Introduction
Apoptosis and necroptosis are two common forms of
programmed cell death that play essential roles in development and
maintaining tissue homeostasis (1). Defects in apoptosis and necroptosis
are strictly connected to the pathogenesis of various human
diseases (2). Apoptosis can be
induced by proteolytic activation of the caspase protease family
(3). Upon caspase activation and
subsequent cleavage of intracellular substrates lead cells to break
into small membrane-wrapped vesicles known as apoptotic bodies
(4). On the other hand,
necroptosis is a caspase-independent form of cell death. Like
apoptosis, it is programmed one and induced by TNF and Fas ligand
but results in organelle swelling and cytoplasmic membrane
breakdown (5,6).
Apoptosis-resistant E3 ubiquitin protein ligase 1
(AREL1) was initially identified as a suppressor of p53-induced
apoptosis (7). However, AREL1
functions as a general inhibitor of apoptosis in p53-positive and
deficient tumor cells because it inhibited apoptosis induced by
various stimuli such as staurosporine, etoposide, and doxorubicin
(7). AREL1 did not affect the
cytosolic release of mitochondrial pro-apoptotic proteins such as
cytochrome c but inhibited caspase-3 activation (7). AREL1 encodes HECT-family E3 ubiquitin
ligase and ubiquitinates IAP antagonists, such as SMAC, HtrA2, and
ARTS, released into the cytosol from mitochondria upon apoptotic
stimulation (7).
This study reports a new target protein of AREL1 E3
ubiquitin ligase and necroptosis-inhibitory function of AREL1. We
found that AREL1 ubiquitinates and promotes ubiquitin-dependent
degradation of Metaxin 2 (MTX2), localized in mitochondria's outer
membrane. However, it has been reported that Metaxin family
proteins are involved in TNF-induced necroptosis. These led us to
find the necroptosis inhibitory function of AREL1 in association
with MTX2.
Materials and methods
Yeast two-hybrid screen
The yeast cell expressing LexA-HECT (Homologous to
E6-AP Carboxyl Terminus, aa. 454-823 of AREL1) was transformed with
the HeLa cell cDNA library fused to the GAL4-AD. Positive clones
were initially selected by their ability to grow on His-deficient
media and produce β-galactosidase activity, as previously
demonstrated (8).
Chemical reagents and Plasmid
construction
Cycloheximide (C-7698), MG132 (Z-Leu-Leu-Leu-al,
C-2211), human TNFα (T-0157), and mouse TNFα (T-7539) were
purchased from Sigma. Blasticidin (R210-01) was purchased from
Invitrogen. Human Metaxin 1 (MTX1) cDNA was cloned by RT-PCR using
total RNA from HeLa cells with a pair of primers: the sense primer
was 5'-CGGAATTCAACATGCTG CTCGG-3' (the EcoRI site is
underlined), and the antisense primer was
5'-CCGCTCGAGAAATCATTCCTCTTCATC-3' (the XhoI site is
underlined). The cDNA of MTX1 was confirmed by DNA sequencing and
cloned into the N-terminally Flag-tagged vector at
EcoRI/XhoI sites. Flag-, GFP-, and GST-tagged MTX2
were generated by subcloning the full-length cDNA of MTX2 into
EcoRI/XhoI sites of pCMV-Tag2B,
EcoRI/ApaI sites of the pEGFP-C2, and
EcoR/XhoI sites of the pGEX-5X-1, respectively. The
MTX2 deletion constructs (1-112, 113-264 aa) were made by PCR
(Forward primer: 5'-GTCGAATTCATGTCTCTAGTGGC GGAAG-3', F-internal
primer: 5'-CGCGAATTCAAAGCTTA CATGGAATTAG-3', Reverse primer:
5'-ACAGTCGACCTAT GACAGCCTGCCTTTAC-3', R-internal primer: 5'-CGCGTC
GACGTAAGCTTTCATTTCTGC-3') and cloned into the
EcoRI/SalI sites of pCMV-Tag2B. AREL1 knockdown was
applied as previously described (7). siRNA oligonucleotides corresponding
to the sequences of AREL1 (5'-AATTGGTC CCTGAGAACCTTT-3') were
generated and used for transfection with Lipofectamine RNAiMAX
(Invitrogen). Scrambled siRNA was obtained from Proligo LLC.
Flag-HtrA2 was previously described (7).
Cell culture and transfection
Human 293T (KCLB, 21573), DLD1 (KCLB, 10221), H1299
(KCLB, 91299), and mouse L929 cells (KCLB, 10001) were cultured in
Dulbecco's modified Eagle's medium (DMEM, Gibco) supplemented with
10% fetal bovine serum (Gibco) at 37˚C in an atmosphere containing
5% CO2. Transfection was performed using the
LipofectamineTM 2000 (Invitrogen) for 90-95% density and
the OptifectTM (Invitrogen) for low density as
previously reported (9).
Co-immunoprecipitation
For co-immunoprecipitation experiments, 293T cells
were lysed in NP40 buffer (10 mM Tris-HCl (pH 7.4), 150 mM NaCl,
10% glycerol, 1% Nonidet P40, 10 µM Na3VO4, 1
mM PMSF, 1 µg/ml pepstatin A, 1 µg/ml leupeptin, 1 µg/ml aprotinin,
1 mM DTT) and lysates were incubated with primary antibodies at 4˚C
for 5 h and then with protein A (for anti-V5, -AREL1 and -MTX2
antibodies) or G (for anti-Flag antibody) agarose for 1.5 h. The
immunoprecipitates were analyzed as described previously (9).
