Introduction
Vascular calcification (VC) refers to the ectopic
deposition of calcium phosphate crystals or hydroxyapatite on the
vascular walls. It is frequently observed during ageing, as well as
in degenerative diseases such as chronic kidney disease (CKD),
diabetes and atherosclerosis, and significantly increases the risk
of cardiovascular disease (CVD) and mortality (1-3).
Contrary to the long-held surmise that calcium and phosphorus are
passively deposited on the vascular walls, recent studies have
indicated that VC is an active and cell-regulated process similar
to the mineralization of osteo/chondrocyte-like cells during bone
formation (4). The incidence of
coronary artery calcification in CKD patients is >50% in the
absence of dialysis and significantly higher at 70-90% among
patients undergoing dialysis (5).
In addition, increased calcium burden of the thoracic aorta
elevates the risk of CVD by 3.7-fold (6), whereas abdominal aorta calcification
increases the relative risk of coronary, cerebrovascular and
cardiovascular events as well as mortality rates (7). VC is a significant risk factor of CVD
in 90% of males and 67% of females above the age of 70 years
(8).
Vascular smooth muscle cells (VSMCs) may
differentiate into the major cell types in the vessel wall in
response to suitable environmental stimuli. There is evidence that
VSMCs lose their contractile phenotype and trans-differentiate into
osteoblast-like cells expressing osteogenic transcription factors
and proteins. These cells may initiate calcification of the
vascular wall by secreting calcium and phosphorous-loaded exosomes
into the extracellular matrix (ECM) (9-11).
However, little is known regarding the specific pathways and
molecular mechanisms underlying VC, which markedly limits the
development of effective drugs.
Non-coding RNAs are a class of transcripts that
regulate the expression of protein-encoding genes through various
mechanisms. MicroRNAs (miRNAs/miRs) are 18-22 nucleotides in length
and regulate target gene expression at the post-transcriptional
level through binding at the 3' untranslated region. Long
non-coding RNAs (lncRNAs) are >200 nucleotides long and exert
their regulatory effects through more complex mechanisms (12). Several lncRNAs have been identified
in recent years that are involved in the progression of various
pathological conditions, including CVD (13-15).
Furthermore, specific miRNAs and lncRNAs have been implicated in
VSMC calcification. For instance, downregulation of miR-204,
miR-29b or miR-30e trigger the osteogenic differentiation and
calcification of VSMCs both in vitro and in vivo,
whereas upregulation of miRNA-128 accelerates cardiovascular
calcification (16-20).
Lin et al (21) demonstrated
that lncRNA-ES3 enhanced hyperglycemia-induced calcification of
VSMCs by suppressing miR-34c-5p expression as a sponge. In
addition, Jeong et al (22)
identified numerous differentially expressed lncRNAs in calcified
rat VSMCs, of which leucine rich repeat containing 75a-antisense
(AS)1 was significantly downregulated and its ectopic expression
attenuated calcium accumulation in VSMCs cultured with inorganic
phosphate. However, the exact mechanistic roles of non-coding RNAs
in VSMC calcification have remained to be elucidated.
In the present study, the differentially expressed
genes (DEGs) and pathways in VSMCs exposed to high and normal
calcium levels for varying durations were identified using
bioinformatics. Given the physiological similarities between VCs
and bone mineralization, the DEGs and pathways common to both
calcifying VSMCs (CVSMCs) and osteoblasts (COs) were also screened
and certain potentially crucial genes were experimentally
validated. The putative regulatory networks of non-coding RNAs and
transcription factors (TFs) in CVSMCs were also predicted. The
present results provide novel insight into the molecular basis of
the pathogenesis of VC.
Materials and methods
Microarray data and identification of
DEGs
The microarray dataset GSE37558 profiled on the
GPL6947 platform (Illumina HumanHT-12 V3.0 expression beadchip) was
downloaded from the National Center for Biotechnology Information
(NCBI)-Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi).
It includes the mRNA expression data of 32 VSMCs and osteoblasts
samples cultured for 0, 2, 8, 12 or 25 days (3-4 replicates per
time-point) in calcified medium containing 1.8 mM Ca2+
(23). The raw data were integrated
and the DEGs between the control and calcified VSMCs were
identified using the GEO2R tool, which is a GEO tool (https://www.ncbi.nlm.nih.gov/geo/geo2r/?acc=GSE37558)
and Morpheus website (https://software.broadinstitute.org) using an adjusted
P<0.05 with |log fold change|>1 as the thresholds. The DEGs
common to VSMCs calcified for varying durations were also
identified using the same criteria. The DEGs in osteoblasts were
similarly screened, and the shared DEGs between CVSMCs and COs were
also defined using the Morpheus website.
Gene ontology (GO) and pathway
enrichment analysis
The DEGs were functionally annotated by GO, Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathway and reactome
analyses using the Database for Annotation, Visualization and
Integrated Discovery version 6.8 (https://david.ncifcrf.gov/). The significantly
enriched genes or pathways were screened on the basis of
P<0.05.
Protein-protein interaction (PPI)
network
The PPI networks of the DEGs were constructed using
a Search Rool for the Retrieval of Interacting Genes and proteins
(STRING) database (http://string.embl.de/; accessed April, 2019)
(24) in order to identify the
interacting and hub genes. Cytoscape software (25) was used for visualizing the networks
and analyzing the degree of connectivity of nodes. Finally, module
analysis of the PPI network was performed using the molecular
complex detection application of Cytoscape software. The module
genes were functionally annotated as described above.
Construction of regulatory
networks
The target miRNAs of calcification-related DEGs were
predicted using the miRWalk (http://zmf.umm.uni-heidelberg.de; accessed April,
2019), miRanda (http://www.microrna.org/microrna/home.do; accessed
April, 2019), miRDB (http://www.mirdb.org/; accessed April, 2019), RNA22
(https://cm.jefferson.-edu/rna22/;
accessed April, 2019) and TargetScan (http://www.targetscan.org/vert_72/; accessed April,
2019) databases. The miRNA-target DEG pairs were established when
predicted by all five databases. The TFs targeting miRNAs in the
miRNA-target gene network were predicted using the TransmiR
(http://www.cuilab.cn/transmir; accessed
April, 2019) database (26) based
on literature-curated TF-miRNA regulation data. The predicted TFs
within the DEGs were defined as differentially expressed TFs
(DETFs) and used to construct the DETF-miRNA-target DEG network. In
addition, the TF-miRNA-hub gene network was also established using
hub genes identified from the PPI network. Finally, the lncRNAs
targeting DETFs in the TF-miRNA-hub gene network were screened from
the LncRNA2Target v2.0 database
(http://123.59.132.21/lncrna2target; accessed April, 2019)
(27) to construct the
lncRNA-DETF-miRNA-target gene network. All regulatory networks were
established and visualized using Cytoscape.
Cell culture and calcification
assay
Human VSMC line (cat. no. CRL-1999) was purchased
from Aolu Biotech and cultured in Dulbecco's modified Eagle's
medium (Gibco; Thermo Fisher Scientific, Inc.) containing 10% fetal
bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin-streptomycin (Beyotime Institute of Biotechnology) at
37˚C under 5% CO2. The cells were harvested at 80-90%
confluency using 0.25% trypsin/0.02% EDTA solution (Beyotime
Institute of Biotechnology) and cells from passages 3-7 were used
in the experiments. For in vitro calcification, VSMCs were
seeded in 6-well plates with 60-70% cell density and cultured in
growth medium containing 1.8 mM CaCl2 (Sigma-Aldrich;
Merck KGaA) for 12 days. The medium was replaced every 2-3
days.
RNA extraction and reverse
transcription-quantitative (RT-q)PCR analysis
TRIzol® (Invitrogen; Thermo Fisher
Scientific, Inc.) was used to extract total RNA from the cultured
VSMCs according to the manufacturer's protocol. The integrity of
the isolated RNA was determined by measuring the absorbance at 260
nm (A260) and the purity in terms of the A260/A280 ratio. Total RNA
was reverse-transcribed into complementary (c)DNA using the
TransScript kit (Takara Biotechnology, Inc.). qPCR was performed
with 2 µl first-strand cDNA as the template and the FastStart SYBR
Green kit (Roche). The primers were purchased from GeneCopoeia
Biotechnology Company and their sequences are presented in Table I. The thermocycling conditions of
qPCR were as follows: 2 min at 50˚C and 10 min at
95˚C, followed by 40 cycles of 15 sec at
95˚C, 30 sec 60˚C and 30 sec at
72˚C (ABI 7500 Fast; Applied Biosystems; Thermo Fisher
Scientific, Inc.). GAPDH and U6 were used as the internal controls
for mRNAs and miRNAs, respectively. The relative expression of
these genes was determined using the comparative CT
(2-ΔΔCq) method (28).
 | Table IPrimer sequences or numbers for
PCR. |
Table I
Primer sequences or numbers for
PCR.
| Gene symbol | Primer sequence
(5'-3') or catalogue number |
|---|
| DANCR | Forward:
CAGCTGACCCTTACCCTGAA |
| | Reverse:
GACCCTGGGGTTGTTAGTCA |
| H19 | Forward:
CAGAGTCCGTGGCCAAGG |
| | Reverse:
CGCCTTCAGTGACTGGCA |
| MELK | Forward:
ACTGCCCTGGAGGAGAGCT |
| | Reverse:
AGCCCTGGCTGTGCACATAA |
| FOXM1 | Forward:
GGAGGAAATGCCACACTTAGCG |
| | Reverse:
TAGGACTTCTTGGGTCTTGGGGTG |
| FOXO1 | Forward:
TACGCCGACCTCATCACCAAG' |
| | Reverse:
GCACGCTCTTCACCATCCACT' |
| SNAI2 | Forward:
CACCTCCTCCAAGGACCA' |
| | Reverse:
GGCCAGCCCAGAAAAAGT |
| GAPDH | Forward:
GTCAGCCGCATCTTCTTT' |
| | Reverse:
CGCCCAATACGACCAAAT' |
| U6 | RQP086799 |
| Hsa-miR-485-3p | HmiRQP0521 |
| Hsa-
miR-181d-5p | HmiRQP0237 |
Statistical analysis
The in vitro experiments were performed in
triplicate and were independently repeated three times. SPSS
version 21.0 (IBM Corp.) was used for statistical analysis. Data
were expressed as the mean ± standard deviation and compared using
an unpaired t-test. P<0.05 was considered to indicate
statistical significance.
Results
Identification of
calcification-related DEGs in the CVSMCs and COs
To identify the genes affected by short- and
long-term high Ca2+ exposure, VSMCs were cultured in the
calcification medium for varying durations and DEGs were detected
(GSE37558). As presented in Fig.
