Introduction
Glioblastoma multiforme (GBM) is an aggressive
malignant human brain tumor (1).
The prognosis for patients with GBM is unfavorable. Despite modern
treatment protocols, the median survival time is 15 months, with
only 27% of patients living longer than 2 years following diagnosis
(2,3). Treatment resistance is commonly
associated with cancer stem cells (CSCs) in GBM (4–6). High
levels of radiation and chemotherapy, including modern cytostatic
agents and targeted drugs, are unable to eliminate CSCs (7). It is therefore, required to develop
new approaches for GBM treatment and identify new molecular targets
that may assist the regulation of CSCs by inhibiting their
proliferative capabilities.
The present study investigated GBM cells that
express the cluster of differentiation 133 (CD133) membrane
antigen, a well-known CSC marker (8). Cells of this type exhibit a high
proliferation rate; therefore, unlike differentiated
CD133− cancer cells, CD133+ CSCs are
susceptible to radiation and chemotherapy (9). In small quantities, CD133+
CSCs have previously been demonstrated to form large tumors when
implanted into the brains or experimental animals (10). More than 60% of intracellular
proteins from GBM CD133+ CSCs are identical to proteins
present in normal neural stem cells of a human brain (11). This suggests that similarities exist
between the main mechanisms regulating the proliferative properties
of these cell types.
Proliferation of all types of stem cells depends on
the activation of the Wnt signaling pathway (12). In different types of malignant
tumors, the activation of this pathway stimulates the proliferation
of CSCs, which promotes tumor relapse and the development of
therapeutic resistance (13–15). A
direct correlation between the aggressive nature of GBM and the
activation of the Wnt signaling pathway in CSCs has previously been
described (16). Another previous
study demonstrated that the suppression of the Wnt cascade
decreased the heterogeneity of GBM cells (17). Therefore, differentially expressed
proteins (DEPs) of the Wnt signaling pathway are prospective
targets for regulating the proliferative properties of CSCs in GBM.
The aim of the present study was to compare the expression levels
of proteins associated with the Wnt signaling pathway in
CD133+ CSCs of human GBM and differentiated
СD133− cancer cells.
Materials and methods
Human GBM cells
The present study used the U-87MG GBM cell line
obtained from the American Type Culture Collection (cat. no.
HTB-14™; ATCC; Manassas, VA, USA). This cell line is not
the original U-87 line established at the University of Uppsala,
but a human GBM of unknown origin (18). However, as demonstrated in our
previous study (11),
CD133+ cells of this cell line exhibit similar proteome
profiles to neural CD133+ human stem cells and exhibit
significant proteomic differences compared with normal mesenchymal
stem cells of the human bone marrow. In our previous study, the
stimulation of GBM U-87MG cells with transforming growth factor
(TGF)-β1 led to a significant increase in the expression of
proteins associated with the epithelial-mesenchymal transition
(EMT) (19), which greatly
increased the invasiveness of the cells. The U-87MG cell line of
GBM possesses a significant amount of CD133+ CSCs that
actively interact with both cancerous and non-cancerous cells
(20). The extensive information
that is available regarding this GBM cell line makes it the optimal
choice for the present study.
The U-87MG cells were cultured in low glucose
Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) with 10% fetal bovine serum
(FBS; Gibco; Thermo Fisher Scientific, Inc.) in standard conditions
(5% CO2 and 37°C). The cells were cultured until they
reached 80% confluence. To obtain glioma spheres the cells were
resuspended in DMEM/F12 (Thermo Fisher Scientific, Inc.) containing
L-glutamine, B27, 20 ng/ml bFGF, 20 ng/ml EGF, 100 U/ml
penicillin/streptomycin and 5 µg/ml heparin. All chemicals were
obtained from Gibco; (Thermo Fisher Scientific, Inc.). Cells were
grown in T75 culture flasks. Every 3 days, fresh growth factors
were added. The extraction of CD133+ cells was performed
via immunosorting using magnetic beads with immobilized antibodies
against CD133 (CD133 MicroBead kit; cat. no. 130-100-857; Miltenyi
Biotec, Inc., San Diego, CA, USA). The purity of the isolated
population was assessed by flow cytometry and CD133/1
(AC133)-VioBright FITC antibodies (cat. no. 130-105-226; Miltenyi
Biotec, Inc.). The dye was diluted at a ratio of 1:11 for 107
cells/100 µl of buffer solution [7.2 pH phosphate-buffered saline
(PBS), 0.5% bovine serum albumin (BSA) and 2 mM EDTA]. The antigen
was labeled by incubation for 10 min in the dark (4°C) to prevent
non-specific cell labeling.
Mass spectrometry of samples
High-performance liquid chromatography-mass
spectrometry was applied and the label-free method was used to
evaluate changes in the protein expression level. Two cell samples
(CD133+ CSCs and cancer CD133− non-stem
cells) were lysed with Mammalian Cell Lysis kit (MCL1-1KT;
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and low-molecular
compounds were removed. Subsequently, enzymatic cleavage was
performed and 4 µl of solution was analyzed by mass spectrometry.
The samples were incubated at 30°С in a Labconco CentriVap
centrifugal concentrator (Labconco Corp., Kansas City, MI, USA) to
remove ammonium bicarbonate. Peptides were diluted during the
mobile stage with 30% acetonitrile, 70% water and 0.1% formic acid
(рН 2.7) and divided into 24 fractions using a Dionex UltiMate 3000
(Dionex Corp., Sunnyvale, CA, USA), equipped with a fraction
collector and cation exchange column MIC-10-CP (Puros 10S; 1 mm ×
10 cm; Thermo Fisher Scientific, Inc.). The obtained fractions were
concentrated at 30°С in the centrifugal concentrator and diluted
with 0.1% formic acid (100 µl).
Peptides were analyzed using Dionex Ultimate 3000
(Dionex Corp.), LTQ Orbitrap XL and Orbitrap Fusion mass
spectrometer (Thermo Fisher Scientific, Inc.) with nanospray
ionization. Peptide division was performed using an Acclaim C18
PepMap 100 column (75 µm х 150 mm; grit size, 3 µm; Dionex Corp.).
Mass spectrometry data were processed using MaxQuant 1.6.1.0
(21–23) and Perseus 1.6.1 software (Max Planck
Institute of Biochemistry, Planegg, Germany). Biological processes,
molecular functions, cell location and protein signaling pathways
were annotated using the following databases: PubMed (http://www.ncbi.nlm.nih.gov/pubmed), Protein
Analysis Through Evolutionary Relationships (http://www.pantherdb.org), Gene Ontology (http://www.geneontology.org), Kyoto Encyclopedia of
Genes and Genomes (http://www.genome.jp/kegg/) and Search Tool for the
Retrieval of Interacting Genes/Proteins v10. (https://string-db.org/).
