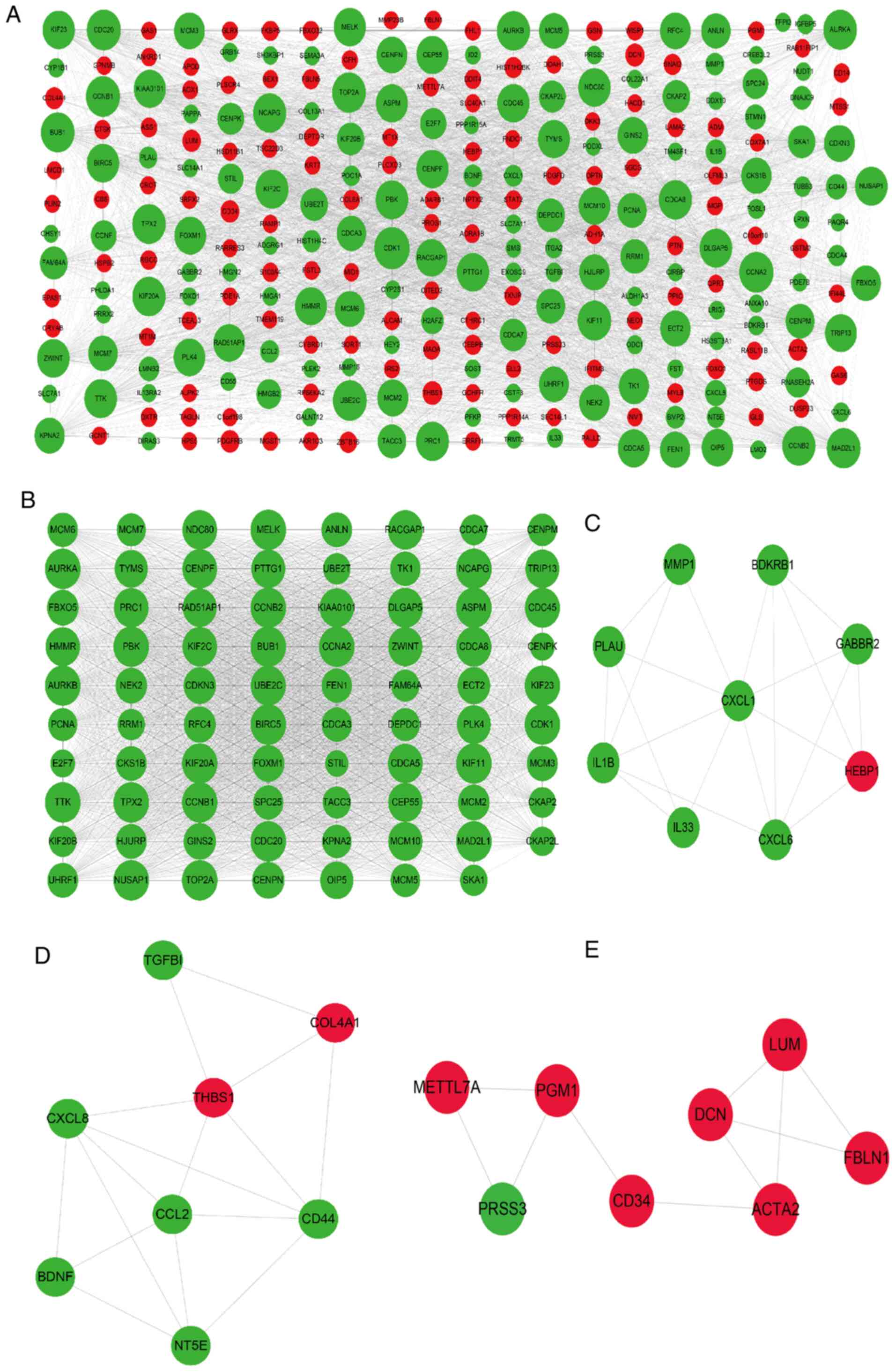
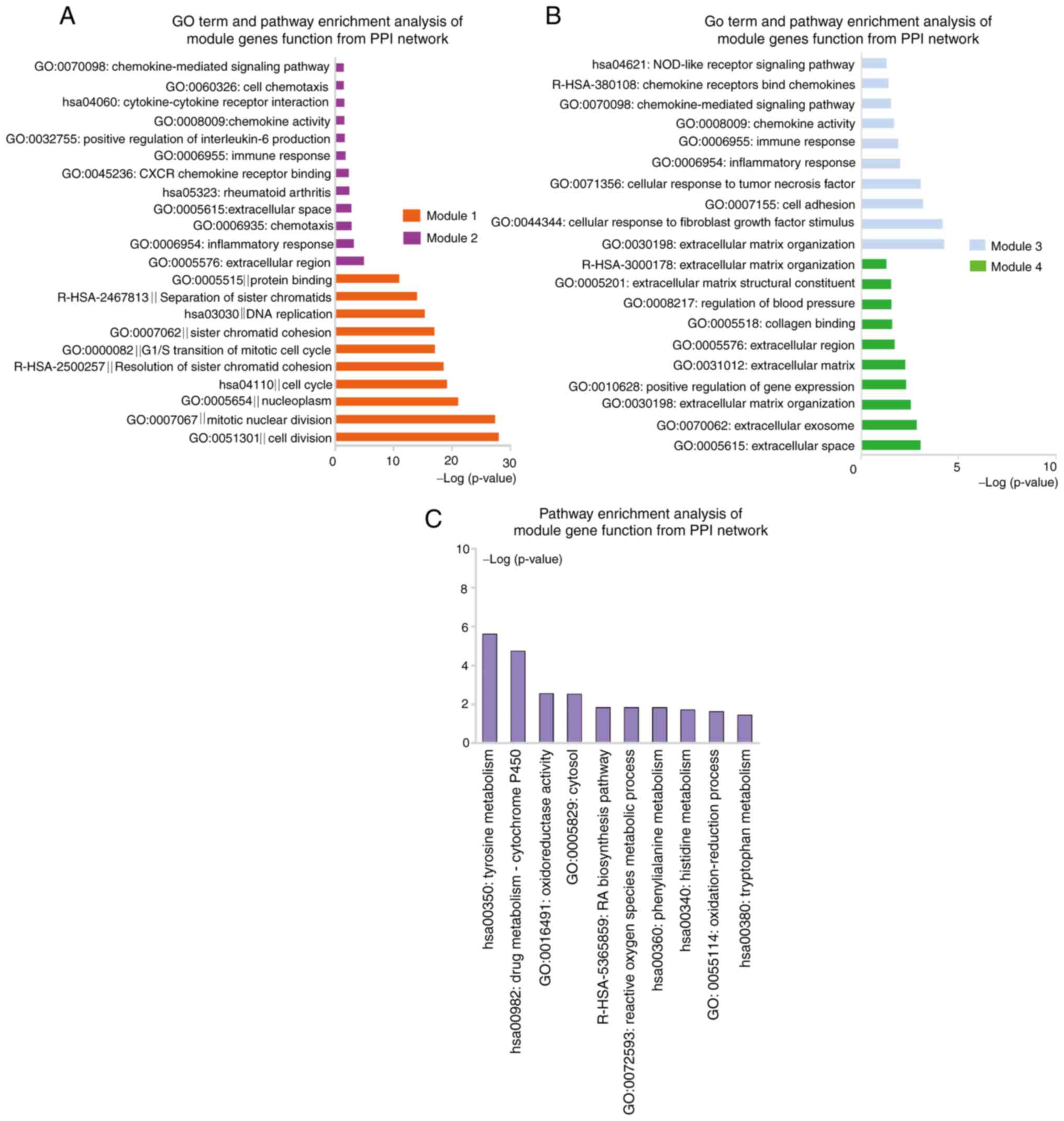
