Introduction
RUVBL1 belongs to the family of AAA+ ATPases
(ATPases associated with various cellular activities) associated
with chromatin-remodelling complexes (1). RUVBL1 localized in the nucleus has
intrinsic ATPase activity (2) and
helicase activity with two ATP binding (Walker) sites (3). RUVBL1 is involved in many cellular
processes that are highly relevant to cancer. RUVBL1 interacts with
the oncogenes c-Myc (4) and
β-catenin (5), and modulate
their transcriptional activities. In cooperation with a member of
the LEF/TCF family, β-catenin activates the transcription of a
number of target genes relevant for cancer progression (6). Nuclear RUVBL1 participates in large
molecular complexes such as the INO80 (7) or the TIP60 (8) complexes that are involved in
chromatin remodeling or DNA damage repair. Nuclear RUVBL1 is also
required for the biogenesis of telomerase (9). Findings from RNA interference (RNAi)
or mutational analyses have indicated that RUVBL1 promotes cell
growth and viability (10,11). Interestingly, an intact RUVBL1
ATPase domain is not essential for all RUVBL1 functions (12). Additionally, recent evidence
indicates that RUVBL1 also has cytosolic functions such as
regulation of nonsense-mediated decay of mRNAs (13). RUVBL1 is reportedly expressed on
the extracellular surface of the plasma membrane of U937 monocytoid
cells and peripheral blood monocytes, where it binds extracellular
plasminogen and promotes activation of plasminogen into plasmin
(14).
Cell motility is critical for a variety of
biological processes in normal and pathological conditions; cell
motility drives cellular development, tissue repair and cancer
invasion and metastasis (15). The
first step in cell motility is the generation of membrane
protrusions in the direction of movement (16). Protrusion formation is driven by
actin polymerization; specifically, monomeric globular-actin
(G-actin) subunits form filamentous actin (F-actin) filaments
(17). Protrusion formation
probably also requires the addition of new membranes at the
protrusion site. In motile processes, cells extend F-actin-rich
protrusion; polarized, branched arrays of actin filaments within
these protrusions are arranged with the fast-growing barbed ends
near the plasma membrane and slow-growing pointed ends toward the
rear (18). A highly polarized,
dendritic network of F-actin polymerizes next to the plasma
membrane of the leading edge; this network probably generates the
forces that push the cell boundary forward (19,20).
Pancreatic ductal adenocarcinoma (PDAC) is among the
deadliest cancers because PDAC cells are highly invasive, they
easily invade surrounding tissues and they metastasize at an early
stage (21). We previously
reported that the formation of additional membrane protrusions
increase the invasive and metastatic properties of the PDAC cells
by regulating the activity of Rho GTPases [Rac1 (22) and RhoA (23)] and a protein kinase C [PKCα
(24)]. The role of RUVBL1 in
migration and invasion of cancer cells, including PDAC cells, has
not been reported. Here, we sought to evaluate the role of RUVBL1
that localized in the cytoplasm in the control of PDAC cell
motility and invasion. In the course of this investigation, we
found that cytoplasmic RUVBL1 accumulated in membrane protrusions
and in the leading edges of PDAC cells. Further investigation
revealed that cytoplasmic RUVBL1 contributed to the formation of
membrane protrusions; specifically, RUVBL1 promoted concentration
of G-actin subunits and polymerization of actin filaments via its
direct binding to F-actin in cell protrusions and results in
increased invasive properties of PDAC cells.
Materials and methods
Antibodies
Anti-RUVBL1 antibody (H00008607-M01) was purchased
from Abnova (Taipei, Taiwan, R.O.C.). JLA20 anti-actin antibody
(MABT219) was purchased from Millipore (Temecula, CA). Anti-vitamin
D-binding protein antibody (ab65636) was purchased from Abcam
(Cambridge, MA).
Cell culture
The human PDAC cell line S2-013, a derivative of
SUIT-2, was obtained from Dr T. Iwamura (Miyazaki Medical College,
Miyazaki, Japan) (25). All cells
were grown in Dulbecco’s modified Eagle’s medium (DMEM; Gibco-BRL,
Carlsbad, CA) supplemented with 10% heat-inactivated fetal calf
serum (FCS) at 37°C in a humid atmosphere saturated with 5%
CO2.
siRNA treatments
A single mixture with four different short hairpin
small interfering RNA (siRNA) oligonucleotides targeting
RUVBL1 was purchased from Qiagen (FlexiTube GeneSolution
GS8607; Valencia, CA) and a single mixture with four different
scrambled negative control siRNA oligo-nucleotides was obtained
from Santa Cruz Biotechnology (37007; Santa Cruz, CA). To examine
the effect of the siRNAs on RUVBL1 expression, S2-013 and
PANC-1 cells that expressed RUVBL1 were plated in 6-well plates.