Statistical analysis
Statistical significance was identified using
Student's t-test [STATISTICA 12 (StatSoft, Moscow, Russia].
P<0.05 was considered to indicate a statistically significant
difference.
Results
General characteristics of the
identified proteins
A total of 1,904 proteins were identified in both
samples of GBM cells. A total of 1,696 proteins were identified in
GBM CD133+ CSCs and 1,435 proteins in GBM
CD133− non-stem cells. The molecular mass of the
proteins was 4–3,995 kDa. A total of 1,227 proteins (64.4% of the
total 1,904 proteins) were detected in all cell lysates, 469
proteins only in CSCs and 208 proteins only in the non-stem cells.
A total of 589 (34.7% of 1,904) DEPs were identified to exhibit
significantly different expression levels in CSCs, as compared with
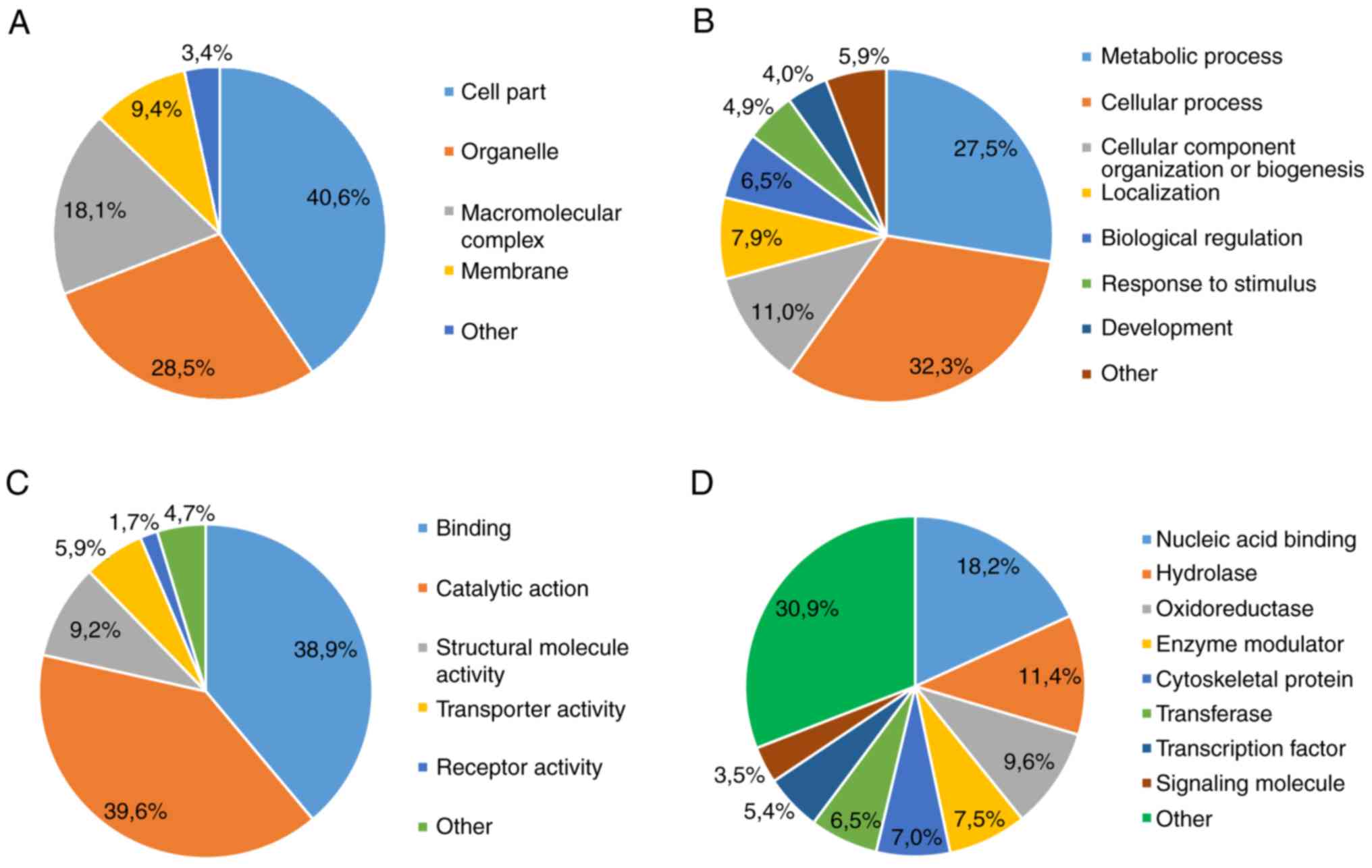
non-stem cells (P<0.05). The majority of DEPs were: i) Localized
intracellularly (Fig. 1A); ii)
associated with metabolic and cellular processes (Fig. 1B); iii) functionally heterogeneous
(Fig. 1C); and iv) associated with
a class of compounds that exhibit active fermentation properties
(Fig. 1D).
Bioinformatics analysis of the
identified proteins
A total of 116 DEPs were found to be associated with
15 signaling pathways (Table I).
Among these 116 DEPs, 88 were upregulated and 28 downregulated in
GBM CSCs. The upregulation was observed in proteins associated with
intracellular signaling pathways, including proteins involved with
the extracellular matrix (cell adhesion molecules, ECM receptor
interaction and focal adhesion) and local microenvironment (tight
and adherens junctions). The majority of upregulated proteins were
associated with the glycolysis pathway and Wnt signaling cascade
(Tables I and II). The majority of the downregulated
proteins were revealed to be associated with the insulin signaling
pathway and MAPK signaling pathway (Table I).
 | Table I.Participation of differentially
expressed proteins of glioblastoma CD133+ stem cells in
the intracellular signaling pathways. |
Table I.
Participation of differentially
expressed proteins of glioblastoma CD133+ stem cells in
the intracellular signaling pathways.
| Signaling
pathway | Total identified
proteins (n) | Upregulated
proteins (n) | Downregulated
proteins (n) |
|---|
| Adherens
junction | 4 | 4 |
|
| Apoptosis | 7 | 4 | 3 |
| Cell adhesion
molecules | 6 | 6 |
|
| Cell cycle | 4 | 4 |
|
| Chemokine signaling
pathway | 5 | 3 | 2 |
| ECM-receptor
interaction | 7 | 7 |
|
| Focal adhesion | 13 | 11 | 2 |
| Gap junction | 5 | 4 | 1 |
|
Glycolysis/gluconeogenesis | 13 | 12 | 1 |
| Insulin signaling
pathway | 6 | 1 | 5 |
| Integrin signaling
pathway | 5 | 2 | 3 |
| MAPK signaling
pathway | 9 | 3 | 6 |
| Regulation of actin
cytoskeleton | 11 | 7 | 4 |
| Tight junction | 9 | 8 | 1 |
| Wnt signaling
pathway | 12 | 12 |
|
 | Table II.Changes in the expression of Wnt
signaling proteins in CD133+ CSCs of glioblastoma. |
Table II.