1A, VSMCs cultured for 2, 8, 12 and 24 h had 393, 684, 892 and
799 DEGs relative to the normal cells (0-day control),
respectively. Furthermore, 318 genes were consistently
differentially expressed across all time-points, including 147
upregulated and 185 downregulated genes (Fig. 1A, Tables SI and SII). Furthermore, there were 206, 441,
565 and 681 DEGs in the osteoblasts cultured for 2, 8, 12 and 25
days, respectively, in the calcification medium compared to normal
osteoblasts, of which 120 DEGs were common to all time-points
(Fig. 1B, Tables SIII and SIV). Furthermore, 43 DEGs were common to
the CVSMCs and COs, including 34 upregulated and 9 downregulated
genes (Fig. 1C).
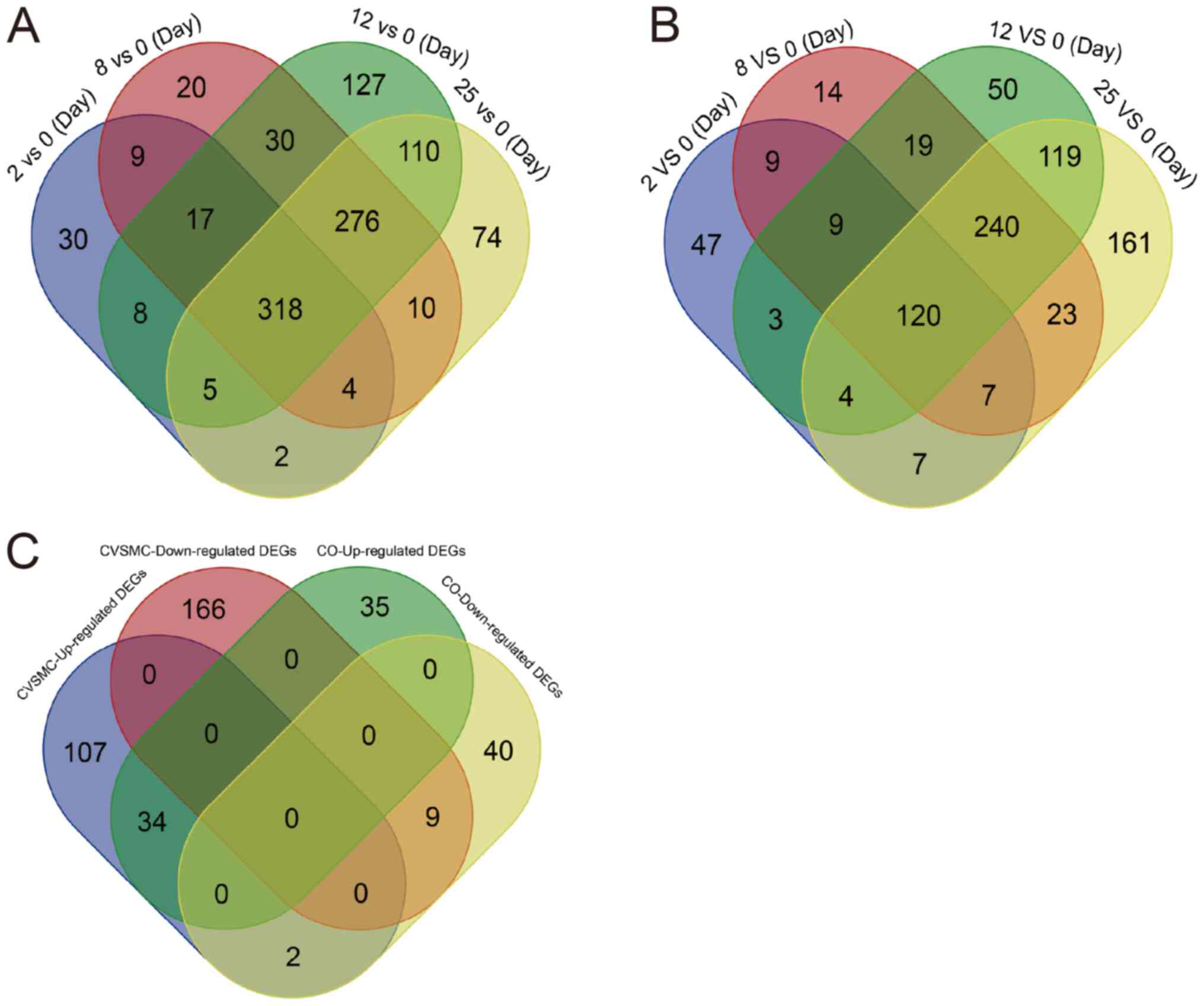 | Figure 1Identification of DEGs in CVSMCs and
COs. (A) VSMCs cultured in high-calcium medium for 2, 8, 12 and 25
days had 318 DEGs relative to the 0-day normal control. The DEGs
observed at the different time-points of calcification are
color-coded. The overlapping regions represent shared DEGs across
the different time-points. (B) A total of 43 DEGs were common
between CVSMCs and COs, including 34 upregulated and 9
downregulated genes, which were color-coded. (C) Osteoblasts
cultured for 2, 8, 12 and 25 days in calcification medium had 206,
441, 565 and 681 DEGs compared to the normal osteoblasts,
respectively, and 120 DEGs were consistent across all time-points.
The DEGs from different groups are color-coded and the overlapping
region represents shared DEGs. VC, vascular calcification; CVSMCs,
calcifying vascular smooth muscle cells; CO, calcifying osteoblast;
DEGs, differentially expressed genes; GO, Gene Ontology. |
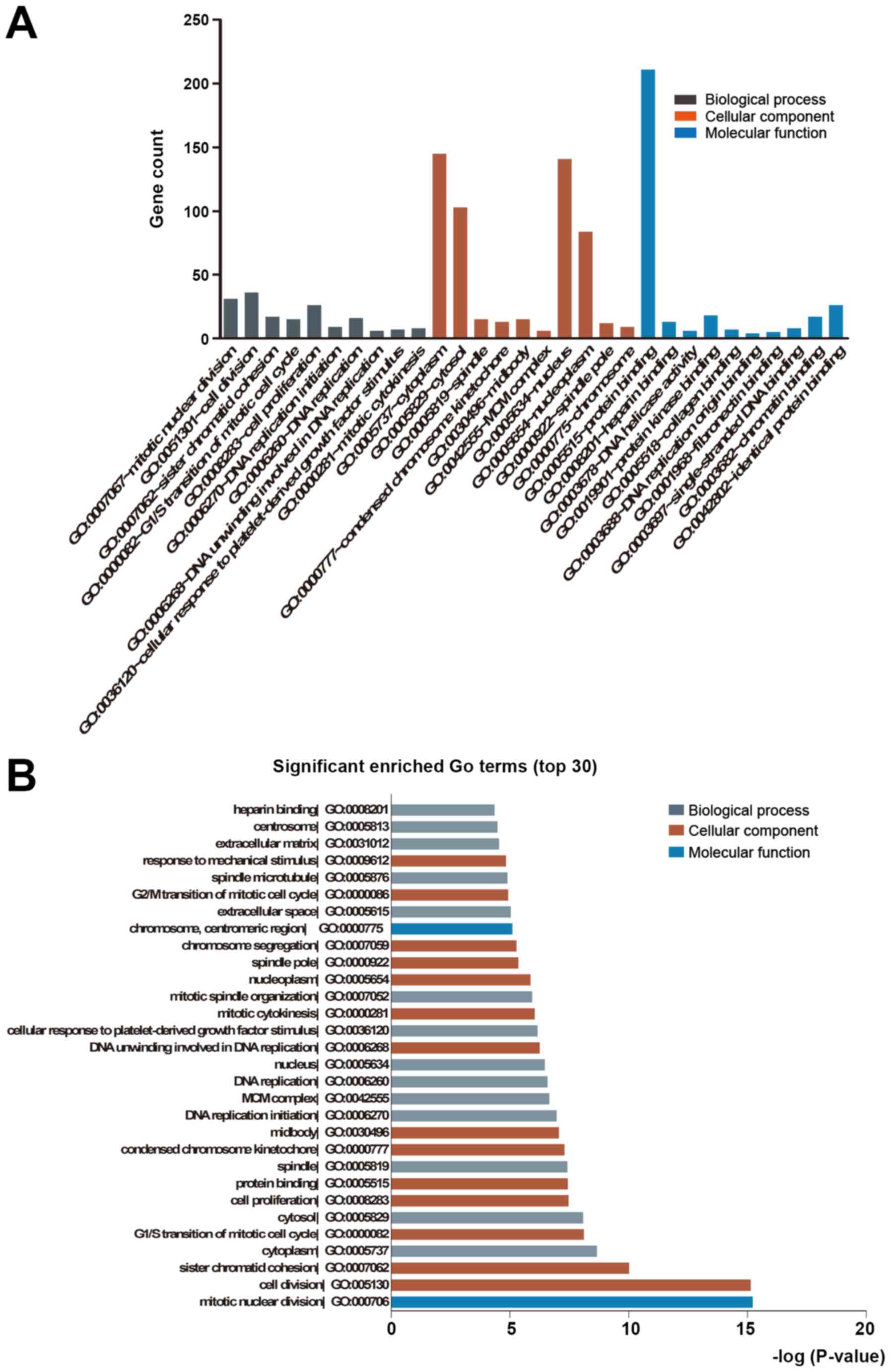
Functional annotation of
calcification-related DEGs
The common DEGs (see above) were functionally
annotated in GO terms and KEGG pathways. The upregulated genes were
significantly enriched in biological process terms of ECM
organization, cell adhesion, positive regulation of apoptotic
process and positive regulation of gene expression, whereas the
