NSCLC diagnosis and therapy. In the
current study, it was identified that compared with adjacent
non-cancerous tissues, Per2 expression was strongly downregulated
in NSCLC samples and reduced expression was associated with
pathological non-differentiation and lymph node metastasis.
Functional studies identified that enhanced Per2 expression in A549
non-small cell lung cancer cells significantly suppressed cell
proliferation, migration and invasion,7 and inhibited NSCLC growth
and metastasis in vivo. Therefore, Per2 may be a novel
molecular target for NSCLC.
Numerous studies have confirmed that circadian
rhythm dysfunction is associated with the pathogenesis of cancer.
Disruption of circadian rhythms may promote tumour progression, and
restored circadian rhythms may potentially have a positive effect
on prognosis (16). For instance,
compared with normal breast tissues, the expression levels of Per1
and Per2 genes may decrease in sporadic and familiar breast tumours
(17). When compared with sporadic
forms, the Per1 gene has lower expression levels, and its form is
similar to that of breast cancer. This means that a potential
deregulation of the circadian clock may result in the inherited
form of the disease (16). The
breast cancer cells may survive due to the methylation of Per1 and
Cry1 gene promoters, inactivating expression of these genes and
disrupting the circadian cell rhythm (18). Furthermore, factors including
patient age, tumour histological grade, invasion depth, lymph node
metastasis and TNM staging are also associated with Per2 protein
expression in patients with breast cancer (19). Although the clinical significance
and certain roles of Per2 in the malignant biological behaviour of
NSCLC have been reported, including chemoresistance and
radioresistance, researchers have not explored the influence of
Per2 on NSCLC metastasis in vitro and in vivo
(20). In the current study, the
roles of Per2 were explored in vitro and a metastasis model
was also established in vivo by tail vein injection. It was
identified that overexpression of Per2 inhibited metastasis of
NSCLC. Consistent with previous studies, the current results
indicated that Per2 expression was markedly downregulated in
~83.87% (26 of 31) of the NSCLC samples compared with their
adjacent control tissues. Furthermore, it was identified that
decreased tumour differentiation and increased lymph node
metastasis were associated with decreased Per2 expression,
indicating that NSCLC is closely associated with the circadian
gene. In addition, these results suggest Per2 deletion or mutation
may result in tumour progression and metastasis.
Numerous studies have reported that Per2 plays an
important role in the generation and maintenance of circadian
rhythms. It controls 2–10% of all mammalian genes (21). For rodents, ~7% of clock-controlled
genes regulate cell proliferation or apoptosis (22,23).
In addition, it has been reported that circadian rhythms change
with human and other mammalian tumour formation (24,25).
Furthermore, studies have suggested that decreased Per2 may disrupt
normal circadian rhythms of behaviour and physiology, and could
significantly increase the chance of tumour and proliferative
phenotypes (26–28). These findings demonstrate that Per2
is of great importance for carcinogenesis. In the current study,
Per2 expression was increased in A549 cells by transducing them
with a Per2-overexpressing lentivirus and then the role of Per2 in
cell proliferation, migration and invasion capacity was evaluated.
It was identified that enhanced Per2 expression strongly suppressed
A549 cell growth in the form of blocking the cell cycle in G1 phase
and restricting cell migration and invasion. The study of NSCLC
xenografts in nude mice also confirmed that tumour growth rates and
volumes were markedly reduced in Per2 lentivirus-transduced A549
NSCLC cells compared with the PBS control group. These findings are
consistent with previous studies and suggest that Per2 may act as
an anti-oncogene in tumour progression.
The antitumour molecular mechanism of Per2 in NSCLC
remains unclear. A previous study reported that the antitumour
effect of the clock gene Per2 is associated with DNA damage
(29). It is now well established
that DNA damage depends on the activation of p53 and p53-associated
transcriptional activation, including p21 (30–32).
Furthermore, hypoxia of tumour cells usually results in VEGF
agonist activity, which may be restrained by Per2; therefore,
tumour angiogenesis may be inhibited (33). It was investigated in the current
study whether the antitumour mechanism of Per2 in NSCLC was
associated the p53-p21-induced DNA damage pathway. In vitro
and in vivo observations indicated that increased Per2
expression in A549 cells not only significantly increased
expression of the tumour anti-oncogenes Bax, P53, and P21, but also
inhibited expression of the pro-oncogenes VEGF, CD44 and c-Myc
(34). These results indicate that
loss of Per2 inhibits tumour cell growth and metastasis by
activating the P53/P21 DNA damage-induced apoptosis pathway in
NSCLC cells. Therefore, it was speculated that reducing Per2 may
promote the proliferation and metastasis of NSCLC. This area will
be investigated further in future studies.