In vivo ubiquitination assay
293T cells were transfected with indicated plasmids
involving MTX2 or AREL1 or both. At 8 h following tsransfection,
cells were treated with or not 4 µM MG132 for 12 h to inhibit
protease activity. Cell lysates were prepared in NP40 lysis buffer
and incubated overnight with an anti-Flag antibody or for 5 h with
an anti-MTX2 antibody. The precipitates were submitted to western
blotting with an anti-Ub antibody.
Immunoblot analysis
For the immunoblotting, cell lysates were obtained
using RIPA buffer (50 mM Tris-HCl (pH 7.4), 5 mM NaCl, 1 µM EGTA,
1% Triton X-100, 50 µM NaF, 10 µM Na3VO4, 1
µg/ml aprotinin, 1 µg/ml leupeptin, 1 µg/ml pepstatin A, 0.1 mM
PMSF, 1 mM DTT), and the soluble protein concentrations were
determined according to a Bradford assay (Bio-Rad Laboratories).
Extracted proteins were separated by SDS-PAGE, electrotransferred,
and probed with the primary antibody. The following antibodies were
used in this study - AREL1, MTX1 (611768, BD Biosciences), MTX2,
GST (554805, Pharmingen), Flag (F-3165, Sigma), V5 (46-0705,
Invitrogen), Actin (SC-1616, Santa Cruz Biotechnology Inc.),
gamma-Tubulin (SC-7396, Santa Cruz Biotechnology Inc.), Ub
(SC-8017, Santa Cruz Biotechnology Inc.). The signal was detected
by the chemiluminescent ECL or ECL-plus system (Amersham
Biosciences).
Necroptosis assay
Mouse L929 cells were used for the necroptosis
assay. After transfection, cells were challenged with TNF, zVAD (10
µM) for 4 h. 10 µM of MG132 was treated to test its effects on cell
death. The trypan blue exclusion assay was performed to detect cell
death following 30 h of treatment. Data are expressed as the
percentage of dead and viable cells by repeated experiments more
than three times. Necroptosis of TNF-treated cells was confirmed by
microscopic observation of necrosis-like morphologic features such
as condensed nuclei and swelled cytoplasm with no apoptotic
blebbing.
Statistical analysis
Data was entered in Microsoft excel spread sheet and
analyzed using SPSS software (version 17). Numerical data were
presented as mean and standard deviation values.
Results
AREL1 interacts with MTX2
For isolating target proteins of AREL1 E3 ubiquitin
ligase involved in cell death, a yeast two-hybrid screen was