downregulated DEGs were significantly enriched in cell division,
mitotic nuclear division, cell proliferation and cell cycle. The
significant molecular function terms for upregulated genes were
integrin, heparin, receptor and fibronectin binding, while those
for downregulated genes were protein, ATP, protein kinase and
chromatin binding. Finally, in the category cellular component, the
upregulated genes were mainly enriched in terms such as cytoplasm,
extracellular space, extracellular exosome, ECM and extracellular
region, while the downregulated genes were mainly related to the
nucleoplasm, nucleus, cytosol and cytoplasm (Fig. 2A and B, Table
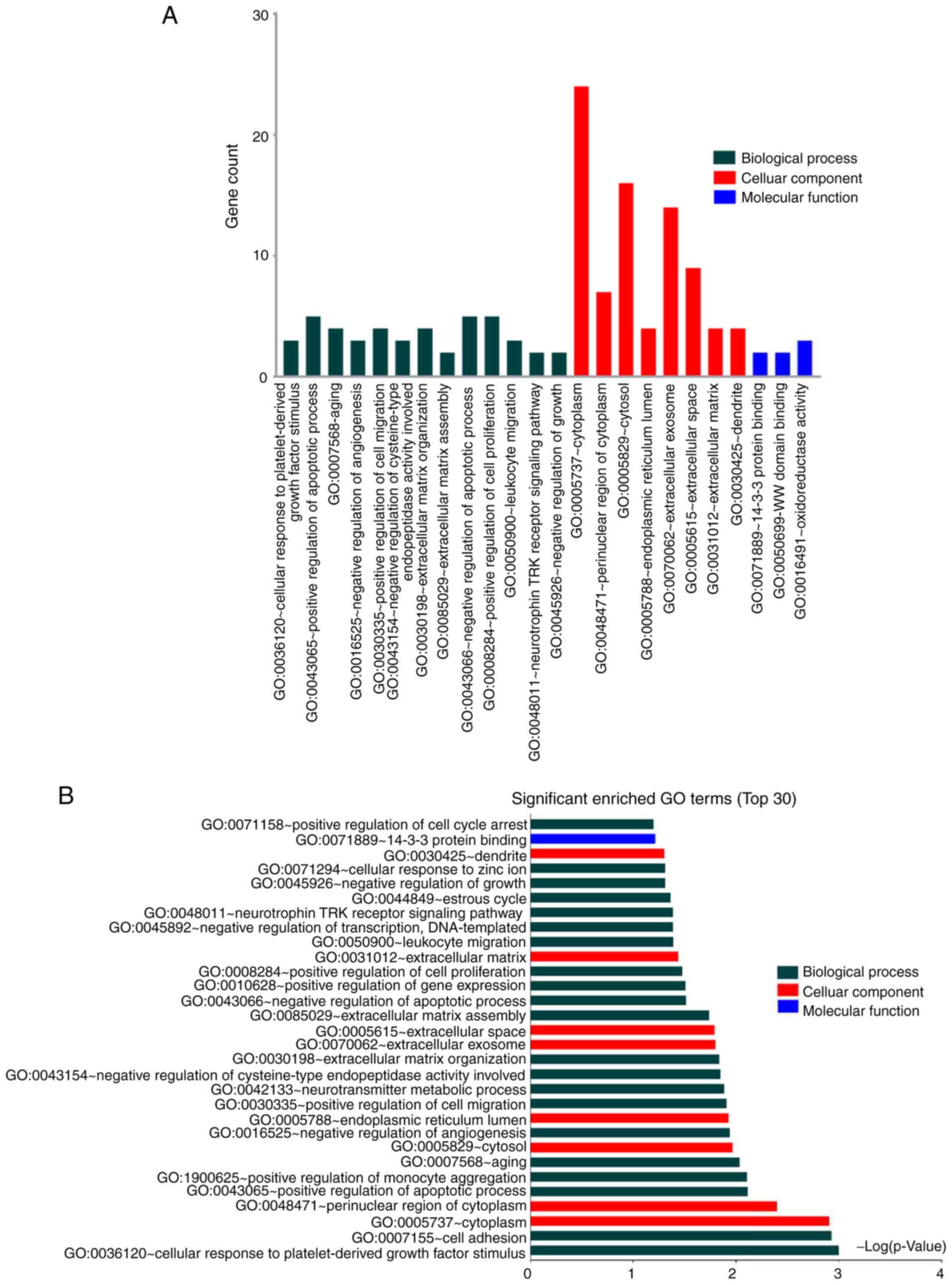
SV). GO analysis of the shared DEGs between CVSMCs and CO
revealed cytoplasm, ECM/exosome/space and tyrosine metabolism as
the enriched terms (Fig. 3A and
B, Table SVI). These results indicate that
the functional clusters of these DEGs are closely related to the
cell cycle, ECM and cell binding. Furthermore, the ECM has an
important role in both VSMC and osteoblast calcification.
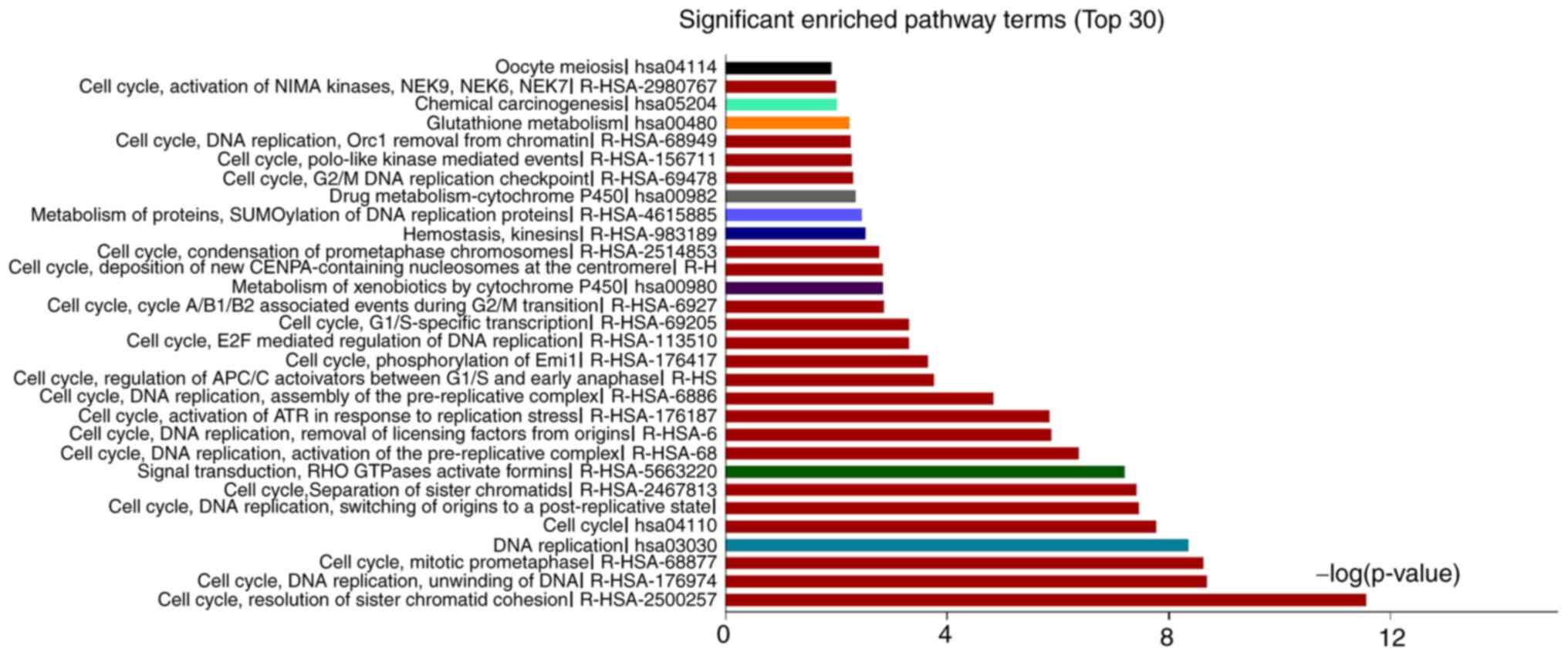
The significantly enriched pathways of upregulated
genes were platelet-derived growth factor, mineral absorption,
vascular smooth muscle contraction, focal adhesion and drug
metabolism-cytochrome P450, and those for downregulated DEGs were
resolution of sister chromatid cohesion, mitotic prometaphase, cell
cycle and separation of sister chromatids (Fig. 4, Table
SVII). Finally, the DEGs common to CVMCs and COs were enriched
in pathways of amino acid metabolism and metallothionein-binding
metals (Table SVIII). Thus, genes
related to distinct pathways were affected during calcification of
VSMCs and the downregulated genes in particular were strongly
associated with cell cycle and proliferation.
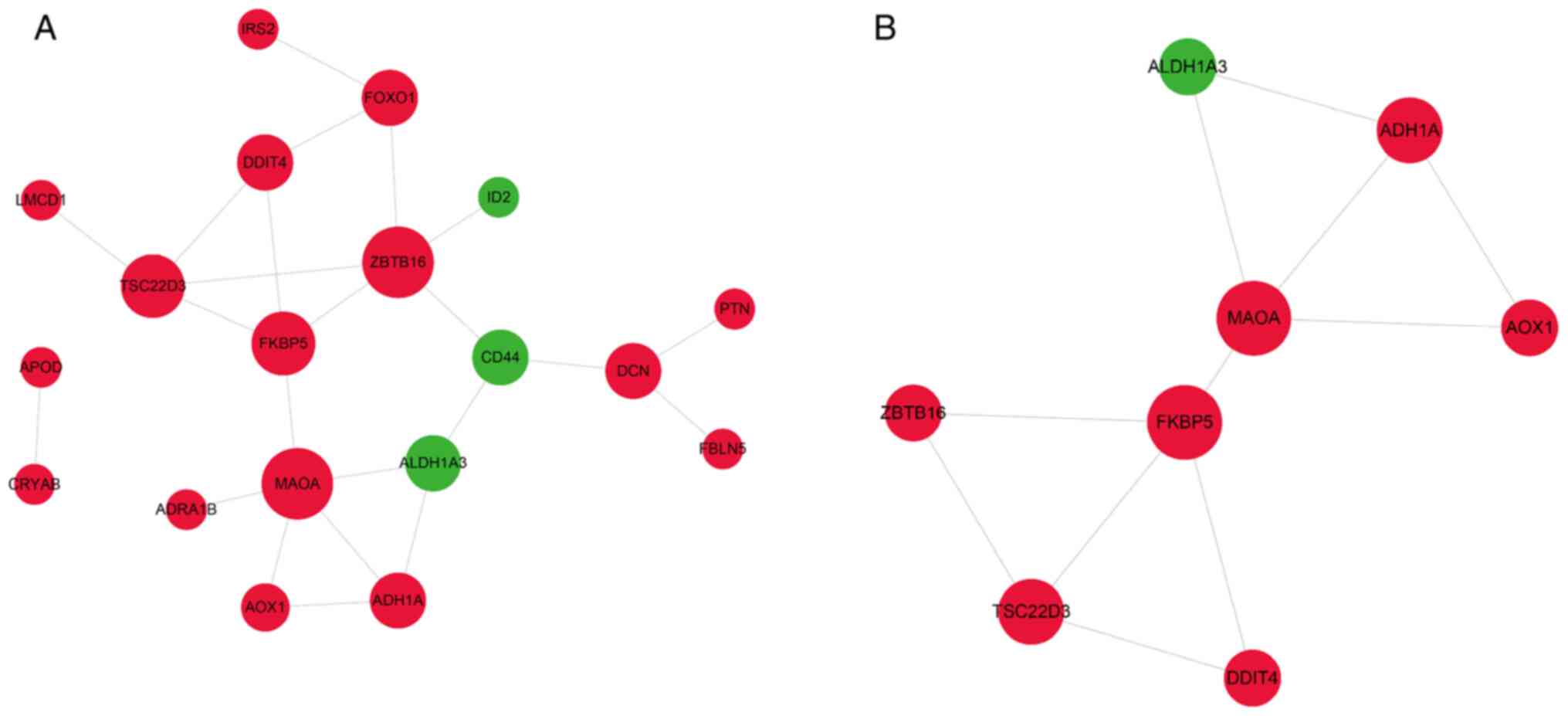
Identification of hub genes involved
in VSMC calcification and osteoblast mineralization
The hub genes involved in the calcification of VSMCs
were next identified by constructing a PPI network of the 318
common DEGs. As presented in Fig.