The Per family may function as a cancer suppressor
in the pathogenesis of NSCLC. However, the detailed mechanisms of
the downregulation of Per still require elucidation. CpG
methylation of promoter sequences may inactivate promoter
functions, particularly in tumour suppressor or related genes, and
may also lead to silencing of these genes. A key factor in
silencing cancer suppressor genes is commonly considered to be
methylation (32). Therefore, it is
speculated that methylation of the promoters of Per genes may
result in the inhibition of Per. Chen et al (35) noted that the abnormal expression of
Period in breast cancer is not caused by genetic mutations but by
the methylation of the Per1 or Per2 promoter. However, Nagel et
al (36) identified that the
microRNA-192/194 cluster is an effective inhibitor of the entire
Per gene family using a forward genetic screen. This introduced a
new mechanism to control the downregulation of Per at the
post-transcriptional level. Based on the aforementioned reported,
the reason for the inhibition of Per genes in cancer is complex,
and not solely due to methylation. There were also certain
limitations to the current study; only A549 cells were available in
our laboratory at that time, therefore Per2 overexpression in A549
was studied according to the laboratory conditions. Further
investigation is required to clarify the effect of Per on NSCLC
development and its underlying mechanisms. In the future, further
experimental studies of Per2 in NSCLC will be performed, and
several cell lines will be selected to conduct a comprehensive
study.
In conclusion, the present study identified that
loss of Per2 may be closely associated with the development of
NSCLC. Recovery of Per2 expression could suppress A549 cell growth
and migration in vitro and in vivo by increasing
apoptosis-associated gene expression and decreasing
metastasis-associated gene expression. These findings suggest that
Per2 may serve as a novel molecular target for NSCLC.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
Scientific Research Foundation of the Technology Department of
Sichuan Province, China (grant nos. 2017SZ0013 and 2018SZ0153).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
RX and YC performed the experiments; JL, TX and YW
provided technical support, critical comments and suggestions. RX,
TX and XY edited the paper; RX, YC and ZW analyzed the data; XY, JL
and QL designed the experiments, analyzed the data and wrote the
paper. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
All patients provided written informed consent, and
the study was approved by the Ethical Committee of Sichuan Cancer
Hospital & Institute (Sichuan, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
NSCLC
|
non-small-cell lung cancer
|
|
Per1
|
Period1
|
|
Per2
|
Period2
|
|
Per3
|
Period3
|
|
Cry1
|
cryptochrome1
|
|
VEGF
|
vascular endothelial growth factor
|
|
NC
|
negative control
|
References
|
1
|
Torre LA, Siegel RL and Jemal A: Lung
Cancer Statistics. Adv Exp Med Biol. 893:1–19. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Schabath MB, Giuliano AR, Thompson ZJ,
Amankwah EK, Gray JE, Fenstermacher DA, Jonathan KA, Beg AA and
Haura EB: TNFRSF10B polymorphisms and haplotypes associated with
increased risk of death in non-small cell lung cancer.
Carcinogenesis. 34:2525–2530. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: GLOBOCAN 2008: Cancer Incidence and
Mortality Worldwide. IARC Cancerbase No, 10International Agency for
Research on Cancer. Lyon: 2010, View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bechet D: Magnesium in health &
disease. Magnesium Res. 29:602016.
|
|
6
|
Cardinali DP: The human body circadian:
How the biologic clock influences sleep and emotion. Neuro
Endocrinol Lett. 21:9–15. 2000.PubMed/NCBI
|
|
7
|
Liu B, Xu K, Jiang Y and Li X: Aberrant
expression of Per1, Per2 and Per3 and their prognostic relevance in
non-small cell lung cancer. Int J Clin Exp Pathol. 7:7863–7871.
2014.PubMed/NCBI
|
|
8
|
Chen-Goodspeed M and Lee CC: Tumor
suppression and circadian function. J Biol Rhythms. 22:291–298.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gery S, Komatsu N, Kawamata N, Miller CW,
Desmond J, Virk RK, Marchevsky A, Mckenna R, Taguchi H and Koeffler
HP: Epigenetic silencing of the candidate tumor suppressor gene
Per1 in non-small cell lung cancer. Clin Cancer Res.
13:1399–1404. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Couto P, Miranda D, Vieira R, Vilhena A,
De Marco L and Bastos-Rodrigues L: Association between CLOCK,
PER3 and CCRN4L with non-small cell lung cancer in
Brazilian patients. Mol Med Rep. 10:435–440. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Retnam L, Chatikavanij P, Kunjara P,
Paramastri YA, Goh YM, Hussein FN, Mutalib AR and Poosala S: Laws,
Regulations, guidelines and standards for animal care and use for
scientific purposes in the countries of Singapore, Thailand,
Indonesia, Malaysia, and India. ILAR J. 57:312–323. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chung S, Son GH and Kim K: Circadian
rhythm of adrenal glucocorticoid: Its regulation and clinical
implications. Biochim Biophys Acta. 1812:581–591. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Savvidis C and Koutsilieris M: Circadian
rhythm disruption in cancer biology. Mol Med. 18:1249–1660. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Winter SL, Bosnoyan-Collins L, Pinnaduwage
D and Andrulis IL: Expression of the circadian clock genes Per1
and Per2 in sporadic and familial breast tumors. Neoplasia.