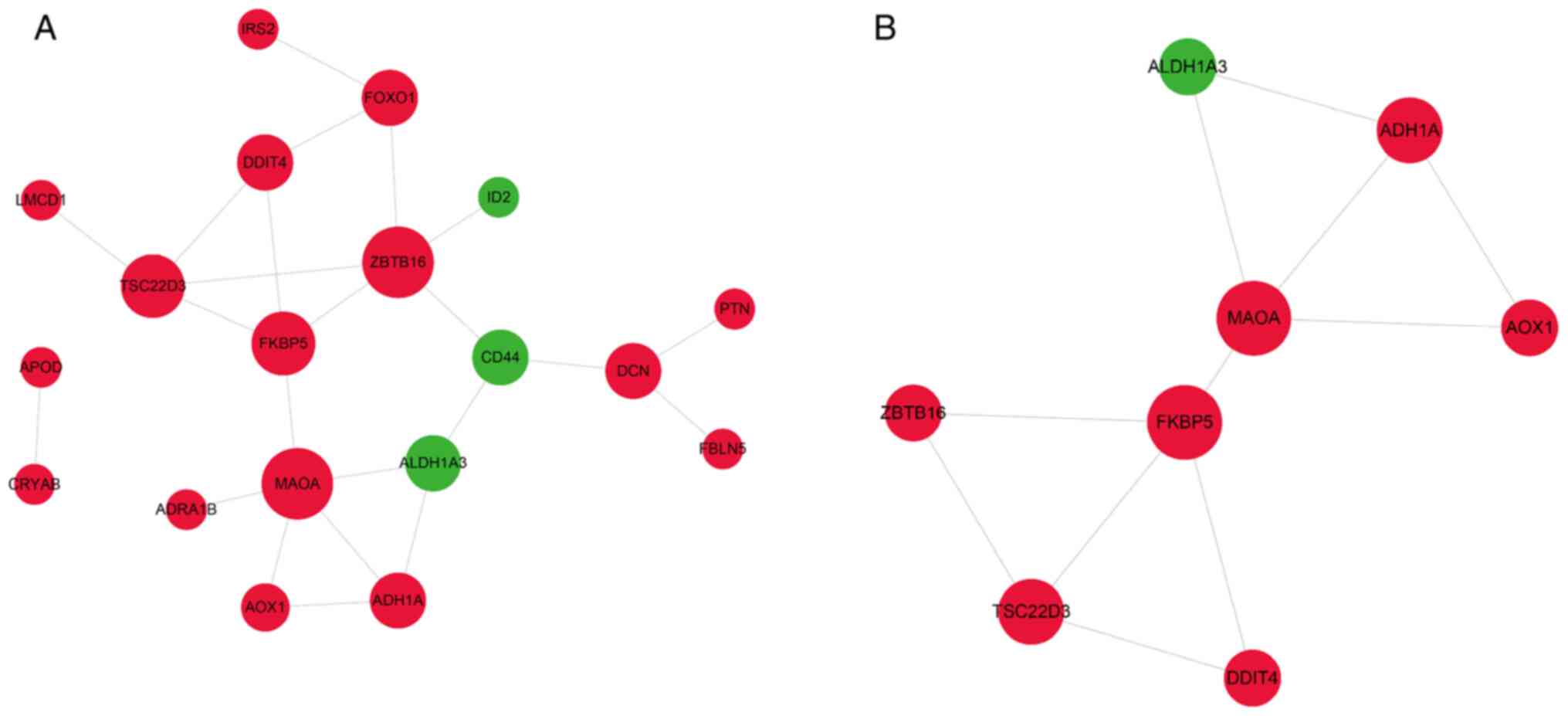
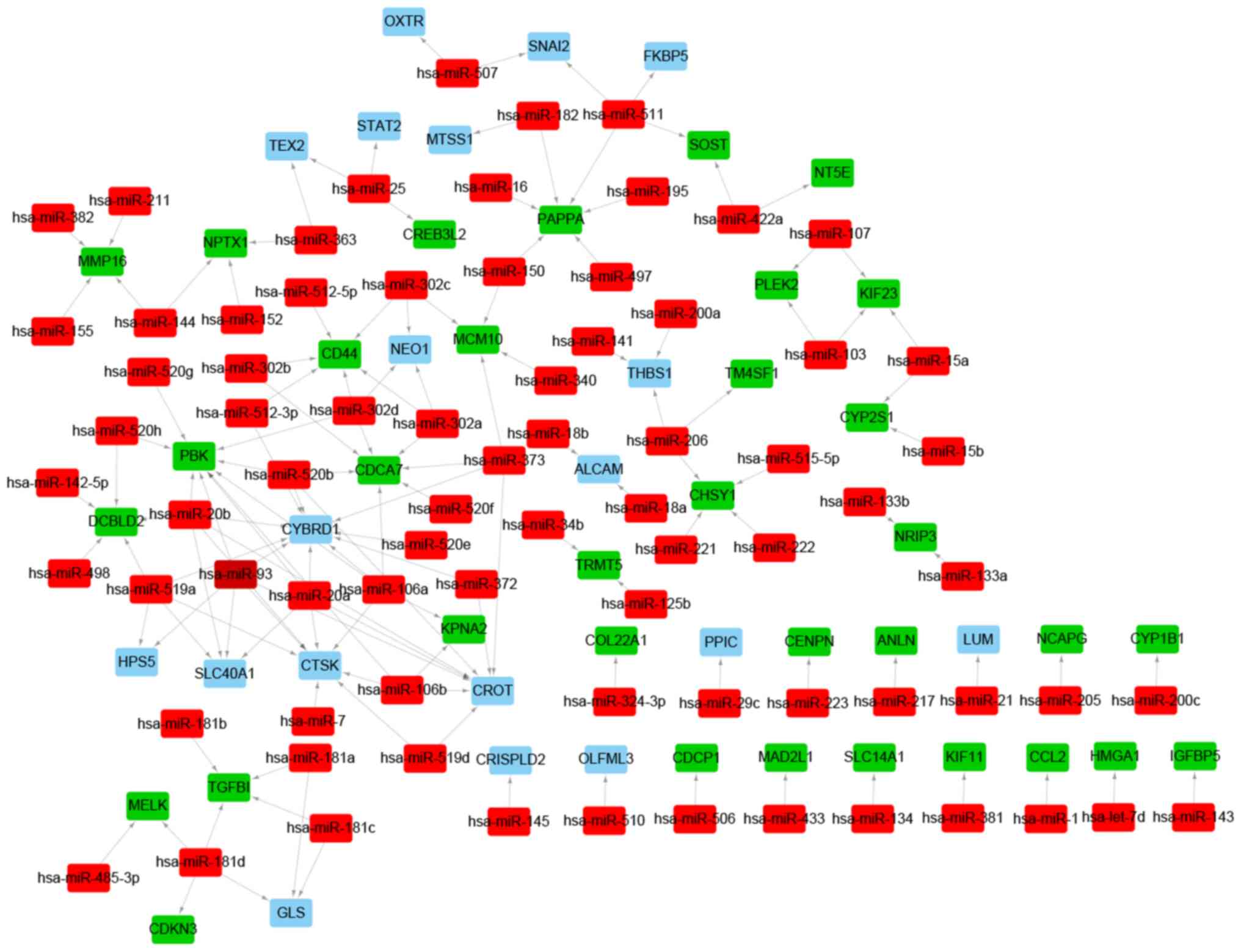
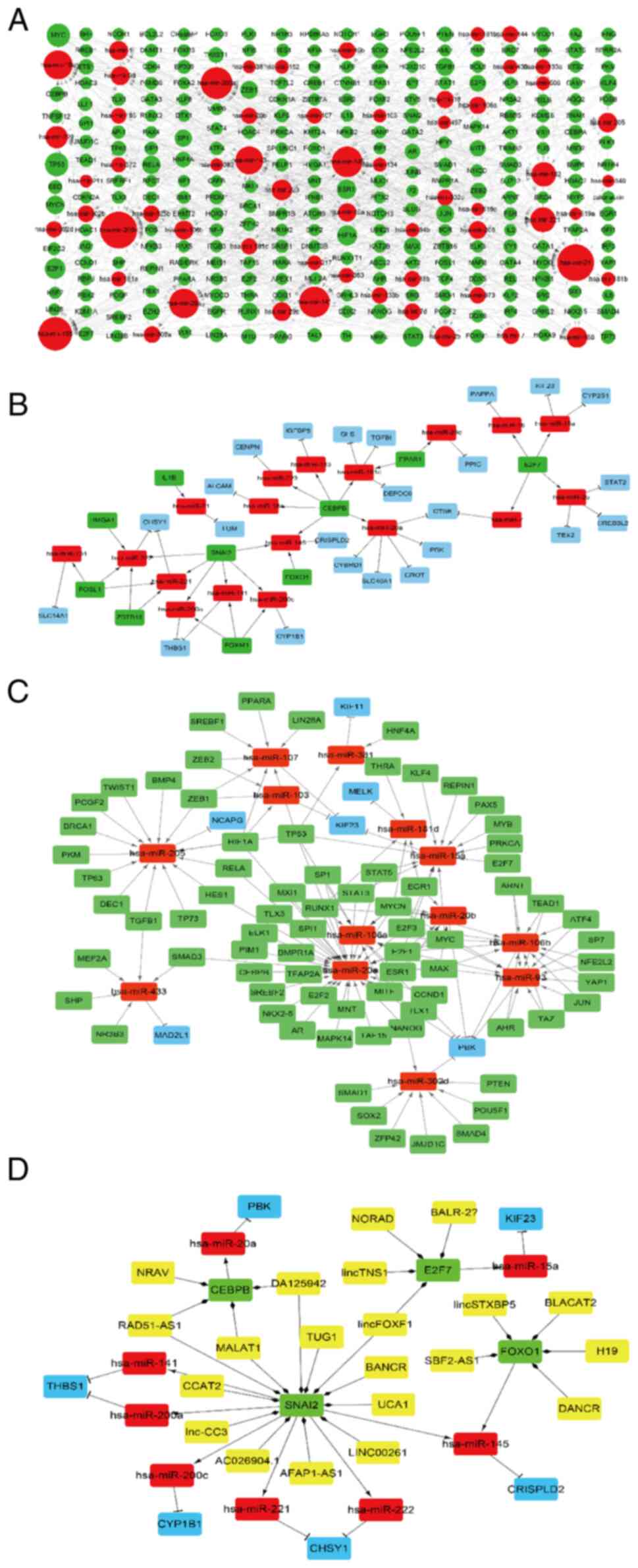