5A, the PPI network consisted of 281 genes, including 143
upregulated and 175 downregulated genes, and 3,650 edges. A total
of 30 genes with a degree of connectivity of >80 in the PPI
network were designated as the hub genes, including cyclin
dependent kinase 1, mitotic arrest deficient 2 like 1 (MAD2L1),
kinesin family member 11 (KIF11), maternal embryonic leucine zipper
kinase (MELK), non-SMC condensin I complex subunit G (NCAPG), PDZ
binding kinase (PBK), kinesin family member 23 (KIF23) and cyclin
A2. Module analysis of the PPI network using the MCODE app further
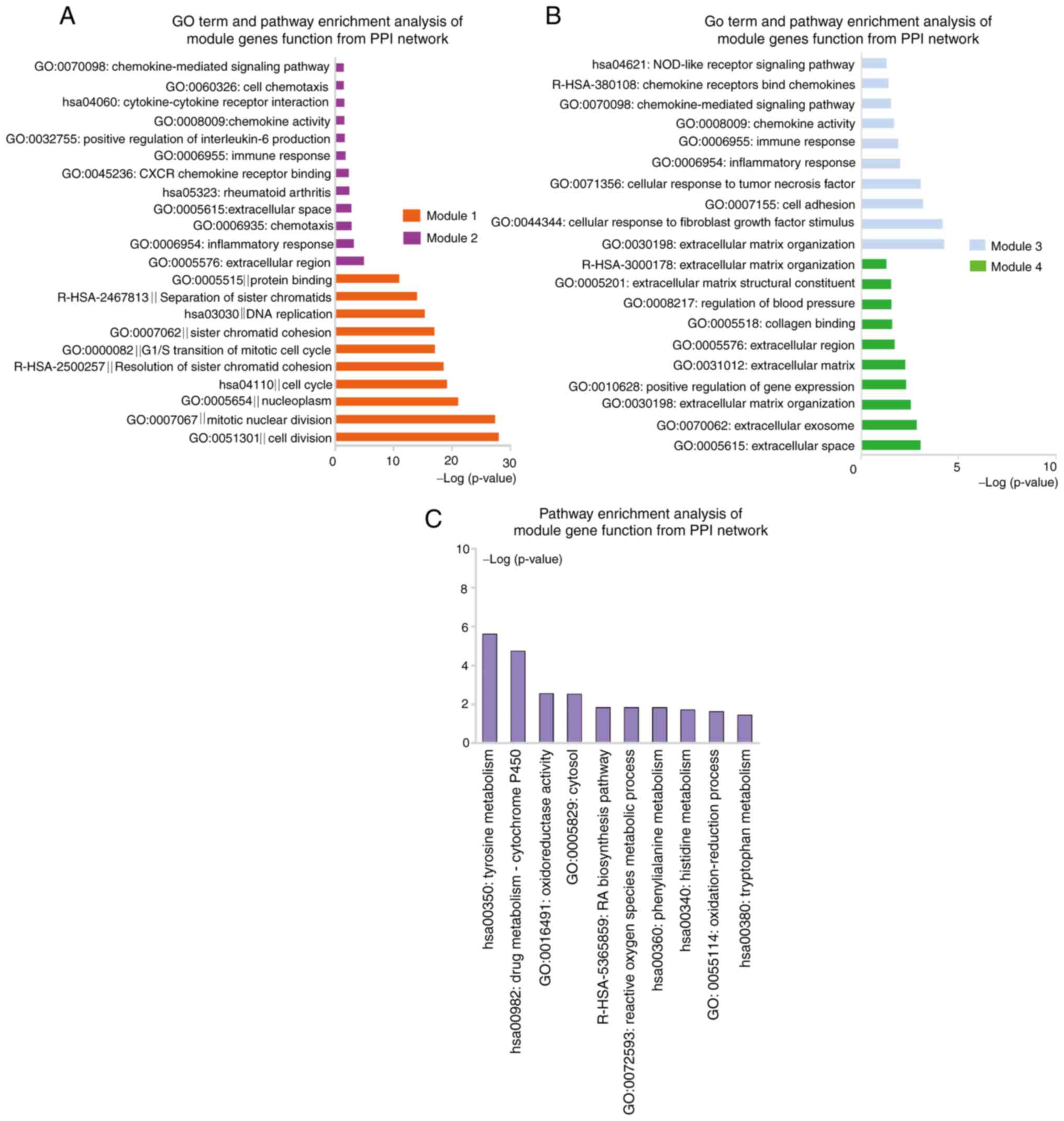
revealed 4 modules (Fig. 5B-E).
Module 1 with 79 nodes and 2,770 edges was closely associated with
the cell cycle, cell division and separation of sister chromatid
pathways. Module 2 consisted of 9 nodes and 20 edges and was
significantly enriched in the pathways of inflammatory response,
immune response, chemokine-mediated signaling and positive
regulation of interleukin (IL)-6 production (Fig. 7A and Table SIX). In addition, module 3 was
comprised of 8 nodes and 16 edges and was mainly involved in ECM
organization, cellular response to fibroblast growth factor
stimulus, cell adhesion, chemokine-mediated signaling and NOD-like
receptor signaling pathways. Finally, module 4 had 8 nodes and 10
edges, and was associated with extracellular space and ECM pathways
(Fig. 7B and Table SIX). These results suggested that
signaling pathways related to the cell cycle, immune response, ECM,
chemotaxis and inflammatory response have a key role in VSMC
calcification.
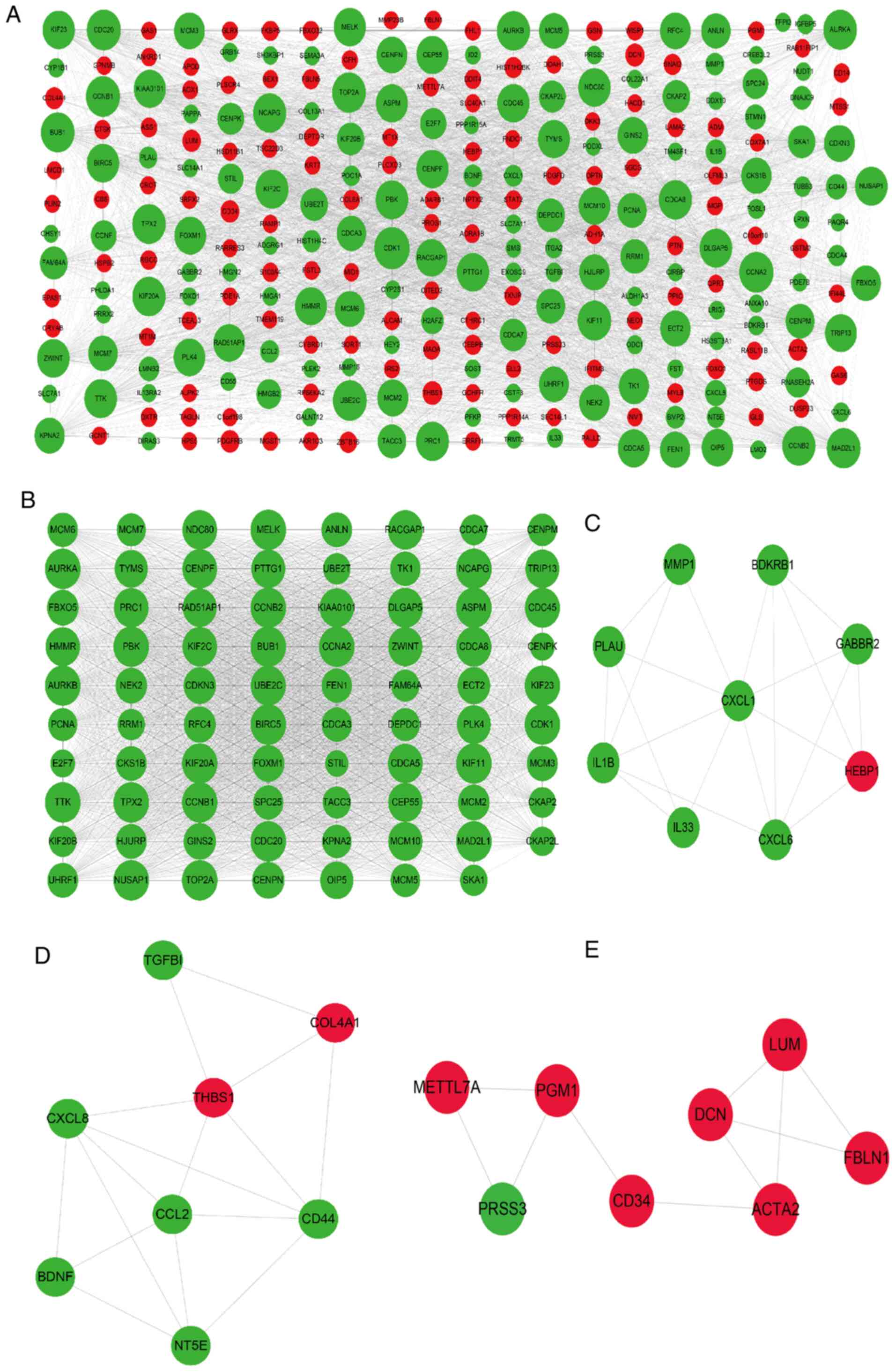 | Figure 5PPI networks and modular analysis of
DEGs. (A) PPI network of DEGs in calcifying vascular smooth muscle
cells, consisting of 281 gene nodes and 3,650 edges. The red and
green gene nodes represent the upregulated and downregulated genes,
respectively. The volume of gene nodes is proportional to the
degree of connectivity. (B) Module 1 consisted of 79 gene nodes and
2,770 edges. (C) Module 2 consisted of 9 gene nodes and 20 edges.
(D) Module 3 consisted of 8 gene nodes and 16 edges. (E) Module 4
consisted of 8 gene nodes and 10 edges. Module analysis utilized
the following cut-off criteria: Degree cutoff, 2; node score
cutoff, 0.2; K-core cutoff, 2; and max depth, 100. DEGs,
differentially expressed genes; PPI, protein-protein
interaction. |
VSMCs are able to differentiate into osteoblast-like
cells expressing osteogenic proteins in response to dysregulated
calcium-phosphate metabolism and thus contribute to VC (9,10).