carried out with the HeLa cell cDNA library to isolate
AREL1-interacting proteins and identified MTX2 as a candidate of
AREL1 target. Immunoprecipitation experiments were performed using
anti-V5 and anti-Flag antibodies. This confirmed the molecular
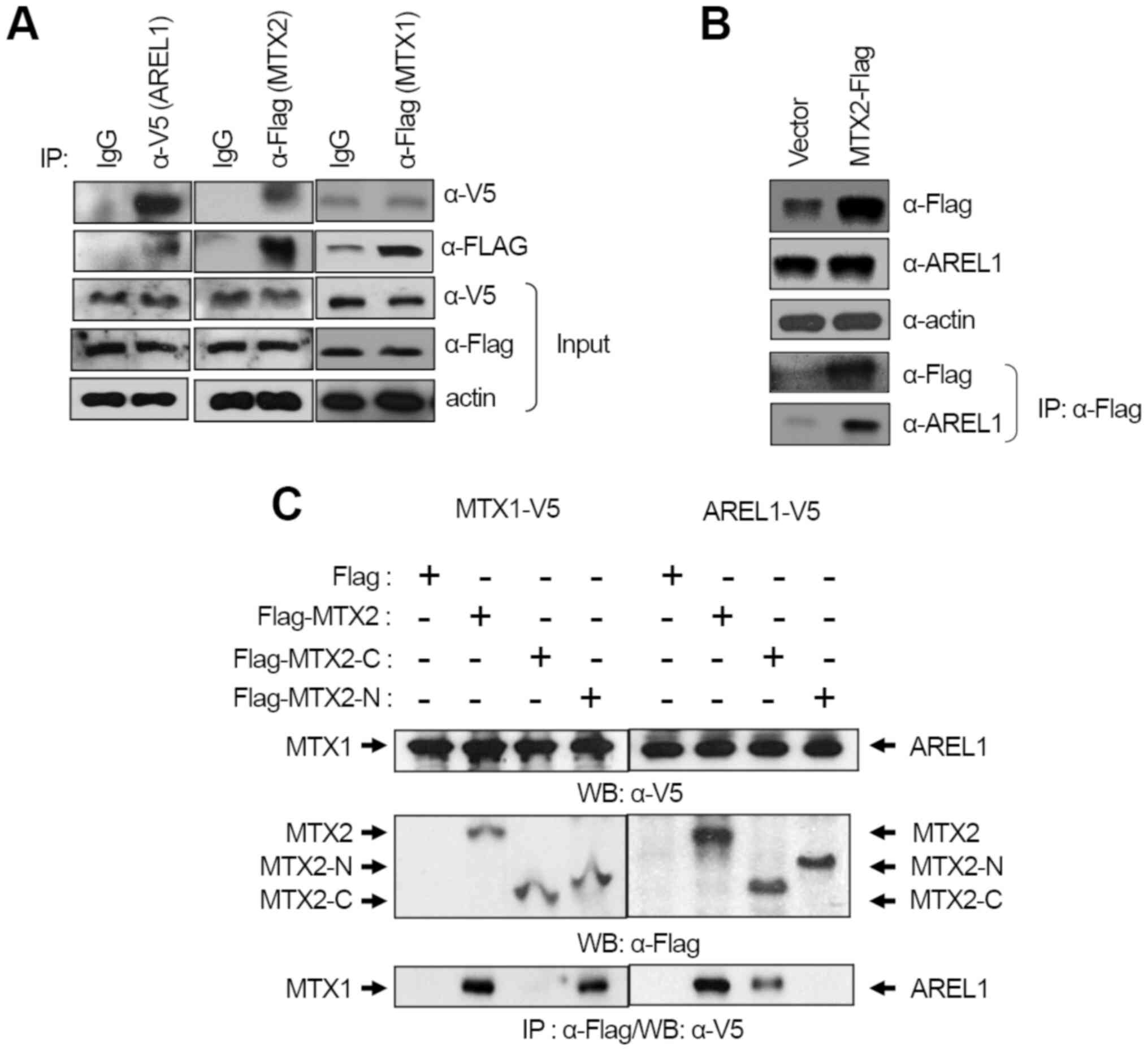
interaction of AREL1 with MTX2, but not MTX1 (Fig. 1A). Endogenous AREL1 was detected
from immunoprecipitates collected from extracts of cells
transfected with Flag-MTX2 using an anti-Flag antibody (Fig. 1B), however, endogenous MTX2 was not
detected in cells expressing AREL1. This observation may correlate
with AREL-1 directed degradation of MTX2 (Fig. 2A). In addition to the
immunoprecipitation experiments, we confirmed co-localization of
AREL1 and MTX2 in the cytosol (Fig.
S1).
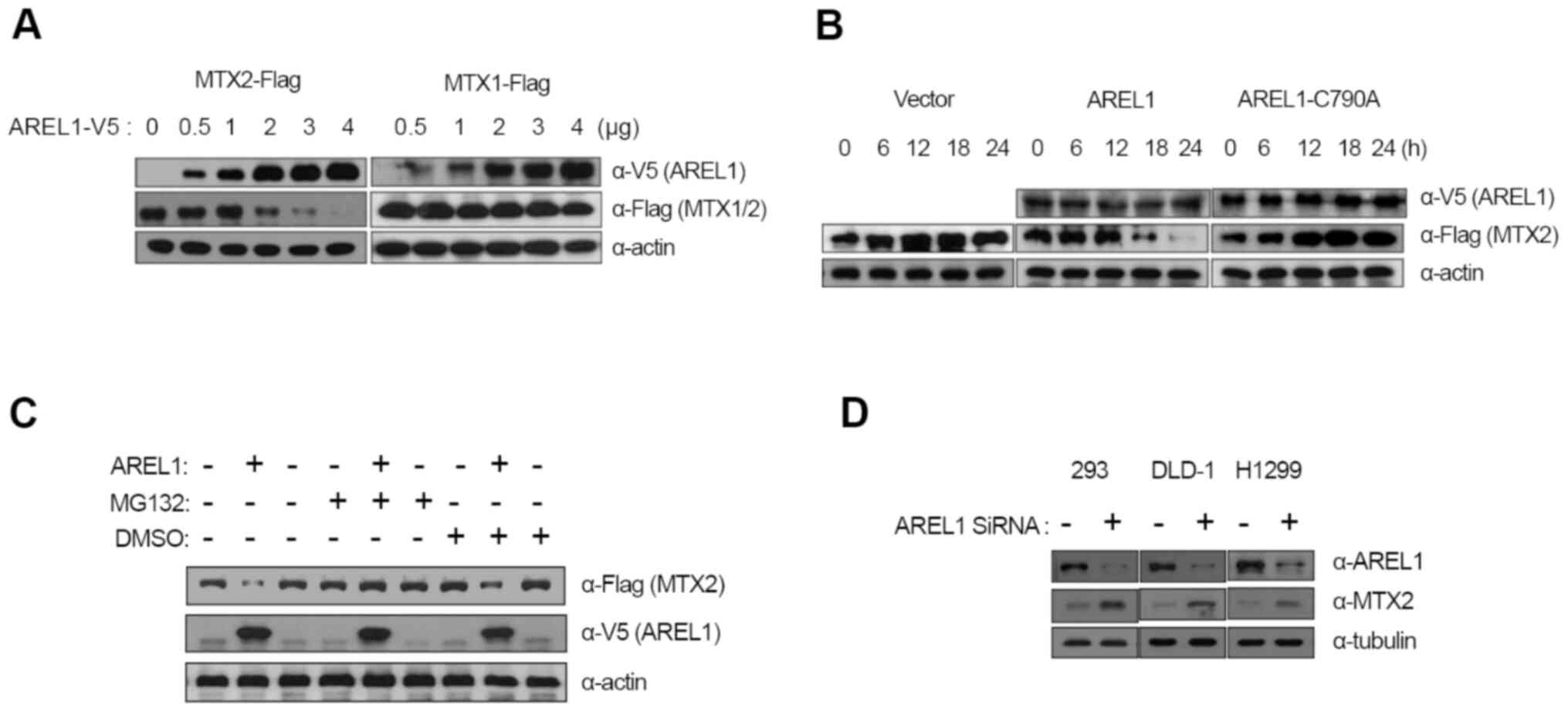 | Figure 2Degradation of MTX2 by AREL1. (A) MTX2
degradation by AREL1. Various amounts of V5-tagged AREL1-expressing
plasmids were transfected into 293T cells with either Flag-tagged
MTX1 or MTX2. WB of the 293T cell extracts was performed with the
indicated antibodies. (B) E3 activity of AREL1 was found to be
essential for MTX2 degradation. 293T cells were transfected with
V5-tagged wild-type AREL1 or its E3-deficient mutant form,
AREL1-C790A, with Flag-tagged MTX2. MTX2 protein was detected via
WB of the cell extracts, which were prepared at the indicated times
after transfection, with the indicated antibodies. (C)
Proteasome-dependent degradation of AREL1-mediated MTX2
degradation. 293T cells were transfected with V5-tagged AREL1 and
cultured with MG132 or DMSO. Endogenous MTX2 proteins were detected
by WB of the cell extracts with the indicated antibodies. (D)
siRNA-mediated knockdown of AREL1. 293T, DLD-1 and H1299 cells were
treated for 48 h with AREL1-directed RNA interference. Next, cell
extracts were generated and WB with anti-AREL1 and anti-MTX2
antibodies was conducted. AREL1, apoptosis-resistant E3 ubiquitin
protein ligase 1; MTX2, Metaxin 2; MTX1, Metaxin 1; siRNA, small
interfering RNA; WB, western blotting |
It has been shown that MTX1 interacts with MTX2,
tethering it into the outer membrane of mitochondria (10), which results in MTX2 bound to the