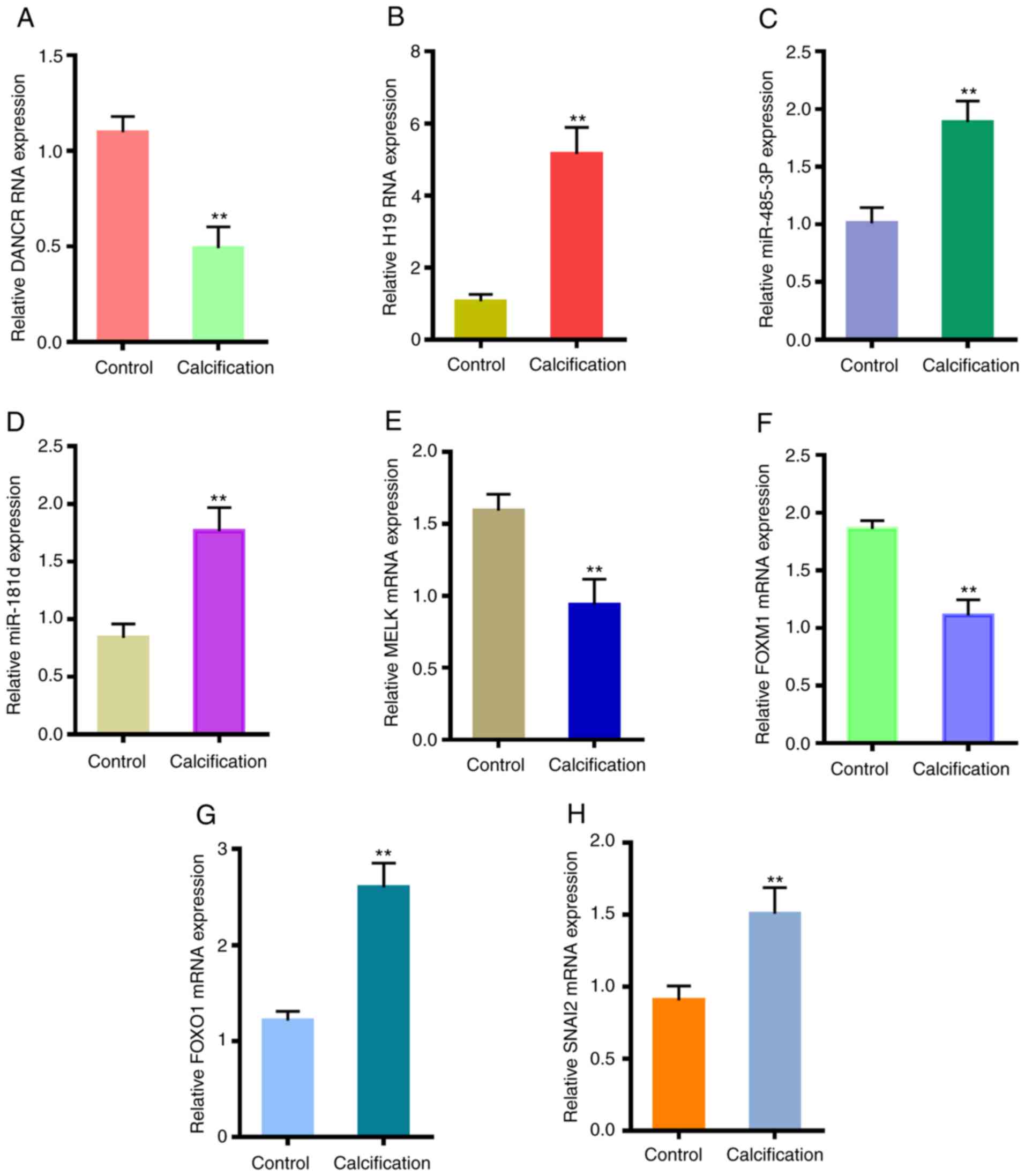
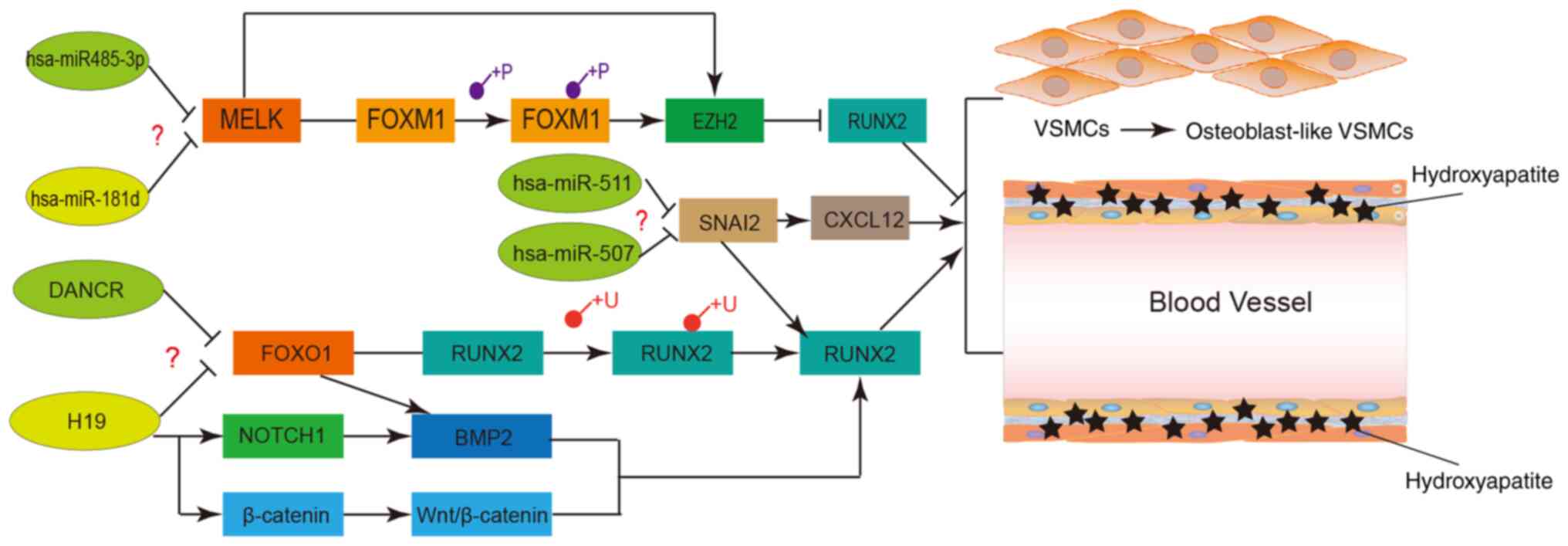
|
1
|
Lanzer P, Boehm M, Sorribas V, Thiriet M,
Janzen J, Zeller T, St Hilaire C and Shanahan C: Medial vascular
calcification revisited: Review and perspectives. Eur Heart J.