Therefore, the PPI network of the 43 DEGs shared by CVSMCs and COs
was also established in order to identify signaling pathways
involved in VSMCs and osteoblast mineralization. As presented in
Fig. 6A, the PPI network contained
19 gene nodes and 23 edges. One module including 8 nodes and 11
edges (Fig. 6B) was identified,
which was associated with amino acid metabolism, drug
metabolism-cytochrome P450, metabolic pathways and oxidoreductase
activity (Fig. 7C and Table SX).
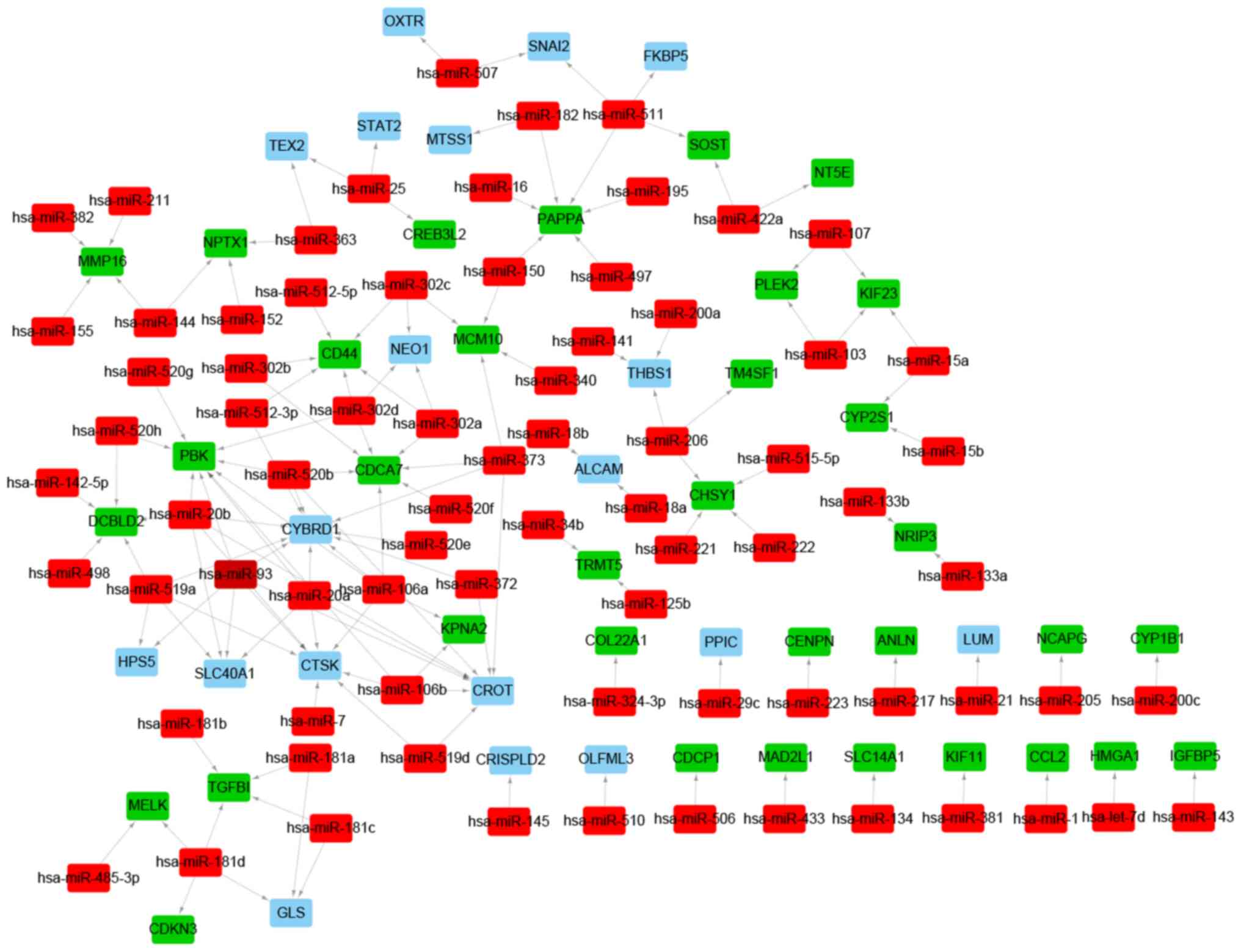
VSMC calcification-related regulatory
networks
To further explore the regulatory mechanisms
involved in VSMC calcification, miRNAs targeting the CVSMC-related
DEGs were predicted by the miRanda, miRDB, miRWalk, RNA22 and
TargetScan databases. A total of 76 putative miRNAs and 140
miRNA-target pairs were identified. A miRNA-target regulatory
network was constructed with 76 miRNAs, 53 DEGs and 140 edges, and
included Homo sapiens (hsa)-miR-20a-PBK, hsa-miR-15a-
kinesin family member 23 (KIF23), hsa-miR-511-snail family
transcriptional repressor 2 (SNAI2), hsa-miR-507-SNAI2,
hsa-miR-181d- maternal embryonic leucine zipper kinase (MELK) and
hsa-miR-485-3p-MELK regulatory pairs (Fig. 8 and Table SXI). Similarly, lncRNA and
TF-mediated regulatory networks were also constructed. A total of
251 TFs were predicted and the TF-miRNA regulatory network
consisted of 754 miRNA-target pairs, 251 TFs and 60 target DEGs
(Fig. 9A and Table SXII). The TFs FOS like 1, AP-1
transcription factor subunit, SNAI2, zinc finger and BTB domain
containing 16, high mobility group AT-hook 1, CCAAT enhancer
binding protein beta (CEBPB), E2F transcription factor 7 (E2F7),
IL1B, forkhead box (FOX)M1, endothelial PAS domain protein 1 and
forkhead box O1 (FOXO1) were differentially expressed and were used
to construct a DETF- miRNA-target network, including 29 DETF-miRNAs
and 27 miRNAs-target DEG pairs, such as FOXO1-hsa-miR-145,
SNAI2-hsa-miR-221/222, SNAI2-hsa-miR-200a and SNAI2-hsa-mir-145
(Fig. 9B and Table SXIII). The hub genes in the PPI
network that were potential targets of the predicted miRNAs, such
as MAD2L1, KIF11, NCAPG, PBK, KIF23 and MELK, were incorporated
into the TF-miRNA-hub gene network. The latter included 135
TF-miRNA and 13 miRNA-hub gene pairs, such as
CEBPB-hsa-miR-20a-PBK, E2F7-hsa-miR-15a-KIF23 and RUNX family
transcription factor 1 (RUNX1)-hsa-mir-181d-MELK (Fig. 9C and Table SXIV). Since CEBPB and E2F7 were
identified as DETFs, the regulatory lncRNAs were next predicted.
The putative CEBPB-targeting lncRNAs were metastasis associated
lung adenocarcinoma transcript 1, chondrogenesis-associated
transcript, RAD51 antisense RNA 1 and negative regulator of
antiviral response, whereas FOXF1, tensin 1, non-coding RNA
activated by DNA damage and CDK6 antisense RNA 1 were predicted to
regulate E2F7. In addition, lncRNAs regulating FOXO1 and SNAI2 were
predicted. The lncRNA-DETF-miRNA-target gene regulatory network is
presented in Fig. 9D.
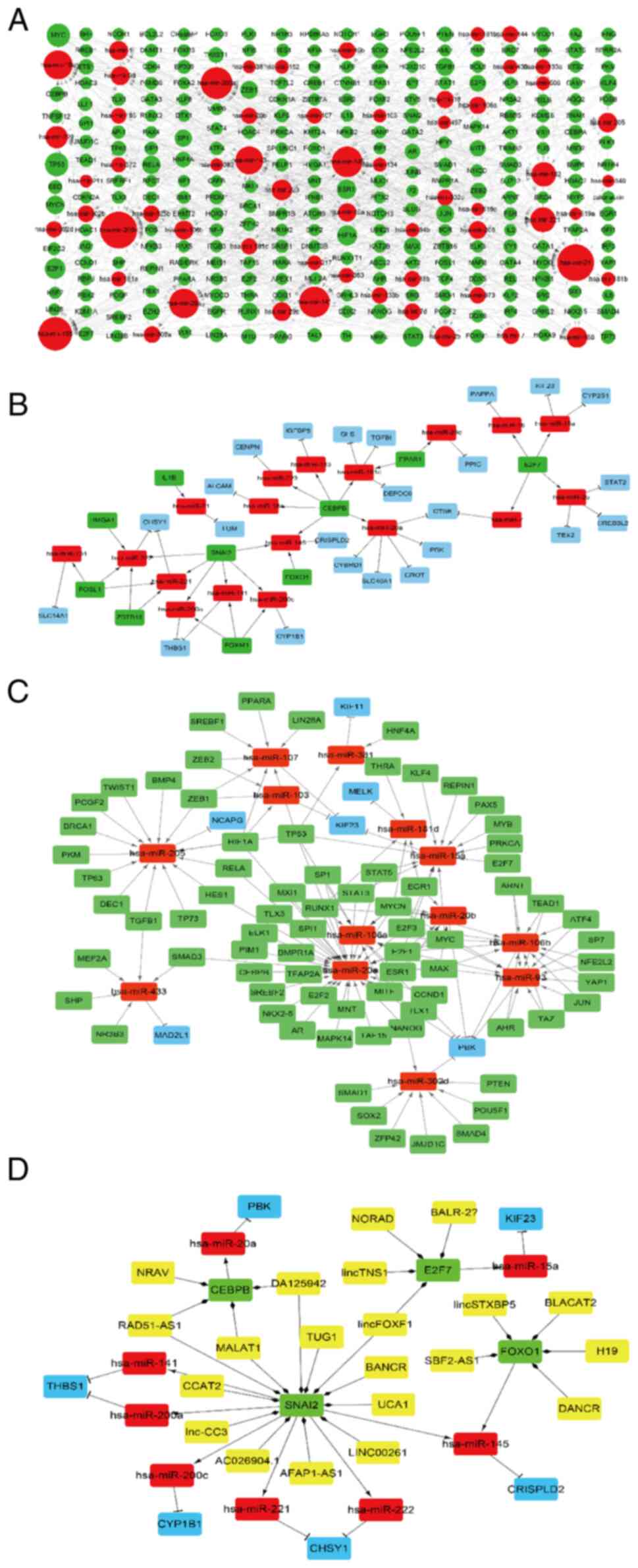 | Figure 9LncRNA-TF-miRNA-target gene
regulatory network. (A) TF-miRNA regulatory network. Red and green
nodes represent the miRNAs and TFs, respectively. The volume of
gene nodes is proportional to the degree of connectivity of the
gene nodes. (B) DETF-miRNA-target gene regulatory network. Red,
green and blue nodes represent miRNAs, DETFs and DEGs,
respectively. (C) TF-target miRNA-target hub gene regulatory
network. Red, green and blue nodes represent miRNAs, TFs and target
hub genes from the PPI network, respectively. (D)
LncRNA-DETF-miRNA-target gene regulatory network. Yellow, green,
red and blue nodes represent LncRNAs, DETFs, miRNAs and DEGs,
respectively. Arrows, diamonds and ‘T’ respectively indicate the
TF-miRNA, lncRNA-target DETF and miRNA-target gene relationships.
