Introduction
Oral squamous cell carcinoma (OSCC) is a common
malignancy of the head and neck (1). It is estimated that ~350,000
individuals are diagnosed with OSCC every year worldwide,
accounting for 2-4% of all malignant tumors (2,3).
Among patients with OSCC, ~50% are diagnosed in the first instance
with advanced-stage cancer. This delay in diagnosis and lack of
specific markers for the prediction of OSCC development and
progression may also explain the high global mortality rate in 2018
(4,5). Despite notable progress made in the
diagnosis and treatment of tumors, surgery, postoperative
radiotherapy and chemotherapy remain the conventional therapeutic
approaches and prognosis of OSCC remains poor (6). Although OSCC, oropharyngeal and
esophageal carcinoma are classified as head and neck squamous cell
carcinomas, the differences in the microenvironment and
pathogenesis between these cancers lead to a large difference in
the gene expression profile between OSCC and other types of
squamous cell carcinoma (7,8).
OSCC carcinogenesis is a global burden that needs to be addressed;
therefore identifying novel effective biomarkers and therapeutic
targets for patients with OSCC should be a priority.
Collagen type IV α1 chain (COL4A1), a type of
collagen that belongs to the type IV collagen family, is a key
component of the basement membrane and several studies have found
that it acts as a cancer-promoting factor in several types of
cancer (9-12).
Recent studies have found that COL4A1 is abnormally expressed in
invasive ductal carcinoma of breast and bladder tumors and is
associated with tumor invasion and metastasis (13,14).
Furthermore, COL4A1 is reported to be upregulated in patients with
OSCC (15). Nevertheless, the
specific role that COL4A1 plays in OSCC progression remains
unclear. Thus, the present study aimed to identify the biological
role and the potential mechanism of COL4A1 in OSCC cells.
Materials and methods
Bioinformatics analysis
The TNMplot database (tnmplot.com) was
used to perform the association analysis between COL4A1 and
Nidogen-1 (NID1) in OSCC. In addition, STRING database (cn.string-db.org/) was used to predict the binding
between COL4A1 and NID1.
Cell culture and treatment
Human OSCC cell lines HN4, HN6, SCC-4 and Cal-27, as
well as human oral keratinocyte (HOK) cells were provided by the
Institute of Basic Medical Sciences, Chinese Academy of Medical
Sciences (Beijing, China). Cells were maintained in DMEM (Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (HyClone;
Cytiva) and 1% penicillin-streptomycin (Gibco; Thermo Fisher
Scientific, Inc.) and cultured at 37˚C in a humified incubator with
5% CO2.
Cell transfection
For knockdown of COL4A1, the specific small
interfering (si)RNAs targeting COL4A1 (si-COL4A1-1,
5'-AGGACAAGCUCAAGUUCAAGA-3'; and si-COL4A1-2,
5'-GGAGCGAGAUGUUCAAGAAGC-3') and the corresponding negative control
siRNA (si-NC, 5'-UUCUCCGAACGUGUCACGU-3') were synthesized by
Shanghai Gene Pharma Co., Ltd. To overexpress NID1, pc-DNA3.1
vector containing the full-length NID1 gene (Oe-NID1) and the empty
vector (Oe-NC) were synthesized by Shanghai Gene Pharma Co., Ltd. A
total of 100 nM plasmids/siRNAs were transfected into SCC-4 cells
using Lipofectamine™ 2000 (Invitrogen; Thermo Fisher Scientific,
Inc.) for 48 h at 37˚C. A total of 48 h post-transfection, cells
were collected for use in subsequent experiments.
Cell Counting Kit-8 (CCK-8) assay
Following transfection, SCC-4 cells were plated into
96-well plates at a density of 5x103 cells/well and
cultured at 37˚C for 24, 48 or 72 h. Subsequently, 10 µl CCK-8
(Beyotime Institute of Biotechnology) was added and cells were
further cultured at 37˚C with 5% CO2 for 4 h. Absorbance
was measured using a microplate reader at a wavelength of 450 nm as
a measure of proliferation (Bio-Rad Laboratories, Inc).
EdU incorporation cell proliferation
assay
SCC-4 cells were plated in a 6-well plate at a
density of 4x105 cells/well and cultured overnight at
37˚C. The following day, cells were fixed in 4% polyformaldehyde at
room temperature for 15 min and permeabilized with 0.5% Triton
X-100 at room temperature for 15 min. Then, cells were stained
using a Cell-Light™ EdU Cell Proliferation Detection Assay (Thermo
Fisher Scientific, Inc.) at room temperature for 30 min according
to the manufacturer's protocol and counterstained with 5 mg/ml DAPI
at room temperature for 10 min. The stained cells were counted
using a fluorescence microscope (Nikon Corporation; magnification,
x100).
Colony formation assay
Transfected cells were plated at a density of 500
cells/well in 6-well plates and incubated in DMEM with 10% bovine
calf serum (Thermo Fisher Scientific, Inc.) at 37˚C. Following 10
days of culture, cells were fixed using 4% paraformaldehyde for 15
min at room temperature and stained with 1.5% crystal violet
(FUJIFILM Wako Pure Chemical Corporation) at room temperature for
10 min. Colonies were counted with ImageJ software (Version 146;
National Institutes of Health) using a light microscope (Olympus
Corporation; magnification, x1). Colonies consisted of ≥50
cells.
Wound healing assay
The migration of SCC-4 cells was assessed using a
wound healing assay. Transfected cells were plated into a 6-well
plate at a density of 5x105 cells/well and cultured at
37˚C until they reached 80-90% confluence. A 20-µl tip was used to
create a scratch in the monolayer, after which cells were cultured
in serum-free DMEM (Gibco; Thermo Fisher Scientific, Inc.). After
24 h incubation at room temperature, the scratched area was
observed using a light microscope (Olympus Corporation;
magnification, x100). The migratory rate (%) was calculated as
follows: (Wound width at 0 h-wound width at 24 h)/wound width at 0
h x 100. Analysis was based on five randomly selected fields of
view.