35:1515–1525. 2014.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Kovacic JC and Fuster V: Vascular
calcification, diabetes, and cardiovascular disease: Connecting the
dots. JACC Cardiovasc Imaging. 5:367–369. 2012.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Chen NX and Moe SM: Pathophysiology of
vascular calcification. Curr Osteoporos Rep. 13:372–380.
2015.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Leopold JA: Vascular calcification:
Mechanisms of vascular smooth muscle cell calcification. Trends
Cardiovasc Med. 25:267–274. 2015.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Block GA, Spiegel DM, Ehrlich J, Mehta R,
Lindbergh J, Dreisbach A and Raggi P: Effects of sevelamer and
calcium on coronary artery calcification in patients new to
hemodialysis. Kidney Int. 68:1815–1824. 2005.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Blomberg BA, de Jong PA, Thomassen A, Lam
MGE, Vach W, Olsen MH, Mali WPTM, Narula J, Alavi A and
Høilund-Carlsen PF: Thoracic aorta calcification but not
inflammation is associated with increased cardiovascular disease
risk: Results of the CAMONA study. Eur J Nucl Med Mol Imaging.
44:249–258. 2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Bastos Gonçalves F, Voûte MT, Hoeks SE,
Chonchol MB, Boersma EE, Stolker RJ and Verhagen HJ: Calcification
of the abdominal aorta as an independent predictor of
cardiovascular events: A meta-analysis. Heart. 98:988–994.
2012.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Bild DE, Detrano R, Peterson D, Guerci A,
Liu K, Shahar E, Ouyang P, Jackson S and Saad MF: Ethnic
differences in coronary calcification: The Multi-Ethnic Study of
Atherosclerosis (MESA). Circulation. 111:1313–1320. 2005.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Zhang MJ, Zhou Y, Chen L, Wang YQ, Wang X,
Pi Y, Gao CY, Li JC and Zhang LL: An overview of potential
molecular mechanisms involved in VSMC phenotypic modulation.
Histochem Cell Biol. 145:119–130. 2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Gomez D and Owens GK: Smooth muscle cell
phenotypic switching in atherosclerosis. Cardiovasc Res.
95:156–164. 2012.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Kapustin AN and Shanahan CM: Calcium
regulation of vascular smooth muscle cell-derived matrix vesicles.
Trends Cardiovasc Med. 22:133–137. 2012.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Yao RW, Wang Y and Chen LL: Cellular
functions of long noncoding RNAs. Nat Cell Biol. 21:542–551.
2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Bar C, Chatterjee S and Thum T: Long
noncoding RNAs in cardiovascular pathology, diagnosis, and therapy.
Circulation. 134:1484–1499. 2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Barwari T, Joshi A and Mayr M: MicroRNAs
in cardiovascular disease. J Am Coll Cardiol. 68:2577–2584.
2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Haemmig S, Simion V, Yang D, Deng Y and
Feinberg MW: Long noncoding RNAs in cardiovascular disease,
diagnosis, and therapy. Curr Opin Cardiol. 32:776–783.
2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Jiang W, Zhang Z, Yang H, Lin Q, Han C and
Qin X: The involvement of miR-29b-3p in arterial calcification by
targeting matrix metalloproteinase-2. Biomed Res Int.
2017(6713606)2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Cui RR, Li SJ, Liu LJ, Yi L, Liang QH, Zhu
X, Liu GY, Liu Y, Wu SS, Liao XB, et al: MicroRNA-204 regulates
vascular smooth muscle cell calcification in vitro and in vivo.
Cardiovasc Res. 96:320–329. 2012.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Ding W, Li J, Singh J, Alif R,
Vazquez-Padron RI, Gomes SA, Hare JM and Shehadeh LA: miR-30e
targets IGF2-regulated osteogenesis in bone marrow-derived
mesenchymal stem cells,aortic smooth muscle cells, and ApoE 2/2
mice. Cardiovasc Res. 106:131–142. 2015.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Du Y, Gao C, Liu Z, Wang L, Liu B, He F,
Zhang T, Wang Y, Wang X, Xu M, et al: Upregulation of a disintegrin
and metalloproteinase with thrombospondin motifs-7 by miR-29
repression mediates vascular smooth muscle calcification.
Arterioscler Thromb Vasc Biol. 32:2580–2588. 2012.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Wang XY, Zhang XZ, Li F and Ji QR:
MiR-128-3p accelerates cardiovascular calcification and insulin
resistance through ISL1-dependent Wnt pathway in type 2 diabetes
mellitus rats. J Cell Physiol. 234:4997–5010. 2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Lin X, Zhan JK, Zhong JY, Wang YJ, Wang Y,
Li S, He JY, Tan P, Chen YY, Liu XB, et al:
lncRNA-ES3/miR-34c-5p/BMF axis is involved in regulating
high-glucose-induced calcification/senescence of VSMCs. Aging
(Albany NY). 11:523–535. 2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Jeong G, Kwon DH, Shin S, Choe N, Ryu J,
Lim YH, Kim J, Park WJ, Kook H and Kim YK: Long noncoding RNAs in
vascular smooth muscle cells regulate vascular calcification. Sci
Rep. 9(5848)2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Alves RD, Eijken M, van de Peppel J and
van Leeuwen JP: Calcifying vascular smooth muscle cells and
osteoblasts: Independent cell types exhibiting extracellular matrix
and biomineralization-related mimicries. BMC Genomics.