PPI, protein-protein interaction; TF, transcription factor; DETF,
differentially expressed TF; lncRNA, long non-coding RNA; miRNA,
microRNA; DEG, differentially expressed gene. |
Expression levels of certain potential
crucial genes in VSMCs
To verify the in-silico results, a VSMC
calcification model was established in vitro and the
expression levels of selected relevant genes were analyzed. As
presented in Fig. 10,
differentiation antagonizing non-protein coding RNA (DANCR), MELK
and FOXM1 were downregulated in the calcified VSMCs, while H19
imprinted maternally expressed transcript (H19), miR-485-3p,
miR-181d, FOXO1 and SNAI2 were upregulated.
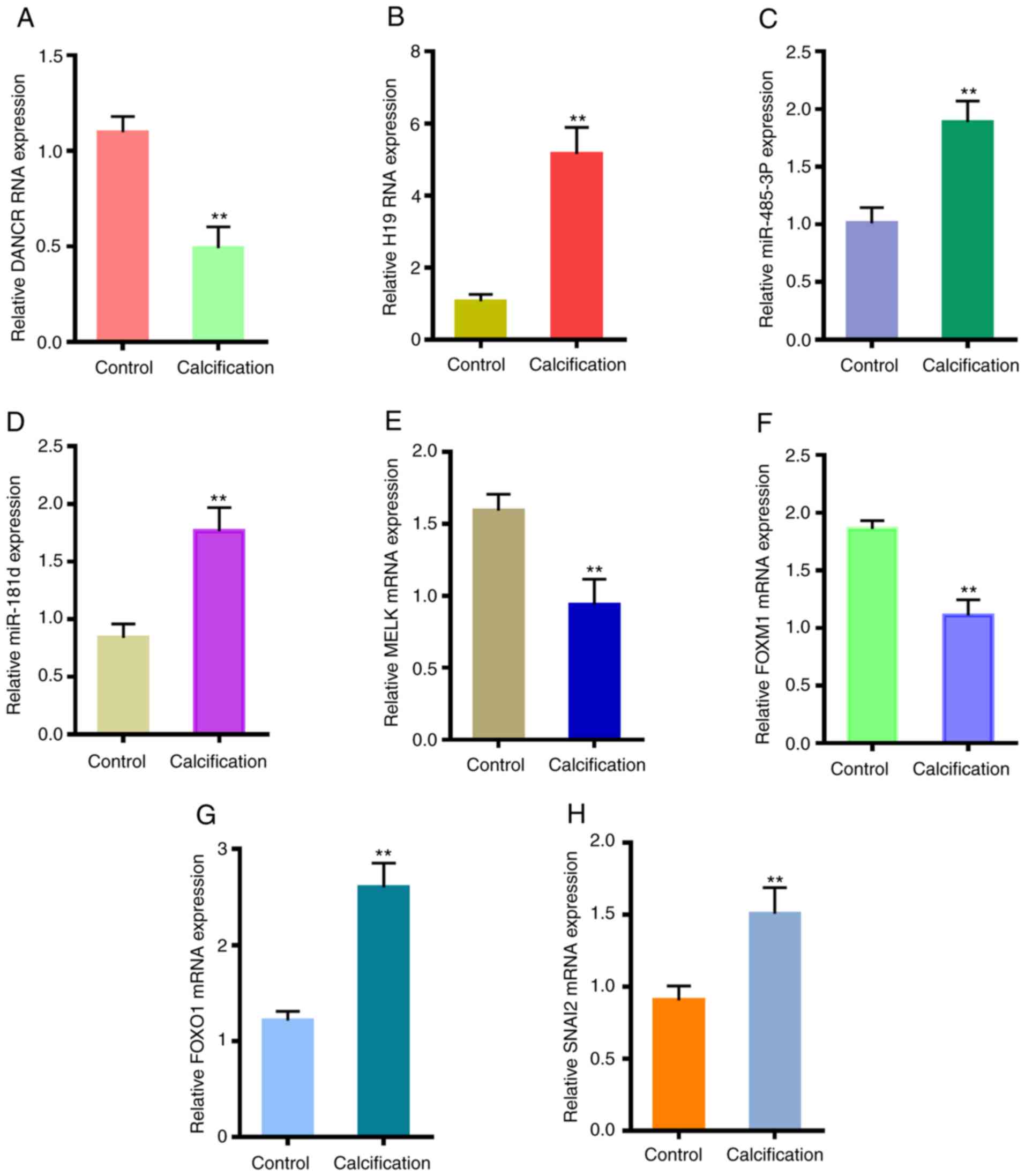 | Figure 10Validation of crucial genes in
CVSMCs. Expression levels of (A) DANCR, (B) H19, (C) miR-485-3p,
(D) miR-181d, (E) MELK, (F) FOXM1, (G) FOXO1 and (H) SNAI2 in the
normal VSMCs and CVSMCs. **P<0.05 vs. control.
CVSMCs, calcifying vascular smooth muscle cells; miR, microRNA;
FOX, forkhead box; DANCR, differentiation antagonizing non-protein
coding RNA; H19, H19 imprinted maternally expressed transcript;
MELK, maternal embryonic leucine zipper kinase; SNAI2, snail family
transcriptional repressor 2. |
Discussion
In the present study, several putative genes,
pathways and non-coding RNAs associated with VSMC calcification
were identified, several of which were common to osteoblast
mineralization. The DEGs in CVSMCs were mainly enriched in the cell
cycle, ECM, inflammation and chemotaxis-mediated signaling
pathways, whereas pathways related to the ECM, cytoplasm and
metabolism were enriched among the DEGs shared between the CVSMCs
and COs. The hub genes in the PPI network were MELK, PBK and KIF23,
as well as DETFs including SNAI2, FOXM1 and FOXO1, and they are
likely to regulate VSMC calcification.
In agreement with the results of the present
bioinformatics analysis, previous studies using DNA microarray
analysis of calcified aortic valve and proteomic analysis of
calcified abdominal and thoracic aorta indicated that the DEGs were
enriched in inflammation, chemokine and immune response signaling
pathways (29,30). In addition, chemokine expression was
upregulated in the sclerotic aortic valves in an apolipoprotein
E-deficient mouse model (31). The
present results indicated that the ECM is similarly affected during
VSMC and osteoblast mineralization, which is consistent with the
increased mineral deposition observed in the ECM during VC. In
addition, studies have revealed significant changes in the ECM
proteins during VSMC calcification. For instance, collagen I and II
content is markedly increased in calcified VSMCs and arteries
(32-34).
Collagen I may induce VSMC transdifferentiation into
osteoblast-like cells and promote calcium crystallization by
interacting with matrix vesicles (32,35,36).
Furthermore, degradation of ECM elastin also increases VSMC
calcification and differentiation into osteoblast-like cells
(37-39).
By contrast, collagen IV expression decreased by 70% in calcified
VSMCs, and collagen IV, collagen XIV and cartilage oligomeric
matrix protein were all able to inhibit VSMC calcification and
osteogenic differentiation through different signaling pathways
(35,36,40,41).
Several hub genes were identified in the PPI
networks, several of which may regulate VSMC calcification. For
instance, inhibition of MELK and its downstream target genes with
the specific inhibitor OTSSP167 was reported to enhance osteoblast
formation and matrix mineralization (42). In line with this, in the present
study, MELK was downregulated in the CVSMCs, indicating that it is
likely inhibited during VSMC calcification. The TFs FOXM1 and EZH2
are the downstream targets of MELK (43), of which FOXM1 was significantly
downregulated in the present study. Dioscin-mediated inhibition of
FOXM1 reduced VSMC proliferation and migration in vitro, as
well as intimal thickening in a rat model with carotid artery
balloon injury (44). EZH2 is also
known to suppress osteogenic differentiation of mesenchymal cells
and its inhibition promoted osteoblast differentiation (45,46).
Therefore, the MELK-FOXM1/EZH2 axis negatively regulates osteogenic
differentiation and mineralization. MELK was also predicted as a
target gene of hsa-miR-485-3p and hsa-miR-181d miRWalk (http://zmf.umm.uni-heidelberg.de; accessed April,
2019), indicating a novel has-485-3p/miR-181d-MELK-FOXM1/EZH2 axis
(Fig. 11) in VSMC calcification
that is worth exploring.