After 20 h, the cells were transfected with 80 pmols of each siRNA
mixture in siRNA transfection reagent (Qiagen) following the
manufacturer’s instructions. After incubation for 48 h, cells were
processed for western blot analysis, transwell motility or Matrigel
invasion assays.
Immunoblot analysis of cell lysates
Each cell pellet was resuspended in 20 mM HEPES (pH
7.4), 100 mM KCl, 2 mM MgCl2, 0.5% Triton X-100,
protease inhibitor cocktail tablets (Roche, Penzberg, Germany) and
phosphatase inhibitor cocktail (Nacalai, Kyoto, Japan). The
bicinchoninic acid (BCA) assay was used to determine protein
concentration in each lysate; an aliquot of each lysate was then
diluted with sample buffer (50 mM Tris, 2% SDS, 0.1% bromophenol
blue and 10% glycerol) to a final concentration of 1–2
μg/μl and analyzed by SDS-PAGE and western blot
analysis.
Confocal immunofluorescence
microscopy
Coverslips were treated with 10 μg/ml
fibronectin (Sigma-Aldrich, St. Louis, MO) for 1 h at room
temperature. Cells were seeded on fibronectin-coated glass
coverslips and incubated for 5 h; cells were then fixed with 4%
paraformaldehyde, permeabilized with 0.1% Triton X-100, covered
with blocking solution (3% BSA/PBS), and then incubated with the
primary antibody for 1 h. Alexa488- or Alexa594-conjugated
secondary antibody (Molecular Probes, Carlsbad, CA) was used with
or without rhodamine-conjugated phalloidin (Cytoskeleton, Denver,
CO). Each specimen was visualized using a Zeiss LSM 510 META
microscope (Carl Zeiss, Gottingen, Germany).
Immunostain wound-healing assay
A plastic pipette tip was used to cut cross-shaped
wounds through a confluent cell monolayer; cells were then allowed
to polarize and migrate into a wounded area. After 4 h, cells were
immunostained with a primary antibody and then incubated with a
fluorophore-conjugated secondary antibody as described above. Each
specimen was examined using a Zeiss LSM 510 META microscope (Carl
Zeiss).
Trans-well motility assay
Cells (3.0×104/chamber) were plated in
the upper chamber of BD BioCoat Control Culture Inserts (24-well
plates, 8-μm pore size; Becton-Dickinson, San Jose, CA).
Serum-free culture medium was added to each upper chamber, and
medium containing 5% FCS was added to each lower chamber. Cells
were incubated on the membranes for 12 h. After this 12-h
incubation, three independent visual fields in each lower chamber
were examined via microscopic observation to count the number of
cells that had moved from the top chamber to the lower chamber.
Matrigel invasion assay
A two-chamber invasion assay was used to assess PDAC
cell invasiveness (24-well plates, 8-μm pore size membrane
coated with a layer of Matrigel extracellular matrix proteins;
Becton-Dickinson). Cells (4.0×104/chamber) suspended in
serum-free medium were seeded into an upper chamber and allowed to
invade towards a 5% FCS chemoattractant in a respective lower
chamber. After 20-h incubation, three independent visual
fields/lower chambers were examined via microscopic observation to
count the number of cells that had moved to the bottom chamber.
Immunoprecipitation and mass
spectrometric analysis of RUVBL1
S2-013 cells were seeded onto fibronectin and
incubated for 5 h. Cells were lysed in lysis buffer [20 mM HEPES
(pH 7.4), 100 mM KCl, 5 mM MgCl2, 0.5% Triton X-100,
protease inhibitor cocktail tablets (Roche), and phosphatase
inhibitor cocktail (Nacalai)]. Lysates were immunoprecipitated with
Dynabeads Protein G (Dynal, Oslo, Norway) and with anti-RUVBL1
antibody or normal mouse IgG (isotype control) for 2 h at 4°C.
Beads were pelleted on a magnetic rack (Dynal).