Changes in the expression of Wnt
signaling proteins in CD133+ CSCs of glioblastoma.
| ID | Protein name |
CD133+/CD133 ratio
(P<0.05) |
|---|
| APC | Adenomatous
polyposis coli | ↑a |
| CacyBP | Calcyclin binding
protein | 2.00 |
| CSNK2A2 | Casein kinase 2 α
2 | 5.24 |
| CSNK2B | Casein kinase 2
β | 12.20 |
| CtBP1 | C-terminal binding
protein 1 | 3.42 |
| CtBP2 | C-terminal binding
protein 2 | 2.24 |
| CTNNB1 | Catenin β-1 | 6.15 |
| Daam1 | Dishevelled
associated activator of morphogenesis 1 | 6.54 |
| CUL1 | Cullin 1 | 3.39 |
| Rac2 | Ras-related C3
botulinum toxin substrate 2 (Rho family, small GTP binding protein
Rac2 | 1.67 |
| RhoA | Ras homolog family
member A | 1.94 |
| RUVBL1
(Pontin52) | RuvB-like AAA
ATPase 1 | 3.22 |
Discussion
Cancer stem cells (CSCs) are an important challenge
in the treatment of glioblastoma multiforme (GBM). The absence of
drugs and medical technologies for the effective elimination of
CSCs in a patient's body has shifted the focus of modern research
to molecular genetics. The discovery of certain GBM isotypes based
on molecular genetic analysis has provided insights into the
pathogenesis of GBM; however, there remains a requirement to
identify mechanisms to regulate and control CSCs (7).
The present study characterized CD133+
CSCs as distinct cells with quantitative and qualitative
differences, when compared with differentiated cells in GBM. A
number of proteins were identified to exhibit a different level of
expression in CSCs, as compared with differentiated cells in GBM.
Furthermore, it was demonstrated herein that the upregulated
proteins in CSCs were associated with the mechanisms of invasion,
survival and proliferation. Both of these findings require further
in-depth analysis. In addition to the aforementioned mechanisms, a
significant change was observed in the expression of proteins
associated with the Wnt signaling pathway, which was the main focus
of the present study.
Embryonic cells with high levels of Wnt synthesis
are known to develop into endodermal and cardiac cells, while cells
with a low level of Wnt synthesis form ectoderm layers. Despite GBM
being a primary neuroectodermal tumor, a number of studies have
suggested its treatment resistance is associated with the
activation of the Wnt pathway in CSCs (16). This statement is supported by the
presence of CD133 on the surface of normal neural and hematopoietic
stem, endothelial progenitor and normal postnatal stem cells, and
progenitor cells of other types (21), in which the Wnt signaling pathway
plays a key role (12,13). In addition, CD133 is present in the
kidneys, mammary glands, trachea, salivary glands, placenta,
intestinal cells and ovaries, where the activation of the
Wnt/β-catenin signaling pathway is associated with drug resistance
and the development of aggressive types of cancer (22–24).
Therefore, it can be hypothesized that these mechanisms are similar
in GBM.
The upregulation of the Wnt cascade in GBM CSCs was
not confirmed in the present study. The expression levels of
established proteins of this cascade, including Wnt ligands,
frizzled receptors and co-receptor LRP5/6, were not identified to
be significantly increased in GBM. However, the upregulation of
components of the Wnt signaling pathway was associated with unusual
characteristics of CSCs.
Adenomatous polyposis coli (APC) protein was
revealed to be significantly upregulated in GBM CSCs (Fig. 2). This finding had previously been
demonstrated in GBM (25). An APC
gene mutation encodes a protein that is a key component of Turcot
syndrome, a known risk factor for GBM (26). The present study demonstrated that
the upregulation of this protein is typical in CSCs, which
indicates that these cells may play a critical role in GBM
biology.
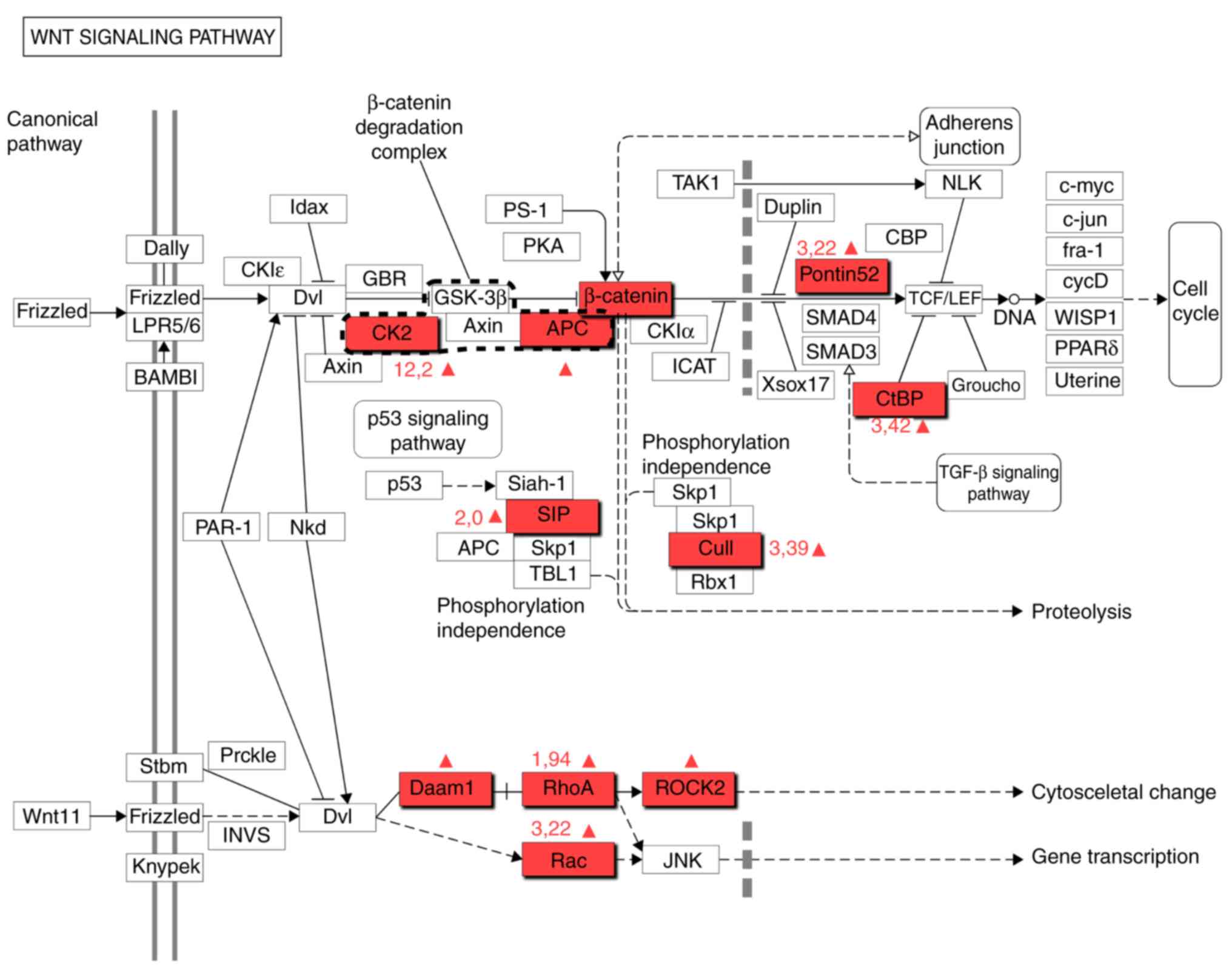 | Figure 2.Wnt signaling pathway according to
the Kyoto Encyclopedia of Genes and Genomes database. The red
marker indicates upregulated proteins in the glioblastoma
CD133+. APC, adenomatous polyposis coli; CK2, Сasein
kinase 2; RhoA, RAS homolog gene family member A; Daam1, disheveled
associated activator of morphogenesis 1; CtBP, C-terminal-binding
proteins; SIP, Siah-1 interacting protein; Rac, Rac family small