cytosolic face of the mitochondrial outer membrane. Since AREL1
protein is localized to the cytosol of cells (7), we examined whether AREL1 and MTX1
compete with each other to interact with MTX2 or interact to
different sites in MTX2, MTX2 with deletion of either N-terminal
112 amino acids or C-terminal 152 amino acids were transfected with
AREL1 or MTX1. Immunoprecipitation and western blotting showed that
the C-terminus of MTX2 interacted with AREL1, whereas it's
N-terminus for MTX1, indicating that AREL1 and MTX1 interact with
distinct domains in MTX2 (Fig.
1C). These results suggest that MTX1 does not bother the
interaction between AREL1 and MTX2 proteins.
Ubiquitination and degradation of MTX2
by AREL1
To examine whether MTX2 is a target of AREL1 E3
ubiquitin ligase, we co-transfected MTX2 with an increasing amount
of AREL1-encoding plasmid into 293T cells and analyzed the protein
levels of both proteins. Transfection of AREL1 results in a
significant reduction in MTX2 proteins in a dose-dependent manner
(Fig. 2A). However, the protein
levels of MTX1 were not affected by AREL1 expression (Fig. 2A).
A role of E3 ubiquitin ligase activity of AREL1 in
MTX2 degradation was examined using a mutant form of AREL1,
AREL1-C790A, in which 790th amino acid cysteine, a
highly conserved residue among HECT family E3 ubiquitin ligase and
essential for forming ubiquitin thioester complex was replaced with
alanine (7). While AREL1
expression led to decrease in MTX2 proteins, AREL1-C790A did not
(Fig. 2B), indicating that E3
ubiquitin ligase activity of AREL1 is essential for MTX2
degradation. Furthermore, the effects of MG132, a potent inhibitor
of the proteasome, was also examined (Fig. 2C). MTX2 degradation in cells
expressing AREL1 was inhibited by treatment with MG132, but not
DMSO (Fig. 2C). We also examined
the effects of AREL1 knockdown on endogenous MTX2 protein levels
(Fig. 2D). SiRNA-mediated
knockdown of AREL1 was applied to three different cell lines such
as 293T, DLD-1, and H1299. Protein levels of MTX2 were
significantly increased in all cell lines tested following
knockdown of AREL1 (Fig. 2D).
Together, these results suggest that the E3 ubiquitin ligase
activity of AREL1 is essential for MTX2 degradation.