Transwell assay
Transwell chambers (Corning, Inc.) were precoated
with 0.1 ml Matrigel (Becton, Dickinson and Company) at 37˚C for 1
h. SCC-4 cells were collected and suspended to a final
concentration of 2x105 cells/ml in DMEM containing 1%
FBS (HyClone; Cytiva). The cell suspensions were placed in the
upper chamber and DMEM supplemented with 10% FBS was added to the
lower chamber. After incubation at 37˚C for 24 h, a cotton swab was
used to remove cells from the upper chamber that had not invaded.
Cells that had invaded were fixed using 70% ethanol at 4˚C for 30
min and stained using 0.5% crystal violet for 10 min at room
temperature. The number of cells that had invaded was counted using
a light microscope (Olympus Corporation; magnification, x200) in
five randomly selected fields of view.
Reverse transcription-quantitative
(RT-q)PCR
Total RNA was extracted from 1x104 SCC-4
cells using TRIzol® (Invitrogen; Thermo Fisher
Scientific, Inc.) and this was used to synthesize cDNA using cDNA
Synthesis kit (Takara Bio, Inc.) according to the manufacturer's
protocol. qPCR was performed using SYBR Premix ExTaq kit (Takara
Bio, Inc.) with amplification performed on an ABI PRISM 7900
Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. The qPCR
thermocycling conditions were as follows: Initial denaturation at
95˚C for 3 min; followed by 40 cycles of denaturation at 95˚C for
30 sec, annealing at 60˚C for 30 sec and extension at 72˚C for 30
sec. The following primer pairs were used for qPCR: COL4A1 forward,
5'-AGGAGTGCCATTGCTTTTTCAA-3' and reverse,
5'-TGGAAACCAGTCCATGCTCG-3'; NID1 forward,
5'-CTTTGAGGTACCCACCGTCC-3' and reverse, 5'-GGGAGGGGCTGCTCATTATC-3'
and GAPDH forward, 5'-GGGAAACTGTGGCGTGAT-3' and reverse,
5'-GAGTGGGTGTCGCTGTTGA-3'. The relative mRNA level was calculated
using the 2-∆∆Cq method and normalized to the internal
reference gene GAPDH (16).
Co-immunoprecipitation (Co-IP)
Total protein was extracted from SCC-4 cells using
IP lysis buffer (20 mM Tris-HCl, 150 mM NaCl and 1% Triton X-100,
pH 7.5). The supernatant was collected after centrifugation at
13,000 x g for 10 min at 4˚C. Protein A agarose beads (0.2 mg; cat.
no. 20366; Thermo Fisher Scientific, Inc.) washed with 100 µl PBS
were added to 500 µg cell lysates and incubated with 2 µg IgG
antibody (cat. no. ab6715; Abcam) or COL4A1 antibody (cat. no.
ab6586; Abcam) overnight at 4˚C. Following rinsing with PBS,
precipitated protein was resuspended and boiled for 5 min at 100˚C.
Finally, the eluates were collected by magnetic separation and
subsequently subjected to western blotting.
Western blotting
Total protein was extracted from SCC-4 cells using
RIPA buffer (Auragene) and quantified using a BCA Protein Assay
kit. Equal amounts of protein (20 µg/lane) were loaded on a 10% SDS
gel (Bio-Rad Laboratories, Inc.), resolved using SDS-PAGE and
transferred to PVDF membranes (MilliporeSigma) at 25 V for 30 min.
Membranes were blocked for 2 h at room temperature using 5% non-fat
milk in 0.1% tris-buffered saline with Tween-20 and incubated with
primary antibodies against COL4A1 (cat. no. ab226485; 1:500;
Abcam), N-cadherin (cat. no. ab76011; 1:5,000; Abcam), Vimentin
(cat. no. ab92547; 1:1,000; Abcam), E-cadherin (cat. no. ab40772;
1:10,000; Abcam), NID1 (cat. no. ab254325; 1:1,000; Abcam) and
GAPDH (cat. no. ab9485; 1:2,500; Abcam) at 4˚C overnight, followed
by incubation with horseradish peroxidase-conjugated secondary
antibodies (cat. no. ab6721; 1:2,000; Abcam) for 2 h at room
temperature. Signals were visualized using an ECL detection system
(Beyotime Institute of Biotechnology) and densitometry analysis was
performed using QuantityOne version 4.5.0 (Bio-Rad Laboratories,
Inc.).
Statistical analysis
All experiments were performed at least three times.
All data were analyzed using SPSS version 22.0 software (IBM Corp)
and are presented as the mean ± standard deviation. Differences
between two groups were compared using unpaired Student's t test
whereas differences between multiple groups were compared using
one-way ANOVA with post hoc Bonferroni's multiple comparison test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
COL4A1 is upregulated in OSCC
cells
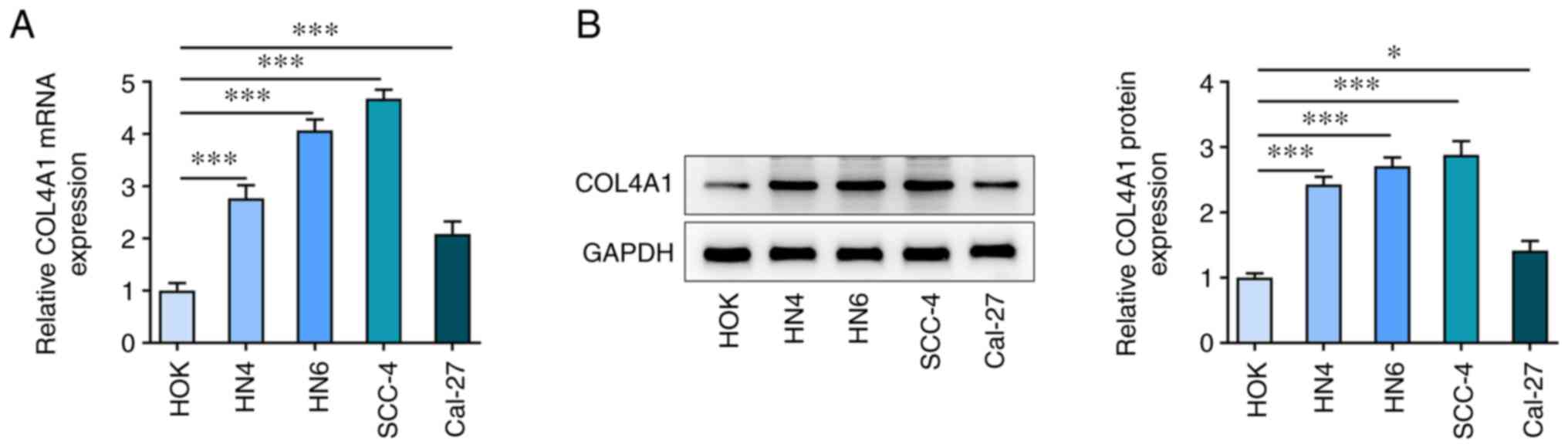
To determine the role that COL4A1 serves in OSCC
progression, COL4A1 expression in OSCC cells was initially
assessed. mRNA expression levels of COL4A1 in OSCC cell lines
including HN4, HN6, SCC-4 and Cal-27 were increased compared with
those in HOK cells (Fig. 1A). In
addition, results from western blot analysis demonstrated that
COL4A1 levels were increased in OSCC cell lines compared with those
in HOK cells (Fig. 1B). Among
these OSCC cell lines, SCC-4 showed the highest expression of
COL4A1, thus this cell line was used for all subsequent
experiments.