GTPase; Cul1, cullin-1; ROCK2, Rho-associated, coiled-coil
containing protein kinase 2. |
The expression of calcyclin-binding and Siah-1
interacting (CacyBP/SIP) proteins was found to be 2-fold higher in
CSCs, as compared with GBM CD133− cells (Fig. 2). This protein is crucial for the
proliferation, differentiation, apoptosis, transcription,
ubiquitination and cytoskeleton organization (27). In addition, it regulates the
degradation of β-catenin. High levels of CacyBP/SIP are typical in
cells of Wnt-associated stomach and colon tumors, as well as
neurons. Furthermore, this protein is a component of the p53
signaling pathway that is important for GBM (7). High levels of CacyBP/SIP are important
for the proliferation of CSCs, as it can act as an antagonist of
p27-selective cyclin-dependent kinase inhibitor 1B (28). The upregulation of CacyBP/SIP has
been revealed to be associated with a high resistance to
doxorubicin and other cytotoxic agents (29). CacyBP/SIP can be used as a marker of
the condition of GBM cell population and serve as a potential
target for the regulation of CSCs. Therefore, inhibiting CacyBP/SIP
expression can increase the efficiency of therapy against GBM.
Сasein kinase 2 (CK2), a component of the canonical
Wnt/β-catenin signaling pathway, regulates the cell cycle,
apoptosis and transcription of stem cells (30). CK2, as well as APC, is a crucial
component of a β-catenin degrading complex (Fig. 2). Furthermore, this protein is
associated with nuclear factor-κB (NF-κB), phosphoinositide
3-kinase/AKT and signal transducer and activation of transcription
signaling in GBM cells (31). CK2
consists of two catalytic subunits, CSNK2A2 and CSNK2B. The present
study demonstrated a significant increase in CSNK2A2 and CSNK2B
expression levels (5.24- and 12.21-fold, respectively) in GBM CSCs.
CK2α is a regulator of the Wnt/β-catenin signaling pathway;
therefore, the expression levels of this protein are increased in
numerous types of tumors.
The expression of CSNK2A2 and CSNK2B has been
identified in GBM CSCs. A treatment option that inhibits CK2 has
been demonstrated to decrease the expression of CSC markers in a
GBM cell culture (32). Compared
with temozolomide monotherapy, CX-4945, a specific CK2 inhibitor,
was revealed to inhibit the generation of glioma spheres in
vitro and significantly increase the survival of animals with
acquired GBM (33). Considering the
aforementioned results, the CSNK2A2 and CSNK2B subunits may serve
as targets for the regulation of CSCs in GBM treatment.
β-catenin is a key element of the Wnt signaling
pathway and is associated with the self-renewal of stem cells
(34,35). According to the present experimental
data, the expression of the β-catenin (CTNNB1) was 6-fold higher in
CD133+ CSCs, when compared with CD133− cells
in GBM. The inhibition of β-catenin in U87 cells decreased the
migration activity of these cancer cells (36). In addition, the expression of this
protein was increased in glial tumor cells, which plays a crucial
role in CSC invasion (37),
survival and proliferation (38).
The pharmacological inhibition of the β-catenin expression reduced
the ability of GBM cells to create glioma spheres in vitro
(39). A combination of the
β-catenin inhibitor tetrandrine, an isoquinoline alkaloid, with
temozolomide has been demonstrated to increase the efficiency of
GBM treatment.
The present study revealed a significant increase in
the expression of key components associated with the degradation of
β-catenin including APC, CK2α and CK2β. In addition, a 6-fold
increase of β-catenin in GBM CSCs was identified, which is a
notable finding due to the key roles of β-catenin in CSCs.
β-catenin is a central element of the Wnt signaling cascade, as it
coordinates and determines the activity of this pathway and other
intracellular signaling mechanisms. An increased production of APC
and CK2 with high levels of β-catenin in GBM cells suggests a lack
of influence of Wnt ligand in glioma spheres rather than a low
productivity of these proteins. Furthermore, the upregulation of
destruction complex proteins suggests that β-catenin may be
associated with the cell membrane, where it coordinates
extracellular interaction in glioma spheres.