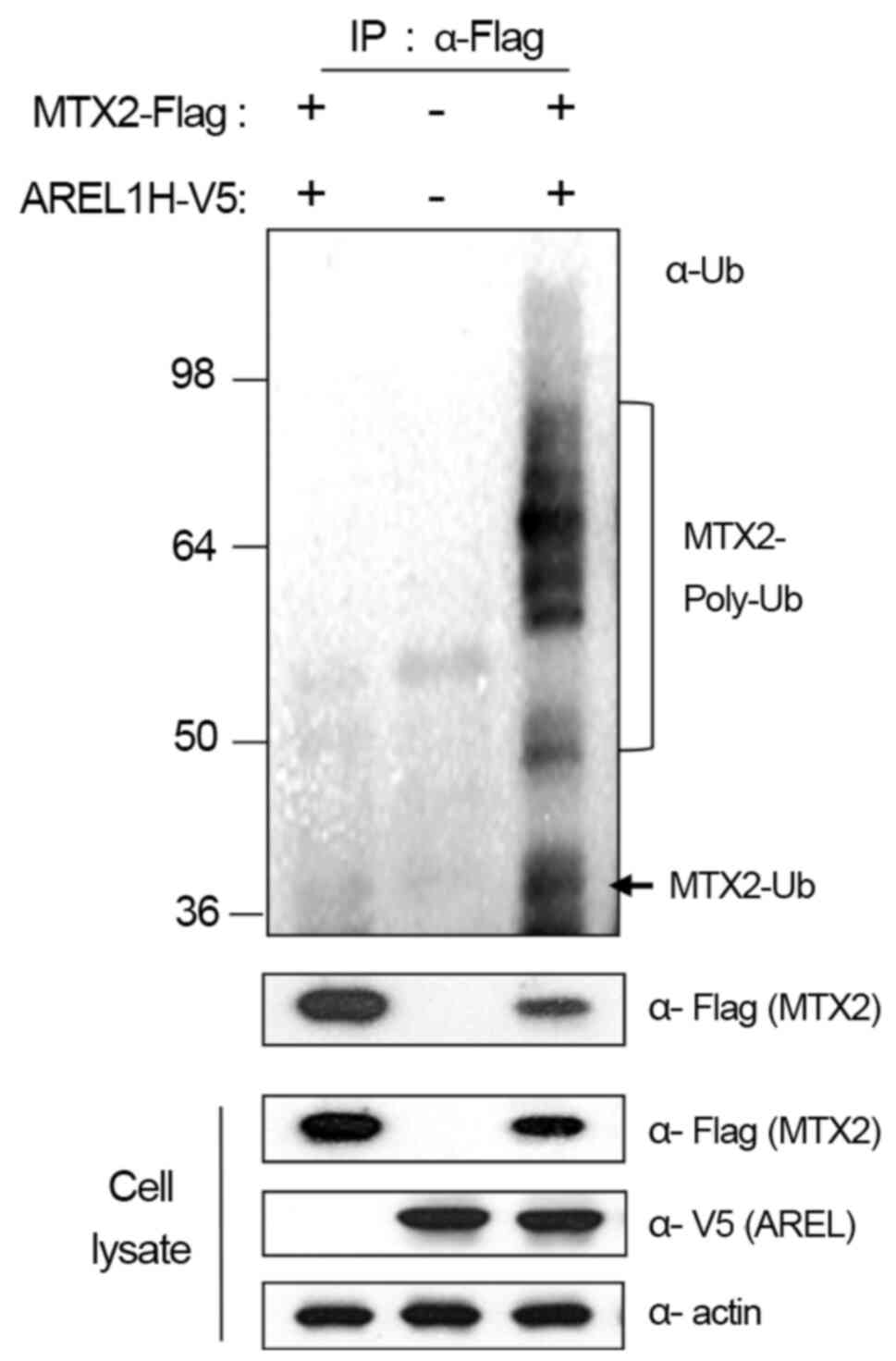
Based on these results, we further examined whether
AREL1 increases the ubiquitination of MTX2 proteins in 293T cells
with AREL1-V5 and MTX2-Flag (Fig.
3). Ubiquitinated MTX2 proteins were barely detected in 293T
cells expressing AREL1 or MTX2 alone but significantly enhanced in
cells expressing both AREL1 and MTX2 (Fig. 3). These results suggest that AREL1
ubiquitinates and promotes the proteasome-dependent degradation of
MTX2.
Inhibition of TNF-induced necroptosis
by AREL1
It was previously reported that Metaxin proteins are
required for TNF-induced cell death (11,12).
Therefore, we examined whether AREL1 also inhibits TNF-induced
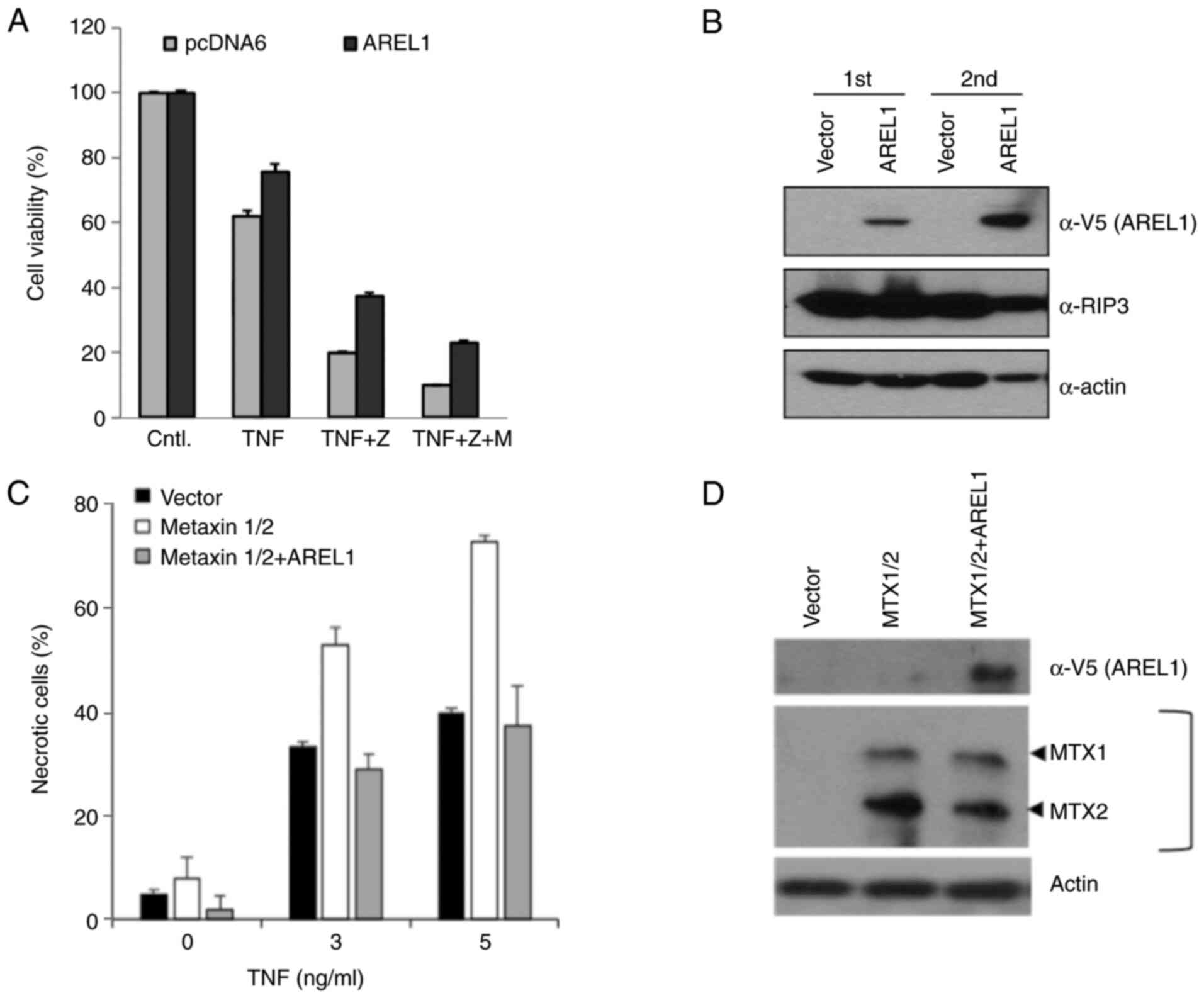
necroptosis as well as apoptosis. To investigate AREL1's function
in TNF-induced necroptosis, we treated AREL1-expressing L929 cells
with TNF and caspase inhibitor, zVAD, to induce necroptotic cell
death. Necroptotic cell death was significantly suppressed in
AREL1-expressing cells compared to control L929 cells (Figs. 4A and S2). However, the toxic effects of MG132
were not specific to AREL1-expressing cells (Fig. 4A). It may be due to broad effects
of MG132 in necroptotic cells (13). AREL1 expression did not affect
protein levels of RIP3 (Fig. 4B),
which is increased and an essential effector during necroptotic
cell death (14), implying that
the anti-necroptosis functions of AREL1 is mediated by down-stream
regulators.
We further examined the effects of AREL1 in
TNF-induced cell death of L929 cells expressing MTX1 and MTX2
(Fig. 4C). Expression of both MTX1
and MTX2 increased susceptibility to TNF-induced necroptotic death
(Fig. 4C), as previously reported
(12). However, this
susceptibility in L929 cells expressing MTX1 and MTX2 was
suppressed by AREL1 expression (Fig.
4C and D). Therefore, these
results suggest that the necroptosis-inhibitory function of AREL1
may relate to its ability to ubiquitinates MTX2 protein.