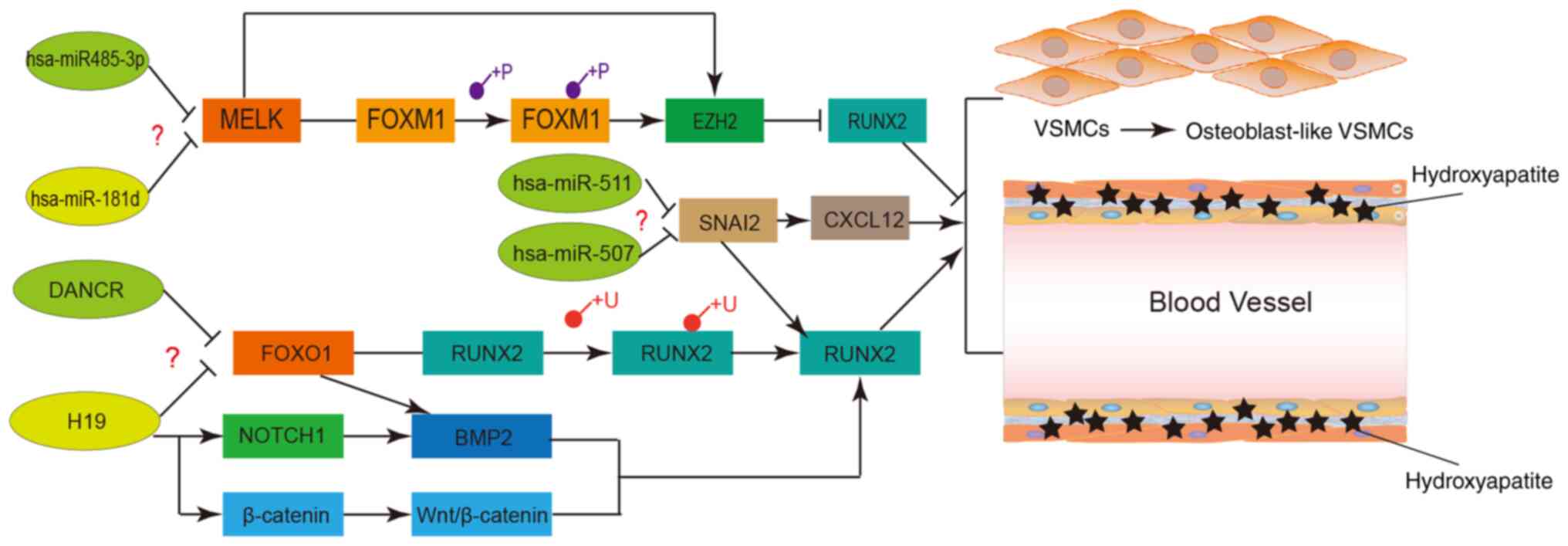 | Figure 11Potential regulatory networks and
pathways of certain crucial long non-coding RNAs, miRNAs and
proteins involved in VSMC calcification. VSMCs, vascular smooth
muscle cells; miRNA/miR, microRNA; FOX, forkhead box; DANCR,
differentiation antagonizing non-protein coding RNA; H19, H19
imprinted maternally expressed transcript; MELK, maternal embryonic
leucine zipper kinase; SNAI2, snail family transcriptional
repressor 2.RUNX2, RUNX family transcription factor 2; CXCL12,
C-X-C motif chemokine ligand 12; NOTCH1, notch receptor 1; EZH2,
enhancer of zeste 2 polycomb repressive complex 2 subunit; BMP2,
bone morphogenetic protein 2; β-catenin, catenin beta interacting
protein 1. |
Other DETFs identified in the present study were
FOXO1 and SNAI2. FOXO1 was previously reported to inhibit the
osteogenic TF RUNX2 in prostate cancer cells (47,48)
and its inhibition in VSMCs prevented RUNX2 ubiquitination, which
increased RNX2 levels and calcification (49). However, FOXO1 was determined to be
upregulated in the present study, suggesting that it may also
facilitate VSMC calcification. Consistent with this finding, FOXO1
levels were previously indicated to be increased in calcified
femoropopliteal arteries from human subjects. Furthermore, five
lncRNAs, including H19, DANCR, SBF2 antisense RNA 1, long
intergenic non-protein coding syntaxin binding protein 5 and long
intergenic non-protein coding RNA 958, were predicted as regulators
of FOXO1 in this study. H19 downregulated FOXO1 in bovine skeletal
muscle satellite cells, induced the osteogenic phenotype in valve
interstitial cells and promoted aortic valve calcification by
upregulating RUNX2 and bone morphogenetic protein (BMP)2, and
inhibiting notch receptor 1 (50,51).
It is also upregulated during osteogenesis of human mesenchymal
stem cells and promotes osteoblast differentiation via the
Wnt/β-catenin pathway (52).
However, the role of the H19-FOXO1 axis in VSMC calcification and
differentiation has remained elusive. DANCR regulates FOXO1
expression by affecting its ubiquitination (53). Zhu and Xu (54) and Jia et al (55) indicated that downregulation of DANC
promoted osteogenic differentiation of human fetal osteoblastic
cells and periodontal ligament stem cells, respectively. Therefore,
it may be hypothesized that the DANCR-FOXO1 axis potentially
regulates osteogenic differentiation of VSMCs. In addition,
hsa-miR-145 is a potential biomarker of VC in chronic kidney
disease (56) and was predicted as
a target miRNA of FOXO1 in the DETF-miRNA-target regulatory
network. Taken together, the H19/DANCR-FOXO1-miRNA-145-target gene
axis has a crucial role during VSMC calcification and should be
experimentally validated (Fig.
11). SNAI2 was predicted as a target of six miRNAs. Previous
studies indicated that SNAI2 promotes osteoblast maturation by
upregulating RUNX2, as well as osteoblast mineralization through
C-X-C motif chemokine ligand 12 signaling (57,58).
In addition, SNAI2 also mediated BMP-dependent transdifferentiation
of mouse non-ciliated aortic endothelial cells into mineralizing
osteogenic cells and promoted atherosclerosis and VC in vivo
(59). In line with this, in the
present study, the upregulation of SNAI2 suggested its stimulatory
effect on VSMC calcification. Furthermore, hsa-miR-511 and
hsa-miR-507, as well as 12 lncRNAs, were predicted to regulate
SNAI2. Most of these lncRNAs were identified in tumors. Therefore,
it was hypothesized that the hsa-miR-511/hsa-miR-507-SNAI2 axis and
the lncRNAs-SNAI2 axis potentially regulate VSMC calcification and
should be explored further (Fig.
11).
In conclusion, the present study identified several
potential regulatory mechanisms of VC and the DEGs and signaling
pathways associated with the calcification of VSCMs. They involve
changes in the inflammatory response, chemotaxis and ECM, and the
latter is characteristic of osteoblast mineralization as well.
Mechanistically, the hsa-485-3p/miR-181d-MELK-FOXM1/EZH2,
H19/DANCR-FOXO1 and SNAI2-mediated pathways possibly regulate VC.
These mechanisms require to be experimentally validated in future
studies.
Supplementary Material
List of DEGs in CVSMCs cultured for 25
days in calcified medium
The list of shared DEGs in CVSMCs
The list of DEGS in COs cultured for
25 days with calcified medium
The list of DEGs in COs from different
times
The enriched GO terms and signaling
pathways of Module 1 from the PPI network
The enriched GO terms of DEGs from
CVSMCs
The enriched GO terms of common DEGs
between CVSMCs and COs
The enriched signaling pathways of
up-regulated-DEGs in CVSMCs using the KEGG PATHWAY and Reactome
online dataset
The enriched signaling pathways of
common DEGs using the KEGG PATHWAY and Reactome online dataset
The enriched GO terms and signaling
pathways of Modules from the PPI network of shared DEGs between
CVSMCs and COs
List of miRNA-target regulatory
pairs
List of TF-miRNAs regulatory
pairs
List of DETF- miRNA-target regulatory
pairs
The list of TF-miRNA-hub gene
regulatory pairs
Acknowledgements
Not applicable.
Funding
Funding: This work was supported in part by the self-financing
research project of the Health and Family Planning Commission of
Guangxi Zhuang Autonomous Region (grant no. Z20180518).
Availability of data and materials
The data sets used and/or analyzed during the
current study are available from the corresponding author on
reasonable request. In addition, the dataset GSE37558 may be
obtained from the GEO database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE37558).
Authors' contributions
XW, YS, QL, ZZ and PH contributed to the study
conception and design. PH, XW and YS checked the associated
databases and analyzed raw data for bioinformatics analysis, cell
culture, PCR. XW, QL and ZZ wrote and revised the manuscript. All
of the authors read and approved the final manuscript. PH and XW
checked and approved the authenticity of the raw data. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lanzer P, Boehm M, Sorribas V, Thiriet M,
Janzen J, Zeller T, St Hilaire C and Shanahan C: Medial vascular
calcification revisited: Review and perspectives. Eur Heart J.
35:1515–1525. 2014.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Kovacic JC and Fuster V: Vascular
calcification, diabetes, and cardiovascular disease: Connecting the
dots. JACC Cardiovasc Imaging. 5:367–369. 2012.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Chen NX and Moe SM: Pathophysiology of
vascular calcification. Curr Osteoporos Rep. 13:372–380.
2015.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Leopold JA: Vascular calcification:
Mechanisms of vascular smooth muscle cell calcification. Trends
Cardiovasc Med. 25:267–274. 2015.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Block GA, Spiegel DM, Ehrlich J, Mehta R,
Lindbergh J, Dreisbach A and Raggi P: Effects of sevelamer and
calcium on coronary artery calcification in patients new to
hemodialysis. Kidney Int. 68:1815–1824. 2005.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Blomberg BA, de Jong PA, Thomassen A, Lam
MGE, Vach W, Olsen MH, Mali WPTM, Narula J, Alavi A and
Høilund-Carlsen PF: Thoracic aorta calcification but not
inflammation is associated with increased cardiovascular disease
risk: Results of the CAMONA study. Eur J Nucl Med Mol Imaging.
44:249–258. 2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Bastos Gonçalves F, Voûte MT, Hoeks SE,
Chonchol MB, Boersma EE, Stolker RJ and Verhagen HJ: Calcification
of the abdominal aorta as an independent predictor of
cardiovascular events: A meta-analysis. Heart. 98:988–994.
2012.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Bild DE, Detrano R, Peterson D, Guerci A,
Liu K, Shahar E, Ouyang P, Jackson S and Saad MF: Ethnic
differences in coronary calcification: The Multi-Ethnic Study of
Atherosclerosis (MESA). Circulation. 111:1313–1320. 2005.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Zhang MJ, Zhou Y, Chen L, Wang YQ, Wang X,
Pi Y, Gao CY, Li JC and Zhang LL: An overview of potential
molecular mechanisms involved in VSMC phenotypic modulation.
Histochem Cell Biol. 145:119–130. 2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Gomez D and Owens GK: Smooth muscle cell
phenotypic switching in atherosclerosis. Cardiovasc Res.
95:156–164. 2012.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Kapustin AN and Shanahan CM: Calcium
regulation of vascular smooth muscle cell-derived matrix vesicles.
Trends Cardiovasc Med. 22:133–137. 2012.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Yao RW, Wang Y and Chen LL: Cellular
functions of long noncoding RNAs. Nat Cell Biol. 21:542–551.
2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Bar C, Chatterjee S and Thum T: Long
noncoding RNAs in cardiovascular pathology, diagnosis, and therapy.
Circulation. 134:1484–1499. 2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Barwari T, Joshi A and Mayr M: MicroRNAs
in cardiovascular disease. J Am Coll Cardiol. 68:2577–2584.
2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Haemmig S, Simion V, Yang D, Deng Y and
Feinberg MW: Long noncoding RNAs in cardiovascular disease,
diagnosis, and therapy. Curr Opin Cardiol. 32:776–783.
2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Jiang W, Zhang Z, Yang H, Lin Q, Han C and
Qin X: The involvement of miR-29b-3p in arterial calcification by
targeting matrix metalloproteinase-2. Biomed Res Int.
2017(6713606)2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Cui RR, Li SJ, Liu LJ, Yi L, Liang QH, Zhu
X, Liu GY, Liu Y, Wu SS, Liao XB, et al: MicroRNA-204 regulates
vascular smooth muscle cell calcification in vitro and in vivo.
Cardiovasc Res. 96:320–329. 2012.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Ding W, Li J, Singh J, Alif R,
Vazquez-Padron RI, Gomes SA, Hare JM and Shehadeh LA: miR-30e
targets IGF2-regulated osteogenesis in bone marrow-derived
mesenchymal stem cells,aortic smooth muscle cells, and ApoE 2/2
mice. Cardiovasc Res. 106:131–142. 2015.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Du Y, Gao C, Liu Z, Wang L, Liu B, He F,
Zhang T, Wang Y, Wang X, Xu M, et al: Upregulation of a disintegrin
and metalloproteinase with thrombospondin motifs-7 by miR-29
repression mediates vascular smooth muscle calcification.