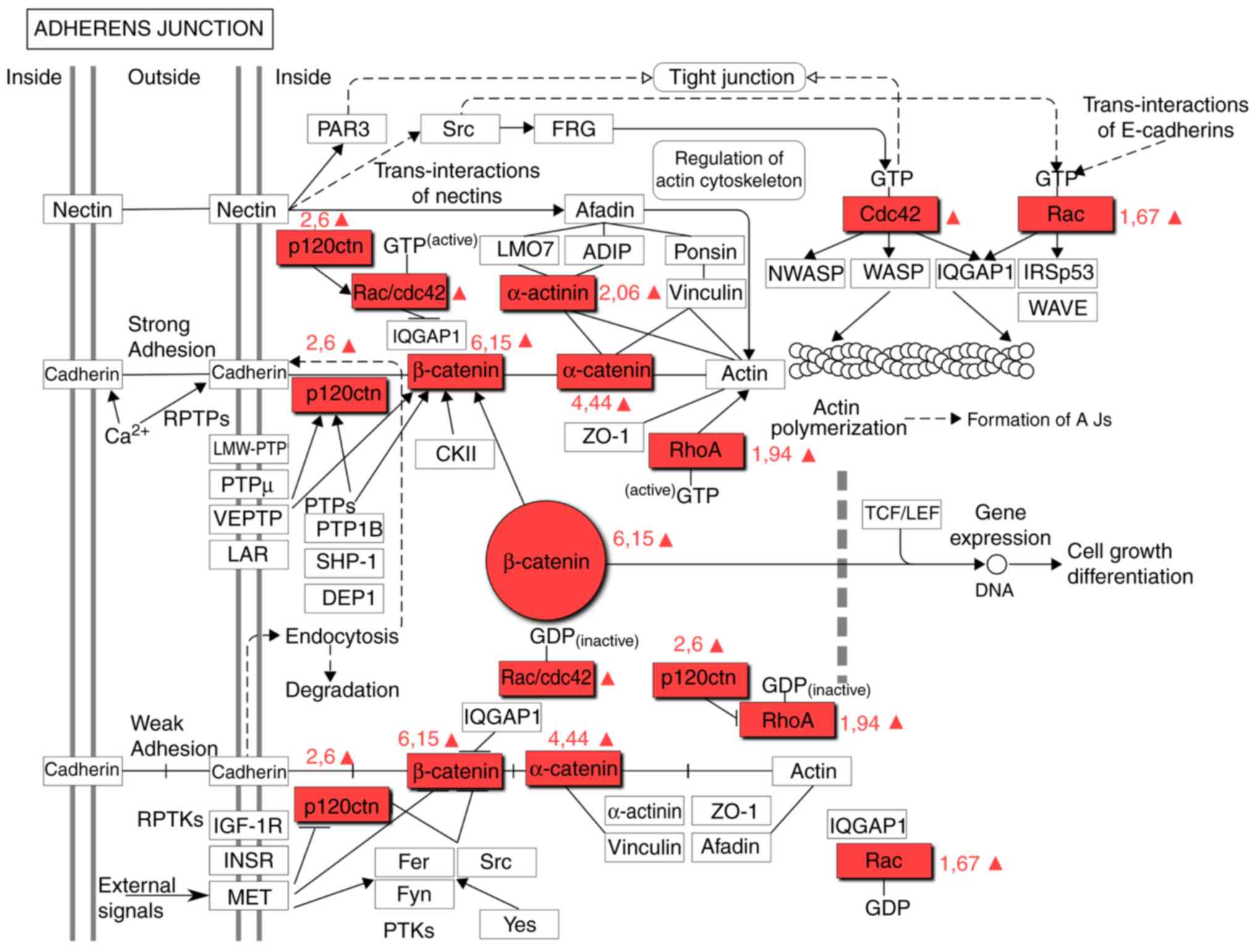
This is supported by the evidence that β-catenin is
a component of the adherens junction pathway (Fig. 3). The upregulation of proteins
associated with the adherens junction pathway has previously been
described as a characteristic of GBM cells (Table III) (25). However, the present results
suggested that this is a characteristic attributed only to
CD133+ CSCs. α-Catenin (CTNNA1) and δ-catenin (CTNND1),
critical components of the adherens junction pathway, were
identified to be upregulated in CSCs (4.44- and 2.6-fold,
respectively), which was consistent with an increase in CTNNB1. The
CTNNA1 tumor-suppressor gene, which encodes α-catenin, is one of
the most frequently deleted or mutated genes in cancer (40). α-Catenin is an essential protein in
adherens junctions, which are critical for maintaining
intercellular adhesion and cellular polarity.
 | Table III.Changes in the expression of adherens
junction signaling proteins in CD133+ CSCs of
glioblastoma. |
Table III.
Changes in the expression of adherens
junction signaling proteins in CD133+ CSCs of
glioblastoma.
| ID | Protein name |
CD133+/CD133 ratio
(P<0.05) |
|---|
| CDC42 | Cdc42
GTPase-activating protein | ↑ |
| RhoA | Ras homolog gene
family, member A | 1,94 |
| RhoC | Ras homolog gene
family, member C | 1,21 |
| ROCK2 | Rho-associated,
coiled-coil containing protein kinase 2 | 1,38 |
| Rac2 | Ras-related C3
botulinum toxin substrate 2 (Rho family, small GTP binding protein
Rac2 | 1.67 |
| CTNNA1 | Catenin
(cadherin-associated protein), α1 | 4,44a |
| CTNNB1 | Catenin β1 | 6.15a |
| CTNND1 | Catenin
(cadherin-associated protein), δ1 | 2,60a |
| ACTN1 | Actinin, α1 | 2,04a |
The fact that α-catenin was also upregulated in
CD133+ CSCs suggests that CSCs exhibit denser
intercellular connections, as compared with CD133−
cells. It is possible that β-catenin is upregulated in GMB cells as
a result of transforming non-CSCs into CSCs. CTNNA1 inhibits the
proliferation and invasion of cancer cells into different tissues
by suppressing the Hippo-Yes-associated protein (YAP) and NF-κB
signaling. The depletion of CTNNA1 and CTNND1 promotes the mobility
and growth of cancer cells (41).
In summary, CTNNA1, CTNNB1 and CTNND1 proteins may be treated as
potential targets for regulating the plasticity of GBM cells.
Cullin-1 (CUL1) is a component of the Wnt signaling
pathway (Fig. 2). The present study
identified that the expression level of CUL1 in CD133+
cells of GBM was 3.39-fold higher than that in CD133−
cells of the common pool. CUL1 is involved in different
intracellular processes, including proliferation, differentiation
and apoptosis. The hyperexpression of CUL1 was revealed to be a
marker of poor prognosis for patients with stomach carcinoma,
breast cancer and non-small-cell lung carcinoma. The expression of
CUL1 has been demonstrated to be significantly higher in glioma
cells than in normal brain tissue (42). Theoretically, the inhibition of CUL1
in GBM cells with CUL1 small interfering RNA may prevent the
migration and invasion of cancer cells due to a lower level of
matrix metallopeptidase (MMP)-2 and MMP-8 expression.