Co-immunoprecipitated proteins were separated on a 4 to 20%
gradient SDS-PAGE gel and then silver stained. Bands precipitated
by the anti-RUVBL1 antibody were excised from the gel; a nano-LC
MS/MS system, which consisted of an Ultimate HPLC system (Agilent
1100; Agilent Technologies, Santa Clara, CA) and a QSTAR XL mass
spectrometer (Applied Biosystems/MDS SCIEX, Concord, ON, Canada)
equipped with a nano-ESI source was used characterize the excised
proteins (Genomine, Inc., Pohang, Korea). MASCOT v1.9.0 (Matrix
Science, Boston, MA) was used to perform database searches with
findings from the MS/MS spectra (Genomine, Inc.).
In vitro actin polymerization assay
Actin exists in equilibrium between monomeric
subunits and polymeric filaments. To accurately quantify these
actin forms, a commercially available actin polymerization assay
(BK003; Cytoskeleton) was used to measure actin polymerization
under defined conditions; specifically, polymerization-dependent
increases in fluorescence of pyrene-conjugated actin were measured.
Each of four concentrations (10, 30, 100 or 300 μg/ml) of
recombinant human RUVBL1 protein (TP301170; Origene, Rockville, MD)
were added to a separate actin polymerization assay. Briefly, the
actin polymerization assays were based on the enhanced fluorescence
of pyrene-conjugated actin that occurs during polymerization. The
enhanced fluorescence was measured by pyrene monomer G-actin formed
polymer pyrene F-actin in a fluorometer at excitation wavelength
365 nm and emission wavelength 407 nm. To quantify changes in
polymerization rate, Boltzmann sigmoidal equations (GraphPad Prism
version 6.0 software; GraphPad Software, Inc., La Jolla, CA) were
used to fit curves to the fluorescence data. Half-maximal saturated