15(965)2014.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Karagoz K, Sevimoglu T and Arga KY:
Integration of multiple biological features yields high confidence
human protein interactome. J Theor Biol. 403:85–96. 2016.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Tong Z, Cui Q, Wang J and Zhou Y: TransmiR
v2.0: An updated transcription factor-microRNA regulation database.
Nucleic Acids Res. 47:D253–D258. 2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Cheng L, Wang P, Tian R, Wang S, Guo Q,
Luo M, Zhou W, Liu G, Jiang H and Jiang Q: LncRNA2Target v2.0: A
comprehensive database for target genes of lncRNAs in human and
mouse. Nucleic Acids Res. 47:D140–D144. 2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Method. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Ohukainen P, Syväranta S, Näpänkangas J,
Rajamäki K, Taskinen P, Peltonen T, Helske-Suihko S, Kovanen PT,
Ruskoaho H and Rysä J: MicroRNA-125b and chemokine CCL4 expression
are associated with calcific aortic valve disease. Ann Med.
47:423–429. 2015.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Matsumoto K, Maniwa T, Tanaka T, Satoh K,
Okunishi H and Oda T: Proteomic analysis of calcified abdominal and
thoracic aortic aneurysms. Int J Mol Med. 30:417–429.
2012.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Tanaka K, Sata M, Fukuda D, Suematsu Y,
Motomura N, Takamoto S, Hirata Y and Nagai R: Age-associated aortic
stenosis in apolipoprotein E-deficient mice. J Am Coll Cardiol.
46:134–141. 2005.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Hodroge A, Trécherel E, Cornu M, Darwiche
W, Mansour A, Ait-Mohand K, Verissimo T, Gomila C, Schembri C, Da
Nascimento S, et al: Oligogalacturonic acid inhibits vascular
calcification by two mechanisms: inhibition of vascular smooth
muscle cell osteogenic conversion and interaction with collagen.
Arterioscler Thromb Vasc Biol. 37:1391–1401. 2017.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Shanahan CM, Cary NR, Salisbury JR,
Proudfoot D, Weissberg PL and Edmonds ME: Medial localization of
mineralization-regulating proteins in association with Mönckeberg's
sclerosis. Circulation. 100:2168–2176. 1999.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Tyson KL, Reynolds JL, McNair R, Zhang Q,
Weissberg PL and Shanahan CM: Osteo/chondrocytic transcription
factors and their target genes exhibit distinct patterns of
expression in human arterial calcification. Arterioscler Thromb
Vasc Biol. 23:489–494. 2003.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Watson KE, Parhami F, Shin V and Demer LL:
Fibronectin and collagen I matrixes promote calcification of
vascular cells in vitro, whereas collagen IV matrix is inhibitory.
Arterioscler Thromb Vasc Biol. 18:1964–1971. 1998.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Freise C, Bobb V and Querfeld U: Collagen
XIV and a related recombinant fragment protect human vascular
smooth muscle cells from calcium-/phosphate-induced
osteochondrocytic transdifferentiation. Exp Cell Res. 358:242–252.
2017.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Lei Y, Sinha A, Nosoudi N, Grover A and
Vyavahare N: Hydroxyapatite and calcified elastin induce
osteoblast-like differentiation in rat aortic smooth muscle cells.
Exp Cell Res. 323:198–208. 2014.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Sinha A and Vyavahare NR: High-glucose
levels and elastin degradation products accelerate osteogenesis in
vascular smooth muscle cells. Diabet Vasc Dis Res. 10:410–419.
2013.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Urry DW: Neutral sites for calcium ion
binding to elastin and collagen: A charge neutralization theory for
calcification and its relationship to atherosclerosis. Proc Natl
Acad Sci USA. 68:810–814. 1971.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Wang L, Zheng J, Du Y, Huang Y, Li J, Liu
B, Liu CJ, Zhu Y, Gao Y, Xu Q, et al: Cartilage oligomeric matrix
protein maintains the contractile phenotype of vascular smooth
muscle cells by interacting with alpha(7)beta(1) integrin. Circ
Res. 106:514–525. 2010.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Du Y, Wang Y, Wang L, Liu B, Tian Q, Liu
CJ, Zhang T, Xu Q, Zhu Y, Ake O, et al: Cartilage oligomeric matrix
protein inhibits vascular smooth muscle calcification by
interacting with bone morphogenetic protein-2. Circ Res.
108:917–928. 2011.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Muller J, Bolomsky A, Dubois S, Duray E,
Stangelberger K, Plougonven E, Lejeune M, Léonard A, Marty C,
Hempel U, et al: Maternal embryonic leucine zipper kinase inhibitor
OTSSP167 has preclinical activity in multiple myeloma bone disease.