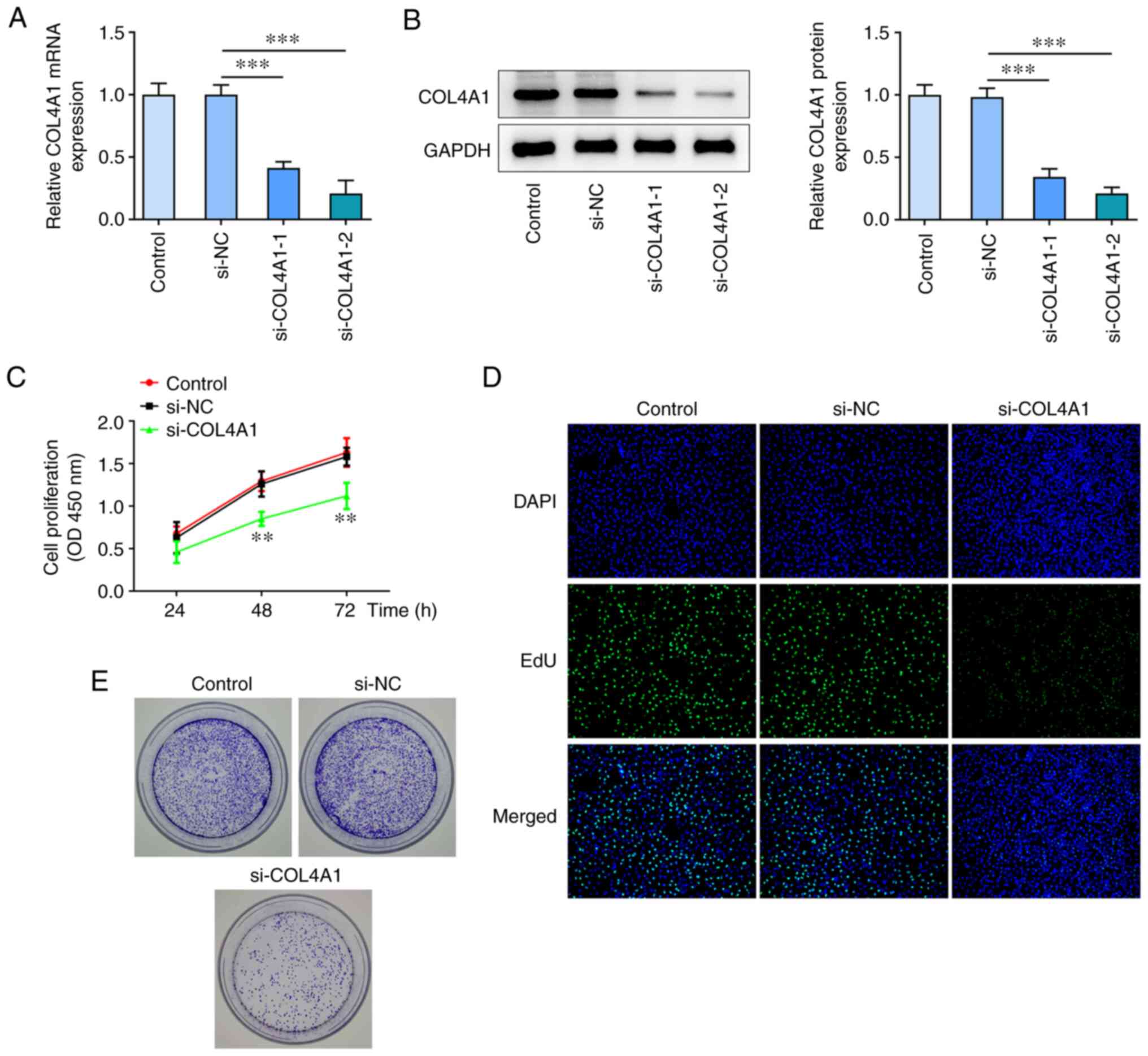
Knockdown of COL4A1 suppresses
proliferation of SCC-4 cells
To determine the biological role that COL4A1 serves
in OSCC cells, si-COL4A1-1/2 was used to knock down COL4A1
expression in SCC-4 cells. RT-qPCR and western blot analysis showed
that si-COL4A1-2 exhibited a greater knockdown efficiency. Thus,
si-COL4A1-2 (henceforth referred to as si-COL4A1) was selected for
all subsequent experiments (Fig.
2A and B). CCK-8 assay showed
that cell proliferation was significantly inhibited following
transfection with si-COL4A1 compared with NC (Fig. 2C). In addition, data from Edu
staining also revealed the number of positive cells in the
si-COL4A1 group was decreased compared with that in the control
(Fig. 2D). As shown in Fig. 2E, the colony-forming ability of
SCC-4 cells was reduced by COL4A1 knockdown compared with that in
the si-NC-transfected cells.