9:797–800. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kuo SJ, Chen ST, Yeh KT, Hou MF, Chang YS,
Hsu NC and Chang JG: Disturbance of circadian gene expression in
breast cancer. Virchows Archiv. 454:467–474. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chu LW, Zhu Y, Yu K, Zheng T, Yu H, Zhang
Y, Sesterhenn I, Chokkalingam AP, Danforth KN, Shen MC, et al:
Variants in circadian genes and prostate cancer risk: A
population-based study in China. Prostate Cancer Prostatic Dis.
11:342–348. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hwang-Verslues WW, Chang PH, Jeng YM, Kuo
WH, Chiang PH, Chang YC, Hsieh TH, Su FY, Lin LC, Abbondante S, et
al: Loss of corepressor PER2 under hypoxia up-regulates
OCT1-mediated EMT gene expression and enhances tumor malignancy.
Proc Natl Acad Sci USA. 110:12331–12336. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Miller BH, McDearmon EL, Panda S, Hayes
KR, Zhang J, Andrews JL, Antoch MP, Walker JR, Esser KA, Hogenesch
JB and Takahashi JS: Circadian and CLOCK-controlled regulation of
the mouse transcriptome and cell proliferation. Proc Natl Acad Sci
USA. 104:3342–3347. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chi C, He ZF, Liu Y, Lin XM and Sun CC:
Expression and clinical significance of circadian gene Per2 in
non-small cell lung cancer. Zhonghua Zhong Liu Za Zhi. 35:129–131.
2013.(In Chinese). PubMed/NCBI
|
|
21
|
McQueen CM, Schmitt EE, Sarkar TR, Elswood
J, Metz RP, Earnest D, Rijnkels M and Porter WW: PER2 regulation of
mammary gland development. Development. 145:pii: dev157966. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fu L and Lee CC: The circadian clock:
Pacemaker and tumour suppressor. Nat Rev Cancer. 3:350–361. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Takahashi JS, Hong HK, Ko CH and McDearmon
EL: The genetics of mammalian circadian order and disorder:
Implications for physiology and disease. Nat Rev Genet. 9:764–775.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Levi F, Okyar A, Dulong S, Innominato PF
and Clairambault J: Circadian timing in cancer treatments. Annu Rev
Pharmacol Toxicol. 50:377–421. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kelleher FC, Rao A and Maguire A:
Circadian molecular clocks and cancer. Cancer Lett. 342:9–18. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fu L and Kettner NM: The circadian clock
in cancer development and therapy. Prog Mol Biol Transl Sci.
119:221–282. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fu L, Pelicano H, Liu J, Huang P and Lee
C: The circadian gene Period2 plays an important role in tumor
suppression and DNA damage response in vivo. Cell. 111:41–50. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Younger ST, Kenzelmann-Broz D, Jung H,
Attardi LD and Rinn JL: Integrative genomic analysis reveals
widespread enhancer regulation by p53 in response to DNA damage.
Nucleic Acids Res. 43:4447–4462. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Garner E and Raj K: Protective mechanisms
of p53-p21-pRb proteins against DNA damage-induced cell death. Cell
Cycle. 7:277–282. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mirzayans R, Andrais B, Scott A and Murray
D: New insights into p53 signaling and cancer cell response to DNA
damage: Implications for cancer therapy. J Biomed Biotechnol.
2012:1703252012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Okabe T, Kumagai M, Nakajima Y, Shirotake
S, Kodaira K, Oyama M, Ueno M and Ikeda M: The impact of HIF1α on
the Per2 circadian rhythm in renal cancer cell lines. PLoS
One. 9:e1096932014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Miyazaki K, Wakabayashi M, Hara Y and
Ishida N: Tumor growth suppression in vivo by overexpression of the
circadian component, PER2. Genes Cells. 15:351–358. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Clark SJ and Melki J: DNA methylation and
gene silencing in cancer: Which is the guilty party? Oncogene.
21:5380–5387. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gotoh T, Vila-Caballer M, Santos CS, Liu
J, Yang J and Finkielstein CV: The circadian factor Period 2
modulates p53 stability and transcriptional activity in unstressed
cells. Mol Biol Cell. 25:3081–3093. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen ST, Choo KB, Hou MF, Yeh KT, Kuo SJ
and Chang JG: Deregulated expression of the PER1, PER2 and
PER3 genes in breast cancers. Carcinogenesis. 26:1241–1246.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Nagel R, Clijsters L and Agami R: The
miRNA-192/194 cluster regulates the Period gene family and
the circadian clock. FEBS J. 276:5447–5455. 2009. View Article : Google Scholar : PubMed/NCBI
|