Rac family small GTPase 2 (Rac2), Ras homolog gene
family member A (RhoA) and disheveled associated activator of
morphogenesis 1 (Daam1) proteins are integrated in the Wnt
signaling pathway (Fig. 2). The
Rac2 and RhoA proteins are also components of the signaling pathway
of adherens junctions (Fig. 3). Rac
proteins are a subfamily of Rho small GTPases. The function of this
family involves modifying the actin cytoskeleton and proliferation,
and regulating key stem cell properties (43). Rac2 is a marker of more aggressive
GBM subtypes (44). The present
study demonstrated that the expression level of Rac2 in
CD133+ cells was 1.67-fold higher. Rho GTPases belong to
the Ras superfamily and these proteins, including RhoA, Rac1 and
cell division control protein 42 homolog, support the
transformation of the cell cytoskeleton during the
epithelial-mesenchymal transition (EMT). RhoA is actively expressed
in GBM cells, which promotes the migration and invasion of cancer
cells. The simultaneous activation of Rac and RhoA was revealed to
increase the invasive properties of CSCs (43). The present study revealed that the
expression level of RhoA was 1.94-fold higher in CD133+
cell than in the non-CSCs. Simvastin, NSC23766 and specific
microRNAs can impact the expression of the aforementioned proteins
(44).
C-terminal-binding proteins (CtBP) have two
isoforms, CtBP1 and CtBP2, which are involved in the Wnt/β-catenin
signaling pathway and other molecular mechanisms determining the
development of malignant tumors, invasion of neoplastic cells,
apoptosis and response to therapy (45). The present study revealed that the
expression levels of CtBP1 and CtBP2 were significantly higher in
GBM CD133+ cells (3.42- and 2.24-fold, respectively).
CtBP inhibits the expression of tumor suppressor genes (46), serves a role in the
epithelial-mesenchymal transition, regulates β-catenin, mediates
the transcription factor 4/lymphoid enhancer-binding factor 1 axis
and activates targeted genes involved in the self-regeneration of
CSCs (47). Combining the CtBP
inhibitor MTOB with temozolomide may serve as a potential treatment
option for GBM.
RuvB-like AAA ATPase 1 (RUVBL1 or Ponti52) is a
component of the Wnt signaling pathway (Fig. 2) that is essential for tumor cell
growth and viability. This study identified that the expression
level of RUVBL1 was significantly higher in GBM CD133+
than in GBM CD133− cells of the common pool (Table II). RUVBL1 plays a key role in the
cell cycle, mitosis, chromatin remodeling, transcription, DNA
repair, apoptosis and regulation of development of normal stem
cells (48,49). It also participates in oncogenic
signaling pathways, including c-Myc and Wnt. RUVBL1 regulates the
activity of glucocorticosteroid and estrogen receptors in the
nucleus, and is essential in choosing the optimum pharmacological
scheme (50). The inhibition of
RUVBL1 activity interferes with the expression of genes responsible
for the response of GBM to hypoxia and is associated with an
aggressive tumor phenotype (51).
The inactivation of this gene with microRNA can increase the
efficiency of GBM therapy.
Of note, the Wnt-associated proteins CtBP and RUVBL1
regulate key gene expression in CSCs of all types of cancer. These
proteins are part of the SMAD signaling cascade, which is mostly
activated by TGF-β and is crucial to GBM pathogenesis (19). It can be suggested that via SMAD
proteins of the TGF-β pathway, CtBP and RUVBL1 generate a closed
system that supports the stemness of GBM cells; however, this
requires further investigation.
In conclusion, the present study attempted to
identify molecular targets that may assist with the regulation of
GBM CSC proliferation. The study focused on selective analysis of
proteins associated with the Wnt signaling pathway in
CD133+ CSCs. In GBM CD133+ CSCs, an increased
expression of 12 proteins that are components of the Wnt signaling
pathway was identified; a number of these proteins, including
CTNNB1, Daam1, Rac2 and RhoA, are also components of the adherens
junction pathway. CacyBP, CSNK2A2, CSNK2B, CtBP1, CtBP2, CUL1 and
RUVBL1 may serve as targets for the pharmaceutical regulation of
CSCs in GBM treatment. Furthermore, the increased expression of
APC, β-catenin, CtBP and RUVBL1 suggested the possibility of
alternative activation of genes in CD133+ CSCs; however,
this requires further investigation.
It is essential to note that suppressing proteins of
the Wnt signaling pathway are required not for producing a
cytostatic and cytotoxic effect on GBM cells of the common pool,
but in order to inhibit the reproductive function of CSCs and, as a
result, extend the remission period. Therefore, conceptually new
methods and techniques need to be developed in order to evaluate
the efficiency of suppressing these targets. For instance, taking
into consideration the fact that β-catenin and certain upregulated
Wnt-proteins belong to the signaling pathway of adherens junctions,
the efficiency of suppressing this target could be evaluated not
just based on the amount of β-catenin in CSCs, but also on the
strength of adherens junctions in a gliomasphere, AFM
investigation, time of gliomasphere creation if cultivated in
serum-free media, speed of cell growth in the standard media with
serum and immunophenotype of these cells' progenitors identified by
flow cytometry. We plan to attempt this in the nearest future,
using CD133+ CSCs from the samples of a human brain with
GBM. Furthermore, the efficiency of the targeted therapy on
extending the life expectancy of patients may not be very high,
since upregulated proteins are involved in several signaling
pathways. That is why suppressing one target may have conflicting
results. The way to solve this issue is a systemic analysis of
survival using the Kaplan-Meier curve for GBM patients with a
significant expression of one of the described proteins (CTNNB1,
Daam1, Rac, RhoA, CacyBP, CSNK2A2, CSNK2B, CtBP1, CtBP2, CUL1 and
RUVBL1) in their CD133+ CSCs. This approach could become
a breakthrough targeted therapy for GBM.
Acknowledgements
Not applicable.
Funding
The present study was funded by the Ministry of
Education and Science of the Russian Federation (grant no.
14.584.21.0027; ID, RFMEFI58417X0030RFMEFI58417X0027).
Availability of data and materials
All data and materials are available upon
request.