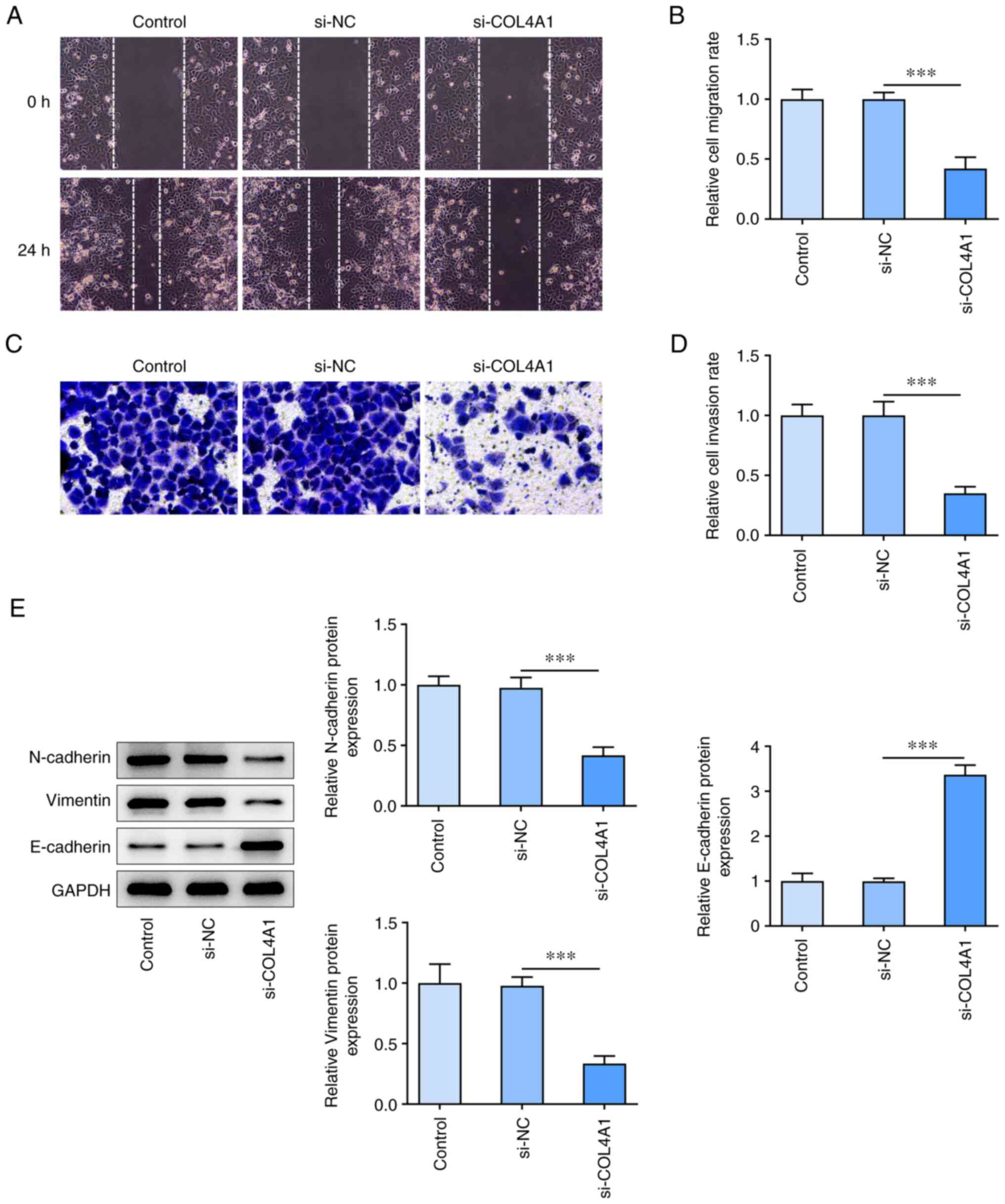
COL4A1 knockdown inhibits migration,
invasion and expression of epithelial-mesenchymal transition
(EMT)-associated protein in SCC-4 cells
Next, the effects of COL4A1 knockdown on migration,
invasion and EMT-associated protein expression in SCC-4 cells were
assessed. Knockdown of COL4A1 decreased cell migration compared
with NC (Fig. 3A and B). Transwell invasion assay results
showed that the invasive ability was decreased in the COL4A1
knockdown cells compared with NC (Fig.
3C and D). Western blot
analysis showed that COL4A1 knockdown resulted in a decrease in the
levels of N-cadherin and vimentin while increasing expression
levels of E-cadherin (Fig.
3E).