Discussion
This study demonstrated the AREL1-dependent
ubiquitination of MTX2 and its involvement in necroptosis
inhibitory functions. AREL1 interacted with the N-terminal domain
of MTX2, whereas MTX1 did the C-terminal part (Fig. 2). We previously reported that AREL1
binds to and ubiquitinates IAP antagonists when released into the
cytosol, but not when they reside in mitochondrial inter-membrane
space (7). MTX2 binds to the
mitochondrial outer membrane's cytosolic face through its
interaction with MTX1, embedded in the mitochondria's outer
membrane by its anchor domain (10,15).
Therefore, MTX1-bound MTX2 and the cytosolic form of MTX2 could be
ubiquitinated by AREL1 E3 ubiquitin ligase, which is mainly
localized in the cytosol (Fig.
S1) (7). Third member of
Metaxin family protein, Metaxin 3, was lately identified from
Zebrafish and Xenopus (16,17).
Although it has been shown that Metaxin 3 is distinct from MTX1 and
MTX2 based on amino acid sequence homology, only limited
information is available for its cellular function.
Metaxin proteins have been reported to be involved
in TNF-induced cell death (11,12).
MTX1 deficiency results in resistance to TNF-induced cell death
(12). Since MTX1 tethers MTX2
into the cytosolic face of the mitochondrial outer membrane
(10), expression of MetaΔTM/C,
which does not contain mitochondrial membrane anchor domain, may
also result in MTX2 deficiency at the outer membrane of
mitochondria. However, the effects of Metaxin-family proteins in
TNF-induced apoptosis and necroptosis is controversial; Chen et
al reported that knockdown of MTX1 and MTX2 had no noted effect
on TNF-induced apoptosis (18).
TNF induces apoptosis in many cancer cells, but necroptosis in
certain cell lines including L929. In our hand, MTX2 knockdown was
not successful by low transfection efficiency of L929 cells and
failed to confirm that MTX2 depletion results in resistance to
TNF-induced necroptosis.