Authors' contributions
VS prepared and analyzed the samples, as well as
performed the cell lysis, chromatography and mass spectrometry, and
contributed to the bioinformatics analysis. NA and MK cultured the
cancer cells and isolated the cancer CD133+ stem cells
of glioblastoma for the experiment. SZ provided and performed the
statistical analysis and was responsible for the mathematical
process of the results. YK and HS discussed, analyzed and
interpreted the results of the study, and also worked on the
manuscript. IB wrote the manuscript, proposed the study idea,
designed the study, offered support with the experiments, organized
the scientific team, provided scientific guidance and contributed
to the bioinformatics analysis. All authors read and approved the
manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the School of Biomedicine, The Far Eastern Federal
University (Vladivostok, Russia) and the Academic Council of the
School of Biomedicine.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Louis DN, Perry A, Reifenberger G, von
Deimling A, Figarella- Branger D, Cavenee WK, Ohgaki H, Wiestler
OD, Kleihues P and Ellison DW: The 2016 World Health Organization
Classification of Tumors of the central nervous system: A summary.
Acta Neuropathol. 131:803–820. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Stupp R, Brada M, van den Bent MJ, Tonn JC
and Pentheroudakis G; ESMO Guidelines Working Group, : High-grade
glioma: ESMO clinical practice guidelines for diagnosis, treatment
and follow-up. Ann Oncol. 25 (Suppl 3):iii93–iii101. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Stupp R, Mason WP, van den Bent MJ, Weller
M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn
U, et al: Radiotherapy plus concomitant and adjuvant temozolomide
for glioblastoma. N Engl J Med. 352:987–996. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rycaj K and Tang DG: Cancer stem cells and
radioresistance. Int J Radiat Biol. 90:615–621. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chang L, Graham P, Hao J, Ni J, Deng J,
Bucci J, Malouf D, Gillatt D and Li Y: Cancer stem cells and
signaling pathways in radio-resistance. Oncotarget. 7:11002–11017.
2016.PubMed/NCBI
|
|
6
|
Friedmann-Morvinski D: Glioblastoma
heterogeneity and cancer cell plasticity. Crit Rev Oncog.
19:327–336. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bryukhovetskiy I, Ponomarenko A, Lyakhova
I, Zaitsev S, Zayats Y, Korneyko M, Eliseikina M, Mischenko P,
Shevchenko V, Shanker Sharma H, et al: Personalized regulation of
glioblastoma cancer stem cells based on biomedical technologies:
From theory to experiment (Review). Int J Mol Med. 42:691–702.
2018.PubMed/NCBI
|
|
8
|
Bradshaw A, Wickremesekera A, Brasch HD,
Chibnall AM, Davis PF, Tan ST and Itinteang T: Cancer stem cells in
glioblastoma multiforme. Front Surg. 3:482016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Brown DV, Filiz G, Daniel PM, Hollande F,
Dworkin S, Amiridis S, Kountouri N, Ng W, Morokoff AP and
Mantamadiotis T: Expression of CD133 and CD44 in glioblastoma stem
cells correlates with cell proliferation, phenotype stability and
intra-tumor heterogeneity. PLoS One. 12:e01727912017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Brown DV, Daniel PM, D'Abaco GM, Gogos A,
Ng W, Morokoff AP and Mantamadiotis T: Coexpression analysis of
CD133 and CD44 identifies proneural and mesenchymal subtypes of
glioblastoma multiforme. Oncotarget. 6:6267–6280. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bryukhovetskiy A, Shevchenko V, Kovalev S,
Chekhonin V, Baklaushev V, Bryukhovetskiy I and Zhukova M: To the
novel paradigm of proteome-based cell therapy of tumors: Through
comparative proteome mapping of tumor stem cells and
tissue-specific stem cells of humans. Cell Transplant. 23 (Suppl
1):S151–S170. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tan SH and Barker N: Wnt signaling in
adult epithelial stem cells and cancer. Prog Mol Biol Transl Sci.
153:21–79. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kahn M: Wnt signaling in stem cells and
cancer stem cells: A tale of two coactivators. Prog Mol Biol Transl
Sci. 153:209–244. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kretzschmar K and Clevers H: Wnt/β-catenin
signaling in adult mammalian epithelial stem cells. Dev Biol.
428:273–282. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mohammed MK, Shao C, Wang J, Wei Q, Wang
X, Collier Z, Tang S, Liu H, Zhang F, Huang J, et al: Wnt/β-catenin
signaling plays an ever-expanding role in stem cell self-renewal,
tumorigenesis and cancer chemoresistance. Genes Dis. 3:11–40. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kim Y, Kim KH, Lee J, Lee YA, Kim M, Lee
SJ, Park K, Yang H, Jin J, Joo KM, et al: Wnt activation is
implicated in glioblastoma radioresistance. Lab Invest. 92:466–473.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kahlert UD, Suwala AK, Koch K, Natsumeda
M, Orr BA, Hayashi M, Maciaczyk J and Eberhart CG: Pharmacologic
Wnt inhibition reduces proliferation, survival, and clonogenicity
of glioblastoma cells. J Neuropathol Exp Neurol. 74:889–900. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Allen M, Bjerke M, Edlund H, Nelander S
and Westermark B: Origin of the U87MG glioma cell line: Good news
and bad news. Sci Transl Med. 8:354re32016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bryukhovetskiy I and Shevchenko V:
Molecular mechanisms of the effect of TGF-β1 on U87 human
glioblastoma cells. Oncol Lett. 12:1581–1590. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bryukhovetskiy IS, Dyuizen IV, Shevchenko
VE, Bryukhovetskiy AS, Mischenko PV, Milkina EV and Khotimchenko
YS: Hematopoietic stem cells as a tool for the treatment of
glioblastoma multiforme. Mol Med Rep. 14:4511–4520. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mizrak D1, Brittan M and Alison M: CD133:
Molecule of the moment. J Pathol. 214:3–9. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shmelkov SV, St Clair R, Lyden D and Rafii
S: AC133/CD133/Prominin-1. Int J Biochem Cell Biol. 37:715–719.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang LS and Lum L: Chemical modulation of
WNT signaling in cancer. Prog Mol Biol Transl Sci. 153:245–269.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gong A and Huang S: FoxM1 and
Wnt/β-catenin signaling in glioma stem cells. Cancer Res.