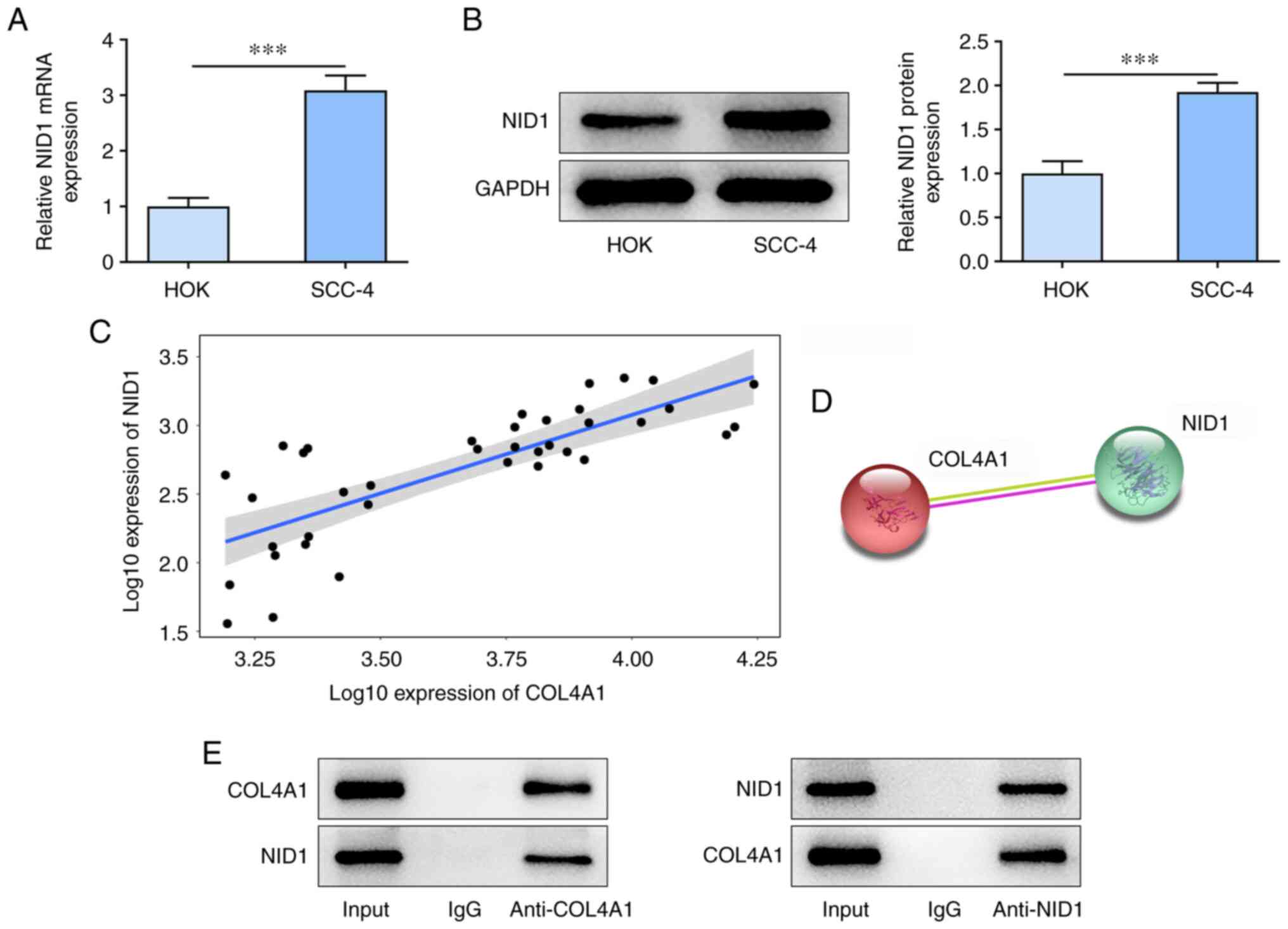
COL4A1 interacts with NID1 in OSCC
cells
mRNA and protein expression levels of NID1 were
significantly higher in SCC-4 cells compared with those in the HOK
cells (Fig. 4A and B). In addition, TNMplot database showed
that COL4A1 was highly positively associated with NID1 in OSCC
(Fig. 4C). STRING database
analysis also showed that COL4A1 and NID1 may form a complex
(Fig. 4D). Co-IP analysis showed
that COL4A1 and NID1 were present in the IP assay with anti-COL4A1
and anti-NID1 antibody but not with control IgG (Fig. 4E).
Upregulation of NID1 reverses the
inhibitory effect of COL4A1 silencing on SCC-4 cells
To identify the role of NID1 in COL4A1 regulation in
SCC-4 cells, NID1 was overexpressed in SCC-4 cells. RT-qPCR and
western blot analysis confirmed that NID1 expression was
successfully increased in the NID1 cells (Fig. 5A and B). CCK-8 assay showed that NID1
overexpression reversed the decrease in cell proliferation compared
with NC (Fig. 5C). Similarly, the
Edu assay showed that transfection with Oe-NID1 notably increased
the number of positive cells (Fig.
5D). Results obtained from the colony formation assays showed
that the number of colonies was higher in NID1-overexpressing cells
(Fig. 5E). Furthermore, cell
migration and invasion were increased following transfection with
Oe-NID1 (Fig. 5F-I). Finally, an
increase in levels of N-cadherin and vimentin and decrease in
E-cadherin levels were observed in the cells transfected with
Oe-NID1 (Fig. 5J).
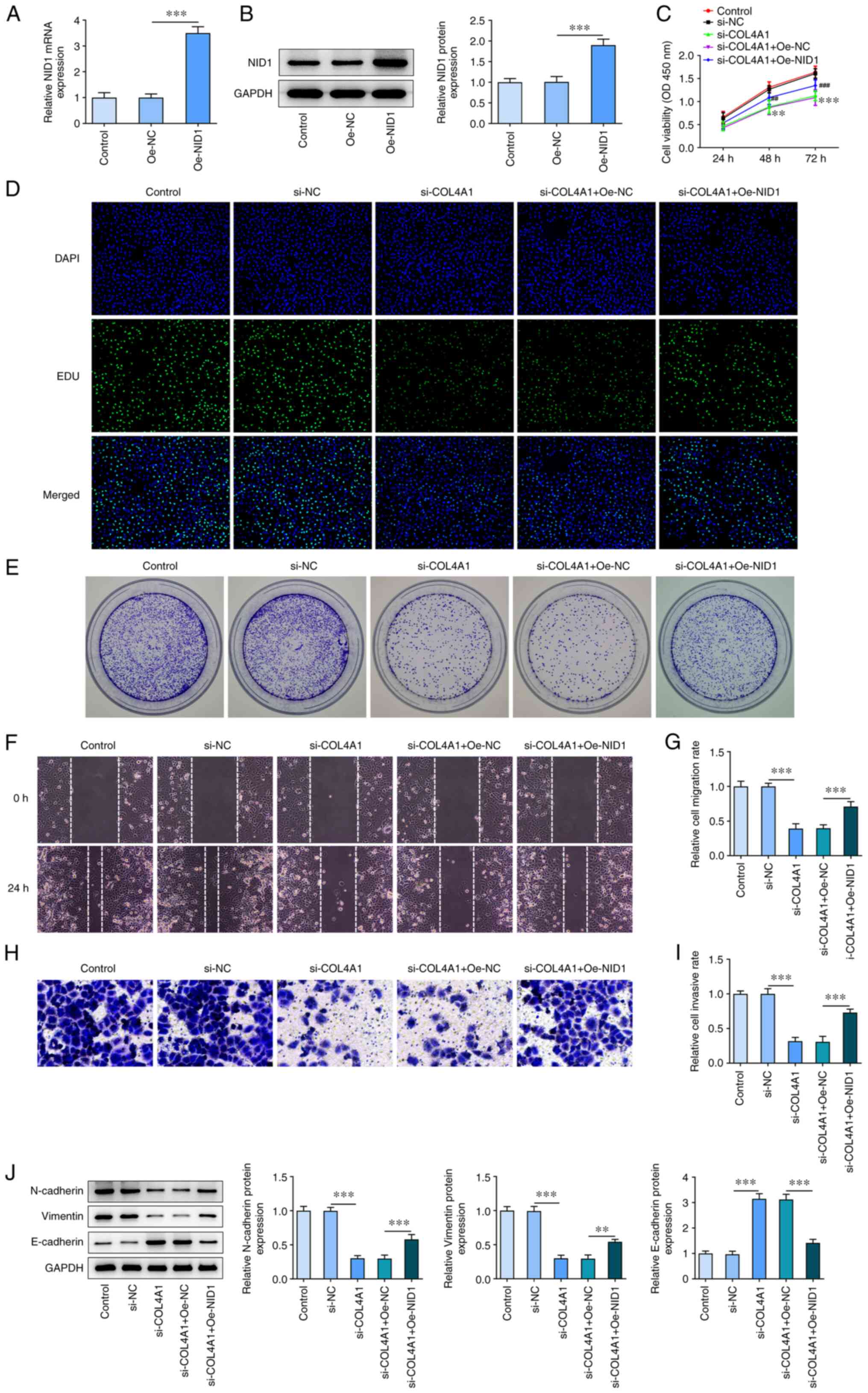 | Figure 5Upregulation of NID1 reverses the
inhibitory effect of COL4A1 silencing on SCC-4 cells. (A) mRNA
expression of NID1 in SCC-4 cells transfected with Oe-NID1 was
measured via reverse transcription-quantitative PCR.