Arterioscler Thromb Vasc Biol. 32:2580–2588. 2012.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Wang XY, Zhang XZ, Li F and Ji QR:
MiR-128-3p accelerates cardiovascular calcification and insulin
resistance through ISL1-dependent Wnt pathway in type 2 diabetes
mellitus rats. J Cell Physiol. 234:4997–5010. 2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Lin X, Zhan JK, Zhong JY, Wang YJ, Wang Y,
Li S, He JY, Tan P, Chen YY, Liu XB, et al:
lncRNA-ES3/miR-34c-5p/BMF axis is involved in regulating
high-glucose-induced calcification/senescence of VSMCs. Aging
(Albany NY). 11:523–535. 2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Jeong G, Kwon DH, Shin S, Choe N, Ryu J,
Lim YH, Kim J, Park WJ, Kook H and Kim YK: Long noncoding RNAs in
vascular smooth muscle cells regulate vascular calcification. Sci
Rep. 9(5848)2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Alves RD, Eijken M, van de Peppel J and
van Leeuwen JP: Calcifying vascular smooth muscle cells and
osteoblasts: Independent cell types exhibiting extracellular matrix
and biomineralization-related mimicries. BMC Genomics.
15(965)2014.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Karagoz K, Sevimoglu T and Arga KY:
Integration of multiple biological features yields high confidence
human protein interactome. J Theor Biol. 403:85–96. 2016.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Tong Z, Cui Q, Wang J and Zhou Y: TransmiR
v2.0: An updated transcription factor-microRNA regulation database.
Nucleic Acids Res. 47:D253–D258. 2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Cheng L, Wang P, Tian R, Wang S, Guo Q,
Luo M, Zhou W, Liu G, Jiang H and Jiang Q: LncRNA2Target v2.0: A
comprehensive database for target genes of lncRNAs in human and
mouse. Nucleic Acids Res. 47:D140–D144. 2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Method. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Ohukainen P, Syväranta S, Näpänkangas J,
Rajamäki K, Taskinen P, Peltonen T, Helske-Suihko S, Kovanen PT,
Ruskoaho H and Rysä J: MicroRNA-125b and chemokine CCL4 expression
are associated with calcific aortic valve disease. Ann Med.
47:423–429. 2015.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Matsumoto K, Maniwa T, Tanaka T, Satoh K,
Okunishi H and Oda T: Proteomic analysis of calcified abdominal and
thoracic aortic aneurysms. Int J Mol Med. 30:417–429.
2012.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Tanaka K, Sata M, Fukuda D, Suematsu Y,
Motomura N, Takamoto S, Hirata Y and Nagai R: Age-associated aortic
stenosis in apolipoprotein E-deficient mice. J Am Coll Cardiol.
46:134–141. 2005.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Hodroge A, Trécherel E, Cornu M, Darwiche
W, Mansour A, Ait-Mohand K, Verissimo T, Gomila C, Schembri C, Da
Nascimento S, et al: Oligogalacturonic acid inhibits vascular
calcification by two mechanisms: inhibition of vascular smooth
muscle cell osteogenic conversion and interaction with collagen.
Arterioscler Thromb Vasc Biol. 37:1391–1401. 2017.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Shanahan CM, Cary NR, Salisbury JR,
Proudfoot D, Weissberg PL and Edmonds ME: Medial localization of
mineralization-regulating proteins in association with Mönckeberg's
sclerosis. Circulation. 100:2168–2176. 1999.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Tyson KL, Reynolds JL, McNair R, Zhang Q,
Weissberg PL and Shanahan CM: Osteo/chondrocytic transcription
factors and their target genes exhibit distinct patterns of
expression in human arterial calcification. Arterioscler Thromb
Vasc Biol. 23:489–494. 2003.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Watson KE, Parhami F, Shin V and Demer LL:
Fibronectin and collagen I matrixes promote calcification of
vascular cells in vitro, whereas collagen IV matrix is inhibitory.
Arterioscler Thromb Vasc Biol. 18:1964–1971. 1998.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Freise C, Bobb V and Querfeld U: Collagen
XIV and a related recombinant fragment protect human vascular
smooth muscle cells from calcium-/phosphate-induced
osteochondrocytic transdifferentiation. Exp Cell Res. 358:242–252.
2017.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Lei Y, Sinha A, Nosoudi N, Grover A and
Vyavahare N: Hydroxyapatite and calcified elastin induce
osteoblast-like differentiation in rat aortic smooth muscle cells.
Exp Cell Res. 323:198–208. 2014.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Sinha A and Vyavahare NR: High-glucose
levels and elastin degradation products accelerate osteogenesis in
vascular smooth muscle cells. Diabet Vasc Dis Res. 10:410–419.
2013.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Urry DW: Neutral sites for calcium ion
binding to elastin and collagen: A charge neutralization theory for
calcification and its relationship to atherosclerosis. Proc Natl
Acad Sci USA. 68:810–814. 1971.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Wang L, Zheng J, Du Y, Huang Y, Li J, Liu
B, Liu CJ, Zhu Y, Gao Y, Xu Q, et al: Cartilage oligomeric matrix
protein maintains the contractile phenotype of vascular smooth
muscle cells by interacting with alpha(7)beta(1) integrin. Circ
Res. 106:514–525. 2010.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Du Y, Wang Y, Wang L, Liu B, Tian Q, Liu
CJ, Zhang T, Xu Q, Zhu Y, Ake O, et al: Cartilage oligomeric matrix
protein inhibits vascular smooth muscle calcification by
interacting with bone morphogenetic protein-2. Circ Res.
108:917–928. 2011.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Muller J, Bolomsky A, Dubois S, Duray E,
Stangelberger K, Plougonven E, Lejeune M, Léonard A, Marty C,
Hempel U, et al: Maternal embryonic leucine zipper kinase inhibitor
OTSSP167 has preclinical activity in multiple myeloma bone disease.
Haematologica. 103:1359–1368. 2018.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Kim SH, Joshi K, Ezhilarasan R, Myers TR,
Siu J, Gu C, Nakano-Okuno M, Taylor D, Minata M, Sulman EP, et al:
EZH2 protects glioma stem cells from radiation-induced cell death
in a MELK/FOXM1-dependent manner. Stem Cell Reports. 4:226–238.
2015.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Fan T, He J, Yin Y, Wen K, Kang Y, Zhao H,
Chen S and Li X: Dioscin inhibits intimal hyperplasia in rat
carotid artery balloon injury model through inhibition of the
MAPK-FoxM1 pathway. Eur J Pharmacol. 854:213–223. 2019.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Dudakovic A, Camilleri ET, Xu F, Riester
SM, McGee-Lawrence ME, Bradley EW, Paradise CR, Lewallen EA, Thaler
R, Deyle DR, et al: Epigenetic control of skeletal development by
the histone methyltransferase Ezh2. J Biol Chem. 290:27604–27617.
2015.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Adamik J, Jin S, Sun Q, Zhang P, Weiss KR,
Anderson JL, Silbermann R, Roodman GD and Galson DL: EZH2 or HDAC1
inhibition reverses multiple myeloma-induced epigenetic suppression
of osteoblast differentiation. Mol Cancer Res. 15:405–417.
2017.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Zhang H, Pan Y, Zheng L, Choe C, Lindgren
B, Jensen ED, Westendorf JJ, Cheng L and Huang H: FOXO1 inhibits
Runx2 transcriptional activity and prostate cancer cell migration
and invasion. Cancer Res. 71:3257–3267. 2011.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Sun Y, Byon CH, Yuan K, Chen J, Mao X,
Heath JM, Javed A, Zhang K, Anderson PG and Chen Y: Smooth muscle
cell-specific Runx2 deficiency inhibits vascular calcification.
Circ Res. 111:543–552. 2012.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Deng L, Huang L, Sun Y, Heath JM, Wu H and
Chen Y: Inhibition of FOXO1/3 promotes vascular calcification.
Arterioscler Thromb Vasc Biol. 35:175–183. 2015.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Xu X, Ji S, Li W, Yi B, Li H, Zhang H and
Ma W: LncRNA H19 promotes the differentiation of bovine skeletal
muscle satellite cells by suppressing Sirt1/FoxO1. Cell Mol Biol
Lett. 22(10)2017.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Hadji F, Boulanger MC, Guay SP, Gaudreault
N, Amellah S, Mkannez G, Bouchareb R, Marchand JT, Nsaibia MJ,
Guauque-Olarte S, et al: Altered DNA methylation of long noncoding
RNA H19 in calcific aortic valve disease promotes mineralization by
silencing NOTCH1. Circulation. 134:1848–1862. 2016.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Liang WC, Fu WM, Wang YB, Sun YX, Xu LL,
Wong CW, Chan KM, Li G, Waye MM and Zhang JF: H19 activates Wnt
signaling and promotes osteoblast differentiation by functioning as
a competing endogenous RNA. Sci Rep. 6(20121)2016.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Tang Z, Gong Z and Sun X: LncRNA DANCR
involved osteolysis after total hip arthroplasty by regulating
FOXO1 expression to inhibit osteoblast differentiation. J Biomed
Sci. 25(4)2018.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Zhu L and Xu PC: Downregulated LncRNA-ANCR
promotes osteoblast differentiation by targeting EZH2 and
regulating Runx2 expression. Biochem Biophys Res Commun.
432:612–617. 2013.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Jia Q, Jiang W and Ni L: Down-regulated
non-coding RNA (lncRNA-ANCR) promotes osteogenic differentiation of
periodontal ligament stem cells. Arch Oral Biol. 60:234–241.
2015.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Massy ZA, Metzinger-Le Meuth V and
Metzinger L: MicroRNAs are associated with uremic toxicity,
cardiovascular calcification, and disease. Contrib Nephrol.
189:160–168. 2017.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Lambertini E, Lisignoli G, Torreggiani E,
Manferdini C, Gabusi E, Franceschetti T, Penolazzi L, Gambari R,
Facchini A and Piva R: Slug gene expression supports human
osteoblast maturation. Cell Mol Life Sci. 66:3641–3653.
2009.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Piva R, Manferdini C, Lambertini E,
Torreggiani E, Penolazzi L, Gambari R, Pastore A, Pelucchi S,
Gabusi E, Piacentini A, et al: Slug contributes to the regulation
of CXCL12 expression in human osteoblasts. Exp Cell Res.
317:1159–1168. 2011.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Sanchez-Duffhues G, de Vinuesa AG,
Lindeman JH, Mulder-Stapel A, DeRuiter MC, Van Munsteren C, Goumans
MJ, Hierck BP and Ten Dijke P: SLUG is expressed in endothelial
cells lacking primary cilia to promote cellular calcification.
Arterioscler Thromb Vasc Biol. 35:616–627. 2015.PubMed/NCBI View Article : Google Scholar
|