72:5658–5662. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nikuseva-Martić T, Beros V, Pećina-Slaus
N, Pećina H and Bulić-Jakus F: Genetic changes of CDH1, APC, and
CTNNB1 found in human brain tumors. Pathol Res Pract. 203:779–787.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dipro S, Al-Otaibi F, Alzahrani A, Ulhaq A
and Al Shail E: Turcot syndrome: A synchronous clinical
presentation of glioblastoma multiforme and adenocarcinoma of the
colon. Case Rep Oncol Med. 2012:7202732012.PubMed/NCBI
|
|
27
|
Shi H, Gao Y, Tang Y, Wu Y, Gong H, Du J,
Zheng B, Hu J, Shi Q and Yu R: CacyBP/SIP protein is important for
the proliferation of human glioma cells. IUBMB Life. 66:286–291.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yan S, Li A and Liu Y: CacyBP/SIP inhibits
the migration and invasion behaviors of glioblastoma cells through
activating Siah1 mediated ubiquitination and degradation of
cytoplasmic p27. Cell Biol Int. 42:216–226. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tang Y, Zhan W, Cao T, Tang T, Gao Y, Qiu
Z, Fu C, Qian F, Yu R and Shi H: CacyBP/SIP inhibits
Doxourbicin-induced apoptosis of glioma cells due to activation of
ERK1/2. IUBMB Life. 68:211–219. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen Y, Zhang K, Wang X, Li Q, Wu Q and
Ning X: Cell cycle-dependent translocation and regulatory mechanism
of CacyBP/SIP in gastric cancer cells. Anticancer Drugs. 29:19–28.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Nitta RT, Gholamin S, Feroze AH, Agarwal
M, Cheshier SH, Mitra SS and Li G: Casein kinase 2α regulates
glioblastoma brain tumor-initiating cell growth through the
β-catenin pathway. Oncogene. 34:3688–3699. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rowse AL, Gibson SA, Meares GP,
Rajbhandari R, Nozell SE, Dees KJ, Hjelmeland AB, McFarland BC and
Benveniste EN: Protein kinase CK2 is important for the function of
glioblastoma brain tumor initiating cells. J Neurooncol.
132:219–229. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ferrer-Font L, Villamañan L, Arias-Ramos
N, Vilardell J, Plana M, Ruzzene M, Pinna LA, Itarte E, Arús C and
Candiota AP: Targeting protein kinase CK2: Evaluating CX-4945
potential for GL261 glioblastoma therapy in immunocompetent mice.
Pharmaceuticals. 10(pii): E242017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chikano Y, Domoto T, Furuta T, Sabit H,
Kitano-Tamura A, Pyko IV, Takino T, Sai Y, Hayashi Y, Sato H, et
al: Glycogen synthase kinase 3β sustains invasion of glioblastoma
via the focal adhesion kinase, Rac1, and c-Jun N-terminal
kinase-mediated pathway. Mol Cancer Ther. 14:564–574. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhao P, Li Q, Shi Z, Li C, Wang L, Liu X,
Jiang C, Qian X, You Y, Liu N, et al: GSK-3β regulates tumor growth
and angiogenesis in human glioma cells. Oncotarget. 6:31901–31915.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang Y, Wen YL, Ma JW, Ye JC, Wang X,
Huang JX, Meng CY, Xu XZ, Wang SX and Zhong XY: Tetrandrine
inhibits glioma stem-like cells by repressing β-catenin expression.
Int J Oncol. 50:101–110. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Náger M, Santacana M, Bhardwaj D, Valls J,
Ferrer I, Nogués P, Cantí C and Herreros J: Nuclear phosphorylated
Y142 β-catenin accumulates in astrocytomas and glioblastomas and
regulates cell invasion. Cell Cycle. 14:3644–3655. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kahlert UD, Mooney SM, Natsumeda M,
Steiger HJ and Maciaczyk J: Targeting cancer stem-like cells in
glioblastoma and colorectal cancer through metabolic pathways. Int
J Cancer. 140:10–22. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kierulf-Vieira KS, Sandberg CJ, Grieg Z,
Günther CC, Langmoen IA and Vik-Mo EO: Wnt inhibition is
dysregulated in gliomas and its re-establishment inhibits
proliferation and tumor sphere formation. Exp Cell Res. 340:53–61.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Buckley CD, Tan J, Anderson KL, Hanein D,
Volkmann N, Weis WI, Nelson WJ and Dunn AR: Cell adhesion. The
minimal cadherin-catenin complex binds to actin filaments under
force. Science. 346:12542112014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ji H, Wang J, Fang B, Fang X and Lu Z:
α-Catenin inhibits glioma cell migration, invasion, and
proliferation by suppression of β-catenin transactivation. J
Neurooncol. 103:445–451. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Xu M, Yang X, Zhao J, Zhang J, Zhang S,
Huang H, Liu Y and Liu J: High expression of Cullin1 indicates poor
prognosis for NSCLC patients. Pathol Res Pract. 210:397–401. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lai YJ, Tsai JC, Tseng YT, Wu MS, Liu WS,
Lam HI, Yu JH, Nozell SE and Benveniste EN: Small G protein Rac
GTPases regulate the maintenance of glioblastoma stem-like cells in
vitro and in vivo. Oncotarget. 8:18031–18049. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Serra N, Rosales R, Masana L and Vallvé
JC: Simvastatin Increases Fibulin-2 expression in human coronary
artery smooth muscle cells via RhoA/Rho-kinase signaling pathway
inhibition. PLoS One. 10:e01338752015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sizemore ST, Zhang M, Cho JH, Sizemore GM,
Hurwitz B, Kaur B, Lehman NL, Ostrowski MC, Robe PA, Miao W, et al:
Pyruvate kinase M2 regulates homologous recombination-mediated DNA
double-strand break repair. Cell Res. 28:1090–1102. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wang Y, Che S, Cai G, He Y, Chen J and Xu
W: Expression and prognostic significance of CTBP2 in human
gliomas. Oncol Lett. 12:2429–2434. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Dcona MM, Morris BL, Ellis KC and Grossman
SR: CtBP- an emerging oncogene and novel small molecule drug
target: Advances in the understanding of its oncogenic action and
identification of therapeutic inhibitors. Cancer Biol Ther.
18:379–391. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Patel J, Baranwal S, Love IM, Patel NJ,
Grossman SR and Patel BB: Inhibition of C-terminal binding protein
attenuates transcription factor 4 signaling to selectively target
colon cancer stem cells. Cell Cycle. 13:3506–3518. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Guo H, Zhang XY, Peng J, Huang Y, Yang Y,
Liu Y, Guo XX, Hao Q, An S and Xu TR: RUVBL1, a novel C-RAF-binding
protein, activates the RAF/MEK/ERK pathway to promote lung cancer
tumorigenesis. Biochem Biophys Res Commun. 498:932–939. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Matias PM, Baek SH, Bandeiras TM, Dutta A,
Houry WA, Llorca O and Rosenbaum J: The AAA+ proteins Pontin and
Reptin enter adult age: from understanding their basic biology to
the identification of selective inhibitors. Front Mol Biosci.
2:172015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Mao YQ and Houry WA: The role of pontin
and reptin in cellular physiology and cancer etiology. Front Mol
Biosci. 4:582017. View Article : Google Scholar : PubMed/NCBI
|