***P<0.001. (B) NID1 protein levels in SCC-4 cells
transfected with Oe-NID1 measured using western blot assay.
***P<0.001. Cell proliferation in SCC-4 cells was
evaluated using (C) Cell Counting Kit-8 (**P<0.01,
***P<0.001 vs. si-NC; ##P<0.01,
###P<0.001 vs. si-COL4A1+ Oe-NC) and (D) EdU
incorporation (magnification, x100). (E) Colony formation assay
(magnification, x1) was used to measure the colony-forming ability
of SCC-4 cells. (F) Representative image of the wound healing assay
of SCC-4 cells transfected with Oe-NID1 (magnification, x100). (G)
Migration rate of SCC-4 cells transfected with Oe-NID1 measured
using a wound healing assay. ***P<0.001. (H)
Representative image of Transwell invasion assay of SCC-4 cells
transfected with Oe-NID1 (magnification, x100). (I) Invasion rate
of SCC-4 cells transfected with Oe-NID1 measured via Transwell
invasion assay. ***P<0.001. (J) Protein levels of
N-cadherin, vimentin and E-cadherin in SCC-4 cells transfected with
Oe-NID1 semi-quantified using western blot assay. Results are
presented as the mean ± standard deviation. **P<0.01
and ***P<0.001. NID1, nidogen-1; Oe, overexpression;
NC, negative control; COL4A1, collagen type IV α1 chain; si, small
interfering; OD, optical density. |
Discussion
Oral cancer is a common malignancy that poses a
notable threat to human health. In the past decade, the occurrence
of oral cancer has increased markedly worldwide (17). OSCC, a common type of oral cancer,
has been characterized by a high degree of malignancy and poor
prognosis (18). It has been shown
that OSCC has high a potential for metastasis (19). The invasion and metastasis of a
tumor is a complex process that requires the involvement of
numerous genes (20). Tumor cells
detach from the primary lesion and invade the basement membrane to
infiltrate the surrounding interstitium, enter the lumen of blood
vessels through the local capillary blood or lymphatic vessel wall,
forming small tumor emboli and are transported in blood or
lymphatic fluid (21-23).
When they are transferred to a target organ, these cells adhere to
endothelial cells of the blood or lymphatic vessels and proliferate
continuously at the secondary site to form a metastasis (24). In the present study, the molecular
mechanism underlying OSCC migration and invasion was explored. It
was demonstrated that COL4A1 may interact with NID1 to promote
proliferation and migration of OSCC cells.
Collagens are the most abundant protein in the
extracellular matrix and are primarily involved in formation of
basement membranes, fibrillar and microfibrillar networks, as well
as other structures in the extracellular matrix (25). A previous study has shown that
aberrantly expressed collagens affect the biological behavior of
cancer cells (26). Biosynthesis
of collagens can be modulated by cancer cells by targeting
transcription factors, mutated genes and receptors, as well as
signaling pathways. Conversely, collagens bind to integrins,
discoidin domain receptors and tyrosine kinase receptors to
influence the behavior of tumor cells (27). COL4A1, which belongs to the
collagen family, serves a tumor-promoting role in several types of
cancer (28,29). Wang et al (30) showed that COL4A1 exerts a promoting
effect on proliferation and metastasis in hepatocellular carcinoma
via the activation of a FAK-Src signaling pathway. Jin et al
(31) used functional enrichment
analysis to screen genes associated with improving invasive ductal
carcinoma treatment and COL4A1 was found to be a key gene that
influenced proliferation and invasion of the invasive ductal
carcinoma cells. Other studies have shown that COL4A1 is
upregulated in head and neck squamous cell carcinoma and may serve
as a novel prognostic biomarker for the recurrence of OSCC
(32,33), indicating that COL4A1 expression is
associated with OSCC. In the present study, COL4A1 was highly
expressed in several OSCC cell lines and COL4A expression was
highest in SCC-4 cells, which may be associated with the high
invasiveness, as well as migration, proliferation and
differentiation, of SCC-4 (34,35).
In addition, COL4A1 knockdown suppressed the ability of SCC-4 cells
to proliferate, migrate and invade. COL4A1 knockdown also
suppressed the expression levels of the EMT-related proteins
N-cadherin and Vimentin, and elevated E-cadherin expression in
SCC-4 cells.
Nidogens are the primary components of the basement
membrane and function as a linker connecting collagen IV and
laminin networks to stabilize the three-dimensional structure of
the basement membrane (36). NID1
and NID2 belong to the nidogen family (37). Studies have shown that NID1 is
involved in promoting tumor development, such as gastric, breast
cancer and colorectal cancer (38-40).