Haematologica. 103:1359–1368. 2018.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Kim SH, Joshi K, Ezhilarasan R, Myers TR,
Siu J, Gu C, Nakano-Okuno M, Taylor D, Minata M, Sulman EP, et al:
EZH2 protects glioma stem cells from radiation-induced cell death
in a MELK/FOXM1-dependent manner. Stem Cell Reports. 4:226–238.
2015.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Fan T, He J, Yin Y, Wen K, Kang Y, Zhao H,
Chen S and Li X: Dioscin inhibits intimal hyperplasia in rat
carotid artery balloon injury model through inhibition of the
MAPK-FoxM1 pathway. Eur J Pharmacol. 854:213–223. 2019.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Dudakovic A, Camilleri ET, Xu F, Riester
SM, McGee-Lawrence ME, Bradley EW, Paradise CR, Lewallen EA, Thaler
R, Deyle DR, et al: Epigenetic control of skeletal development by
the histone methyltransferase Ezh2. J Biol Chem. 290:27604–27617.
2015.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Adamik J, Jin S, Sun Q, Zhang P, Weiss KR,
Anderson JL, Silbermann R, Roodman GD and Galson DL: EZH2 or HDAC1
inhibition reverses multiple myeloma-induced epigenetic suppression
of osteoblast differentiation. Mol Cancer Res. 15:405–417.
2017.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Zhang H, Pan Y, Zheng L, Choe C, Lindgren
B, Jensen ED, Westendorf JJ, Cheng L and Huang H: FOXO1 inhibits
Runx2 transcriptional activity and prostate cancer cell migration
and invasion. Cancer Res. 71:3257–3267. 2011.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Sun Y, Byon CH, Yuan K, Chen J, Mao X,
Heath JM, Javed A, Zhang K, Anderson PG and Chen Y: Smooth muscle
cell-specific Runx2 deficiency inhibits vascular calcification.
Circ Res. 111:543–552. 2012.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Deng L, Huang L, Sun Y, Heath JM, Wu H and
Chen Y: Inhibition of FOXO1/3 promotes vascular calcification.
Arterioscler Thromb Vasc Biol. 35:175–183. 2015.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Xu X, Ji S, Li W, Yi B, Li H, Zhang H and
Ma W: LncRNA H19 promotes the differentiation of bovine skeletal
muscle satellite cells by suppressing Sirt1/FoxO1. Cell Mol Biol
Lett. 22(10)2017.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Hadji F, Boulanger MC, Guay SP, Gaudreault
N, Amellah S, Mkannez G, Bouchareb R, Marchand JT, Nsaibia MJ,
Guauque-Olarte S, et al: Altered DNA methylation of long noncoding
RNA H19 in calcific aortic valve disease promotes mineralization by
silencing NOTCH1. Circulation. 134:1848–1862. 2016.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Liang WC, Fu WM, Wang YB, Sun YX, Xu LL,
Wong CW, Chan KM, Li G, Waye MM and Zhang JF: H19 activates Wnt
signaling and promotes osteoblast differentiation by functioning as
a competing endogenous RNA. Sci Rep. 6(20121)2016.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Tang Z, Gong Z and Sun X: LncRNA DANCR
involved osteolysis after total hip arthroplasty by regulating
FOXO1 expression to inhibit osteoblast differentiation. J Biomed
Sci. 25(4)2018.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Zhu L and Xu PC: Downregulated LncRNA-ANCR
promotes osteoblast differentiation by targeting EZH2 and
regulating Runx2 expression. Biochem Biophys Res Commun.
432:612–617. 2013.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Jia Q, Jiang W and Ni L: Down-regulated
non-coding RNA (lncRNA-ANCR) promotes osteogenic differentiation of
periodontal ligament stem cells. Arch Oral Biol. 60:234–241.
2015.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Massy ZA, Metzinger-Le Meuth V and
Metzinger L: MicroRNAs are associated with uremic toxicity,
cardiovascular calcification, and disease. Contrib Nephrol.
189:160–168. 2017.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Lambertini E, Lisignoli G, Torreggiani E,
Manferdini C, Gabusi E, Franceschetti T, Penolazzi L, Gambari R,
Facchini A and Piva R: Slug gene expression supports human
osteoblast maturation. Cell Mol Life Sci. 66:3641–3653.
2009.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Piva R, Manferdini C, Lambertini E,
Torreggiani E, Penolazzi L, Gambari R, Pastore A, Pelucchi S,
Gabusi E, Piacentini A, et al: Slug contributes to the regulation
of CXCL12 expression in human osteoblasts. Exp Cell Res.
317:1159–1168. 2011.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Sanchez-Duffhues G, de Vinuesa AG,
Lindeman JH, Mulder-Stapel A, DeRuiter MC, Van Munsteren C, Goumans
MJ, Hierck BP and Ten Dijke P: SLUG is expressed in endothelial
cells lacking primary cilia to promote cellular calcification.
Arterioscler Thromb Vasc Biol. 35:616–627. 2015.PubMed/NCBI View Article : Google Scholar
|