polymerization values [T1/2max (s)] were
calculated from raw data.
Statistical analysis
GraphPad Prism version 6.0 software was used for all
statistical analyses. Statistical significance was determined using
a two-tailed Student’s t-test and standard deviations. For all
analyses, p<0.05 was considered significant.
Results
RUVBL1-knockdown reduces cell motility
and invasion
To investigate whether RUVBL1 regulates cell growth,
motility and invasion, RUVBL1 expression was suppressed by a single
mixture with four different siRNA oligonucleotides against RUVBL1
in moderately differentiated PDAC cells (line S2-013) and the
poorly differentiated PDAC cell line PANC-1 that endogenously
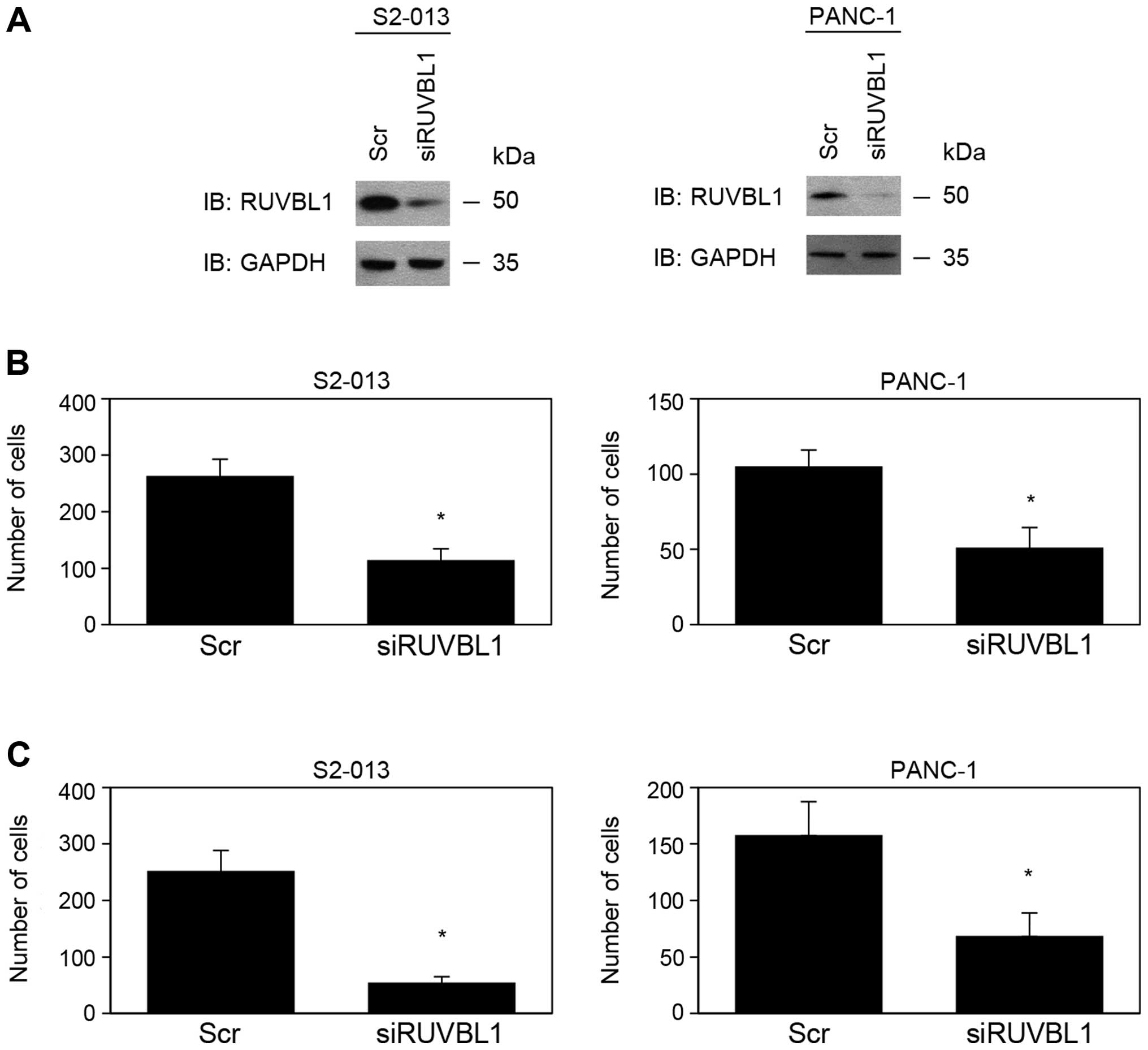
expressed high levels of RUVBL1. Based on western blot data, RUVBL1
expression was markedly lower in RUVBL1-RNAi cells than in
control cells 72 h after transfection of the respective siRNAs
(Fig. 1A). RUVBL2 expression was
not changed in RUVBL1-RNAi cells, compared to control cells
(data not shown). These results indicated that RUVBL1-siRNAs
specifically suppressed endogenous expression of RUVBL1 in S2-013
and PANC-1 cells. RNAi-mediated suppression of RUVBL1 did not
affect cell growth in an in vitro MTT assay of S2-013 and
PANC-1 (data not shown). Transwell motility and Matrigel invasion
assays were used to examine the effect of RUVBL1 on cell motility
and invasiveness. In Transwell motility assays, motility of S2-013
and PANC-1 cells was significantly lower in RUVBL1-RNAi
cells than in control cells (Fig.
1B). In two-chamber invasion assays, invasiveness of
RUVBL1-RNAi cells of S2-013 and PANC-1 was significantly
lower than that of control cells (Fig.
1C). These results indicated that RUVBL1 promoted the motility
and invasiveness of PDAC cells.