Xu et al (41) found that
microRNA-1298-3p suppresses the ability of glioma cells to
proliferate and invade via downregulation of NID1. Yuan et
al (42) reported that NID1 is
upregulated in ovarian cancer cells and NID1 overexpression
reverses the inhibitory effect of long non-coding RNA-ATB silencing
on progression of ovarian cancer. Hsu et al (43) showed that NID1 expression is
enhanced in OSCC tissues and that it could be used as a biomarker
for OSCC. Based on TNMplot and STRING databases, the present
analysis found that NID1 was highly positively associated with
COL4A1 and bound to COL4A1. Thus, co-IP was used to verify the
binding between NID1 and COL4A1. Moreover, the present study
indicated that NID1 overexpression reversed the suppressive effects
of COL4A1 knockdown on OSCC cell proliferation, migration and
invasion, as well as on EMT-associated protein expression. There
are several limitations in the present study. Only one OSCC cell
line was used to explore the biological role of COL4A1; the effects
of COL4A1 in other OSCC cell lines and other potential mechanisms
are currently being investigated. In addition, the results of the
present would be further supported by NID1 silencing experiments as
no significant difference was demonstrated for cell proliferation
in the Oe-NID1 group.
In conclusion, the molecular mechanisms by which
COL4A1 exerts its effects in OSCC were determined in the present
study. The results revealed that COL4A1 increased the proliferation
and migration of OSCC cells by interacting with NID1, highlighting
a potential novel avenue for therapeutic targeting in the
management of OSCC.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
XT and KZ designed the research and drafted and
revised the manuscript. XT, JS, CL and KZ performed the
experiments. JS and CL searched the literature and analyzed the
data. KZ guided the experiments. XT and KZ confirm the authenticity
of all the raw data All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Perera M, Al-Hebshi NN, Perera I, Ipe D,
Ulett GC, Speicher DJ, Chen T and Johnson NW: Inflammatory
bacteriome and oral squamous cell carcinoma. J Dent Res.
97:725–732. 2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Almangush A, Mäkitie AA, Triantafyllou A,
de Bree R, Strojan P, Rinaldo A, Hernandez-Prera JC, Suárez C,
Kowalski LP, Ferlito A and Leivo I: Staging and grading of oral
squamous cell carcinoma: An update. Oral Oncol.
107(104799)2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Satgunaseelan L, Allanson BM, Asher R,
Reddy R, Low HTH, Veness M, Gopal Iyer N, Smee RI, Palme CE, Gupta
R and Clark JR: The incidence of squamous cell carcinoma of the
oral tongue is rising in young non-smoking women: An international
multi-institutional analysis. Oral Oncol.
110(104875)2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Thomson PJ: Perspectives on oral squamous
cell carcinoma prevention-proliferation, position, progression and
prediction. J Oral Pathol Med. 47:803–807. 2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Mumtaz M, Bijnsdorp IV, Böttger F, Piersma
SR, Pham TV, Mumtaz S, Brakenhoff RH, Akhtar MW and Jimenez CR:
Secreted protein markers in oral squamous cell carcinoma (OSCC).
Clin Proteomics. 19(4)2022.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Brands MT, Brennan PA, Verbeek ALM, Merkx
MAW and Geurts SME: Follow-up after curative treatment for oral
squamous cell carcinoma. A critical appraisal of the guidelines and
a review of the literature. Eur J Surg Oncol. 44:559–565.
2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Ezhilarasan D, Lakshmi T, Subha M, Deepak
Nallasamy V and Raghunandhakumar S: The ambiguous role of sirtuins
in head and neck squamous cell carcinoma. Oral Dis. 28:559–567.
2022.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Solomon B, Young RJ and Rischin D: Head
and neck squamous cell carcinoma: Genomics and emerging biomarkers
for immunomodulatory cancer treatments. Semin Cancer Biol.
52:228–240. 2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Labelle-Dumais C, Schuitema V, Hayashi G,
Hoff K, Gong W, Dao DQ, Ullian EM, Oishi P, Margeta M and Gould DB:
COL4A1 mutations cause neuromuscular disease with tissue-specific
mechanistic heterogeneity. Am J Hum Genet. 104:847–860.
2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Shannon P, Hum C, Parks T, Schauer GM,
Chitayat D, Chong K, Shinar S, Blaser S, Moore G and Van Mieghem T:
Brain and placental pathology in fetal COL4A1 related disease.
Pediatr Dev Pathol. 24:175–186. 2021.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Li F, Wang NN, Chang X, Wang SL, Wang LS,
Yao J, Li ZS and Bai Y: Bioinformatics analysis suggests that
COL4A1 may play an important role in gastric carcinoma recurrence.
J Dig Dis. 20:391–400. 2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Wang H, Liu Z, Li A, Wang J, Liu J, Liu B,
Lian X, Zhang B, Pang B, Liu L and Gao Y: COL4A1 as a novel
oncogene associated with the clinical characteristics of malignancy
predicts poor prognosis in glioma. Exp Ther Med.
22(1224)2021.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Wang SM, Chen PM, Sung YW, Huang WC, Huang
HS and Chu PY: Effect of COL4A1 expression on the survival of
neoadjuvant chemotherapy breast cancer patients. J Oncol.
2020(5209695)2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Xie X, He H, Zhang N, Wang X, Rui W, Xu D
and Zhu Y: Overexpression of DDR1 promotes migration, invasion,
though EMT-related molecule expression and COL4A1/DDR1/MMP-2
signaling axis. Technol Cancer Res Treat.
19(1533033820973277)2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Chen C, Méndez E, Houck J, Fan W,
Lohavanichbutr P, Doody D, Yueh B, Futran ND, Upton M, Farwell DG,
et al: Gene expression profiling identifies genes predictive of
oral squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev.
17:2152–2162. 2008.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Wong T and Wiesenfeld D: Oral cancer. Aust
Dent J. 63 (Suppl 1):S91–S99. 2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Fan T, Wang X, Zhang S, Deng P, Jiang Y,
Liang Y, Jie S, Wang Q, Li C, Tian G, et al: NUPR1 promotes the
proliferation and metastasis of oral squamous cell carcinoma cells
by activating TFE3-dependent autophagy. Signal Transduct Target
Ther. 7(130)2022.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Bugshan A and Farooq I: Oral squamous cell
carcinoma: metastasis, potentially associated malignant disorders,
etiology and recent advancements in diagnosis. F1000Res.