Here, we focused on roles of AREL1 and MTX2 in
TNF-induced necroptosis using L929 cells (Fig. 4). It is noteworthy that AREL1 has
anti-necroptosis effects in addition to the anti-apoptosis effects
that have been previously reported (7). AREL1 seems to function as a
downstream regulator of cell death because it inhibits apoptosis
induced by various stimuli including p53, staurosoporine, and DNA
damaging agents (7). Necroptosis
can be induced by not only TNF but also various stimuli such as
ceramide, lonidamine, and sodium nitroprusside (19,20).
Since AREL1 did not affect RIP3, upstream regulator of necroptosis,
it seems to be a downstream regulator of necroptosis induced by
various necroptosis stimuli.
Although it is still unclear how Metaxin deficiency
in the outer membrane affects TNF-induced necroptosis, the role of
Metaxin proteins has been implicated to serve in the pre-protein
import of mitochondria (15).
Metaxin proteins have been identified recently as components of the
sorting and assembly machinery (SAM)/translocase of the
outer-membrane β-barrel protein (TOB) complex (SAM/TOB complex) in
the mitochondrial outer membrane (21,22).
It is recently reported that SAM/TOB complex is a component of the
mitochondrial intermembrane space bridging (MIB) complex that
maintains cristae morphology (23). MIB complex likely contains third
member of Metaxin family protein, Metaxin 3(23). The depletion of MTX2 leads to the
reduction of MTX1 and inhibits the import and assembly of a
voltage-dependent anion-selective channel (VDAC) located in the
outer mitochondrial membrane (22,24);
however, the role of these proteins in import has yet to be clearly
defined. VDAC mediates metabolite exchange between the cytosol and
mitochondria and is involved in apoptosis (25-28).
These results imply that MTX2 depletion will affect cell death via
the integrity of the outer membrane of mitochondria.
Although necroptosis involves the active destruction
of mitochondrial and plasma membranes, our current knowledge has
been limited in plasma membrane-proximal protein complex, including
TNFR and its associated proteins (29,30).
Therefore, it is interesting to identify proteins located in the
cytosol and outer membrane of mitochondria and involved in
necroptosis. AREL1 did not affect RIP3, an essential component of
the membrane-proximal necroptosis complex (Fig. 4A). Therefore, mitochondrial MTX2
protein will be a key target to elucidate the
necroptosis-inhibitory function of AREL1 and the cytosolic process
of necroptosis including mitochondrial membrane disintegration.
Supplementary Material
Localization of AREL1 and MTX2. 293T
cells were transfected with both V5-tagged AREL1 and GFP-tagged
MTX2. After 36 h, cells were fixed and visualized by indirect
immunofluorescence using the anti-Flag antibody followed by
rhodamine-conjugated secondary antibody. DAPI was used to visualize
nuclei. Normal IgG was used as a negative control. Each column
represents the same field of cells as visualized under the green
(MTX2-GFP, top left), red (Flag-AREL1, top right) and blue (DNA,
bottom left) filters. Scale bar, 5 μm. AREL1, apoptosis-resistant
E3 ubiquitin protein ligase 1; MTX2, Metaxin 2.
Necrotic death of L929 cells after
treatment with TNF. L929 cells were transfected with AREL1 and then
treated with TNF and caspase inhibitors Z for 4 h. Scale bar, 100
μm AREL1, apoptosis-resistant E3 ubiquitin protein ligase 1; Cntl,
control; Z, zVAD-fmk.
Acknowledgements
Not applicable.
Funding
Funding: This work was supported by research grants from Dankook
University (Cheonan, Korea) (grant no. R-2019-00586).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
DS made substantial contributions to the conception
and design of the present study, performed data interpretation, was
involved in drafting and revising the manuscript, and also gave
final approval of the version to be published. YJ and BK were
involved in designing the experiments, and contributed to the
acquisition, analysis and interpretation of data. DS, YJ and BK
confirm the authenticity of all the raw data. All authors have read
and approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Festjens N, Cornelis S, Lamkanfi M and
Vandenabeele P: Caspase-containing complexes in the regulation of
cell death and inflammation. Biol Chem. 387:1005–1016.
2006.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Lockshin RA and Zakeri Z: Cell death in
health and disease. J Cell Mol Med. 11:1214–1224. 2007.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Cohen GM: Caspases: The executioners of
apoptosis. Biochem J. 326:1–16. 1997.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Kerr JF, Wyllie AH and Currie AR:
Apoptosis: A basic biological phenomenon with wide-ranging
implications in tissue kinetics. Br J Cancer. 26:239–257.
1972.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Festjens N, Vanden Berghe T and
Vandenabeele P: Necrosis, a well-orchestrated form of cell demise:
Signalling cascades, important mediators and concomitant immune
response. Biochim Biophys Acta. 1757:1371–1387. 2006.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Newton K: RIPK1 and RIPK3: Critical
regulators of inflammation and cell death. Trends Cell Biol.