9(229)2020.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Valastyan S and Weinberg RA: Tumor
metastasis: molecular insights and evolving paradigms. Cell.
147:275–292. 2011.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Yeung KT and Yang J:
Epithelial-mesenchymal transition in tumor metastasis. Mol Oncol.
11:28–39. 2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Zeeshan R and Mutahir Z: Cancer
metastasis-tricks of the trade. Bosn J Basic Med Sci. 17:172–182.
2017.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Majidpoor J and Mortezaee K: Steps in
metastasis: An updated review. Med Oncol. 38(3)2021.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Burr R, Gilles C, Thompson EW and
Maheswaran S: Epithelial-mesenchymal plasticity in circulating
tumor cells, the precursors of metastasis. Adv Exp Med Biol.
1220:11–34. 2020.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Gelse K, Pöschl E and Aigner T:
Collagens-structure, function, and biosynthesis. Adv Drug Deliv
Rev. 55:1531–1546. 2003.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Gilkes DM, Chaturvedi P, Bajpai S, Wong
CC, Wei H, Pitcairn S, Hubbi ME, Wirtz D and Semenza GL: Collagen
prolyl hydroxylases are essential for breast cancer metastasis.
Cancer Res. 73:3285–3296. 2013.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Rømer AMA, Thorseth ML and Madsen DH:
Immune modulatory properties of collagen in cancer. Front Immunol.
12(791453)2021.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Hu YZ, Hu ZL, Liao TY, Li Y and Pan YL:
LncRNA SND1-IT1 facilitates TGF-β1-induced
epithelial-to-mesenchymal transition via miR-124/COL4A1 axis in
gastric cancer. Cell Death Discov. 8(73)2022.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Zhang H, Wang Y and Ding H: COL4A1,
negatively regulated by XPD and miR-29a-3p, promotes cell
proliferation, migration, invasion and epithelial-mesenchymal
transition in liver cancer cells. Clin Transl Oncol. 23:2078–2089.
2021.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Wang T, Jin H, Hu J, Li X, Ruan H, Xu H,
Wei L, Dong W, Teng F, Gu J, et al: COL4A1 promotes the growth and
metastasis of hepatocellular carcinoma cells by activating FAK-Src
signaling. J Exp Clin Cancer Res. 39(148)2020.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Jin R, Shen J, Zhang T, Liu Q, Liao C, Ma
H, Li S and Yu Z: The highly expressed COL4A1 genes contributes to
the proliferation and migration of the invasive ductal carcinomas.
Oncotarget. 8:58172–58183. 2017.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Jin Y and Qin X: Comprehensive analysis of
transcriptome data for identifying biomarkers and therapeutic
targets in head and neck squamous cell carcinoma. Ann Transl Med.
8(282)2020.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Reis PP, Tokar T, Goswami RS, Xuan Y,
Sukhai M, Seneda AL, Móz LES, Perez-Ordonez B, Simpson C, Goldstein
D, et al: A 4-gene signature from histologically normal surgical
margins predicts local recurrence in patients with oral carcinoma:
Clinical validation. Sci Rep. 10(1713)2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Kuo CL, Lai KC, Ma YS, Weng SW, Lin JP and
Chung JG: Gallic acid inhibits migration and invasion of SCC-4
human oral cancer cells through actions of NF-κB, Ras and matrix
metalloproteinase-2 and -9. Oncol Rep. 32:355–361. 2014.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Pineiro NM, Carneiro A and Crema VO: Rho
GTPases are involved on regulation of cytodifferentiation of SCC-4
oral squamous cell carcinoma cell line: A preliminary study. Asian
Pac J Cancer Prev. 21:3–6. 2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Timpl R and Brown JC: Supramolecular
assembly of basement membranes. Bioessays. 18:123–132.
1996.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Fox MA, Ho MS, Smyth N and Sanes JR: A
synaptic nidogen: developmental regulation and role of nidogen-2 at
the neuromuscular junction. Neural Dev. 3(24)2008.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Zhou S, Zhang S, Wang L, Huang S, Yuan Y,
Yang J, Wang H, Li X, Wang P, Zhou L, et al: BET protein inhibitor
JQ1 downregulates chromatin accessibility and suppresses metastasis
of gastric cancer via inactivating RUNX2/NID1 signaling.
Oncogenesis. 9(33)2020.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Urooj T, Wasim B, Mushtaq S, Haider G,
Shah SNN, Ghani R and Qureshi MFH: Increased NID1 expression among
breast cancer lung metastatic women; A comparative analysis between
naive and treated cases. Recent Pat Anticancer Drug Discov.
15:59–69. 2020.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Vaes N, Schonkeren SL, Rademakers G,
Holland AM, Koch A, Gijbels MJ, Keulers TG, de Wit M, Moonen L, Van
der Meer JRM, et al: Loss of enteric neuronal Ndrg4 promotes
colorectal cancer via increased release of Nid1 and Fbln2. EMBO
Rep. 22(e51913)2021.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Xu X, Ban Y, Zhao Z, Pan Q and Zou J:
MicroRNA-1298-3p inhibits proliferation and invasion of glioma
cells by downregulating Nidogen-1. Aging (Albany NY). 12:7761–7773.
2020.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Yuan D, Qian H, Guo T, Ye J, Jin C, Liu X,
Jiang L, Wang X, Lin M and Yu H: LncRNA-ATB promotes the
tumorigenesis of ovarian cancer via targeting miR-204-3p. Onco
Targets Ther. 13:573–583. 2020.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Hsu CW, Chang KP, Huang Y, Liu HP, Hsueh
PC, Gu PW, Yen WC and Wu CC: Proteomic profiling of paired
interstitial fluids reveals dysregulated pathways and salivary NID1
as a biomarker of oral cavity squamous cell carcinoma. Mol Cell
Proteomics. 18:1939–1949. 2019.PubMed/NCBI View Article : Google Scholar
|