25:347–353. 2015.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Kim JB, Kim SY, Kim BM, Lee H, Kim I, Yun
J, Jo Y, Oh T, Jo Y, Chae HD, et al: Identification of a novel
anti-apoptotic E3 ubiquitin ligase that ubiquitinates antagonists
of inhibitor of apoptosis proteins SMAC, HtrA2, and ARTS. J Biol
Chem. 288:12014–12021. 2013.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Chae HD, Kim SY, Park SE, Kim J and Shin
DY: p53 and DNA-dependent protein kinase catalytic subunit
independently function in regulating actin damage-induced
tetraploid G1 arrest. Exp Mol Med. 44:236–240. 2012.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Jo Y and Shin DY: Repression of the F-box
protein Skp2 is essential for actin damage-induced tetraploid G1
arrest. BMB Rep. 50:379–383. 2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Armstrong LC, Saenz AJ and Bornstein P:
Metaxin 1 interacts with metaxin 2, a novel related protein
associated with the mammalian mitochondrial outer membrane. J Cell
Biochem. 74:11–22. 1999.PubMed/NCBI
|
|
11
|
Ono K, Wang X, Kim SO, Armstrong LC,
Bornstein P and Han J: Metaxin deficiency alters mitochondrial
membrane permeability and leads to resistance to TNF-induced cell
killing. Protein Cell. 1:161–173. 2010.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Wang X, Ono K, Kim SO, Kravchenko V, Lin
SC and Han J: Metaxin is required for tumor necrosis factor-induced
cell death. EMBO Rep. 2:628–633. 2001.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Cho YS, Challa S, Moquin D, Genga R, Ray
TD, Guildford M and Chan FK: Phosphorylation-driven assembly of the
RIP1-RIP3 complex regulates programmed necrosis and virus-induced
inflammation. Cell. 137:1112–1123. 2009.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Armstrong LC, Komiya T, Bergman BE, Mihara
K and Bornstein P: Metaxin is a component of a preprotein import
complex in the outer membrane of the mammalian mitochondrion. J
Biol Chem. 272:6510–6518. 1997.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Adolph KW: Characterization of the cDNA
and amino acid sequences of Xenopus Metaxin 3, and
relationship to Xenopus Metaxins 1 and 2. DNA Seq.
16:252–259. 2005.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Adolph KW: The zebrafish metaxin 3 gene
(mtx3): cDNA and protein structure, and comparison to zebrafish
metaxins 1 and 2. Gene. 330:67–73. 2004.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Ross K, Rudel T and Kozjak-Pavlovic V:
TOM-independent complex formation of Bax and Bak in mammalian
mitochondria during TNFalpha-induced apoptosis. Cell Death Differ.
16:697–707. 2009.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Chen J, Kos R, Garssen J and Redegeld F:
Molecular Insights into the Mechanism of Necroptosis: The Necrosome
As a Potential Therapeutic Target. Cells. 8(E1486)2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Magtanong L, Ko PJ and Dixon SJ: Emerging
roles for lipids in non-apoptotic cell death. Cell Death Differ.
23:1099–1109. 2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Kim SK, Kim WJ, Yoon JH, Ji JH, Morgan MJ,
Cho H, Kim YC and Kim YS: Upregulated RIP3 Expression Potentiates
MLKL Phosphorylation-Mediated Programmed Necrosis in Toxic
Epidermal Necrolysis. J Invest Dermatol. 135:2021–2030.
2015.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Cartron PF, Petit E, Bellot G, Oliver L
and Vallette FM: Metaxins 1 and 2, two proteins of the
mitochondrial protein sorting and assembly machinery, are essential
for Bak activation during TNF alpha triggered apoptosis. Cell
Signal. 26:1928–1934. 2014.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Kozjak-Pavlovic V, Ross K, Benlasfer N,
Kimmig S, Karlas A and Rudel T: Conserved roles of Sam50 and
metaxins in VDAC biogenesis. EMBO Rep. 8:576–582. 2007.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Huynen MA, Mühlmeister M, Gotthardt K,
Guerrero-Castillo S and Brandt U: Evolution and structural
organization of the mitochondrial contact site (MICOS) complex and
the mitochondrial intermembrane space bridging (MIB) complex.
Biochim Biophys Acta. 1863:91–101. 2016.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Ott C, Ross K, Straub S, Thiede B, Götz M,
Goosmann C, Krischke M, Mueller MJ, Krohne G, Rudel T, et al: Sam50
functions in mitochondrial intermembrane space bridging and
biogenesis of respiratory complexes. Mol Cell Biol. 32:1173–1188.
2012.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Cheng EH, Sheiko TV, Fisher JK, Craigen WJ
and Korsmeyer SJ: VDAC2 inhibits BAK activation and mitochondrial
apoptosis. Science. 301:513–517. 2003.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Hodge T and Colombini M: Regulation of
metabolite flux through voltage-gating of VDAC channels. J Membr
Biol. 157:271–279. 1997.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Shimizu S, Narita M and Tsujimoto Y and
Tsujimoto Y: Bcl-2 family proteins regulate the release of
apoptogenic cytochrome c by the mitochondrial channel VDAC.
Nature. 399:483–487. 1999.PubMed/NCBI View
Article : Google Scholar
|
|
28
|
Shore GC: Apoptosis: It's BAK to VDAC.
EMBO Rep. 10:1311–1313. 2009.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Micheau O and Tschopp J: Induction of TNF
receptor I-mediated apoptosis via two sequential signaling
complexes. Cell. 114:181–190. 2003.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Wang L, Du F and Wang X: TNF-alpha induces
two distinct caspase-8 activation pathways. Cell. 133:693–703.
2008.PubMed/NCBI View Article : Google Scholar
|