RUVBL1 localizes in cell protrusion of
migrating PDAC cells
Endogenous RUVBL1 localizes with transcription
factors mainly to the nucleus, and regulates its target genes
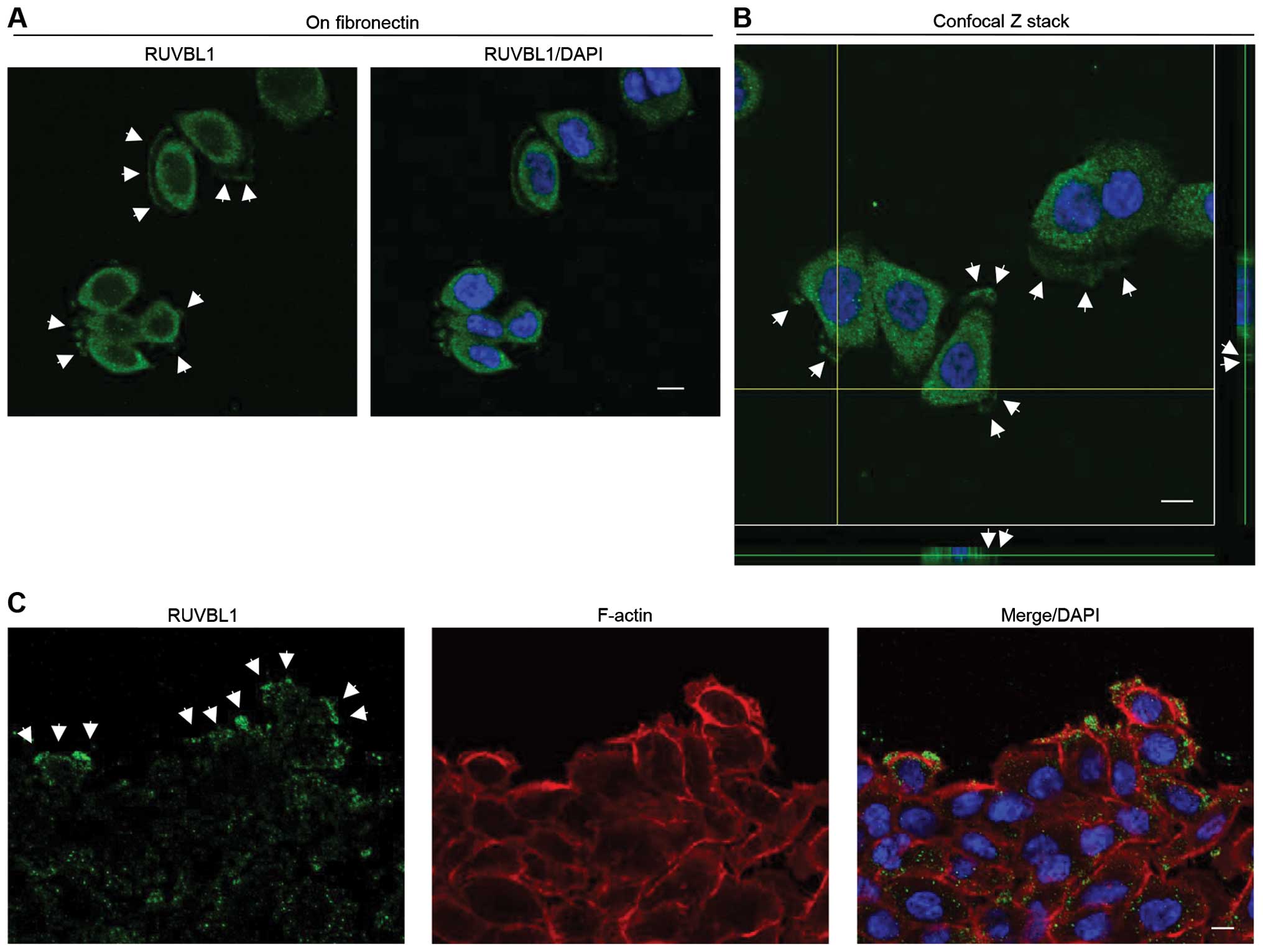
including p53 in colon cancer cells (26). We used immunocytochemistry to
determine the subcellular localization of RUVBL1 in S2-013 cells.
Notably, when S2-013 cells that were initially in suspension attach
to an immobilized fibronectin substrate, nascent membrane
protrusions (de novo formation of actin patches at the cell
periphery) form, and as these protrusions mature, they promote cell
motility and invasion (22).
Therefore, we analyzed the subcellular distribution of RUVBL1 in
PDAC cells cultured on fibronectin. Spreading of S2-013 cells on
fibronectin promoted accumulation of cytoplasmic RUVBL1 in membrane
protrusions (Fig. 2A). These
results indicated that the function of cytoplasmic RUVBL1 may be
different from that of nuclear RUVBL1 in PDAC cells. Z stack panels
substantiated this result in S2-013 cells cultured on fibronectin
(Fig. 2B). Additionally, an
immunostaining wound-healing assay was used to analyze localization
of RUVBL1 in polarized migrating S2-013 cells; results from this
assay showed that RUVBL1 was recruited to the leading edges, in
which peripheral actin structures were abundant, during wound
healing (Fig. 1C). Additionally,
an immunostaining wound-healing assay was used to analyze
localization of RUVBL1 in polarized migrating S2-013 cells
(Fig. 2C); results from this assay
showed that cytoplasmic RUVBL1 was recruited to the leading edges
of S2-013 cells during wound healing. Based on these results, we
reasoned that localization of RUVBL1 in membrane protrusions may be
important to cell motility and invasiveness.
RUVBL1 associates with actin
filaments
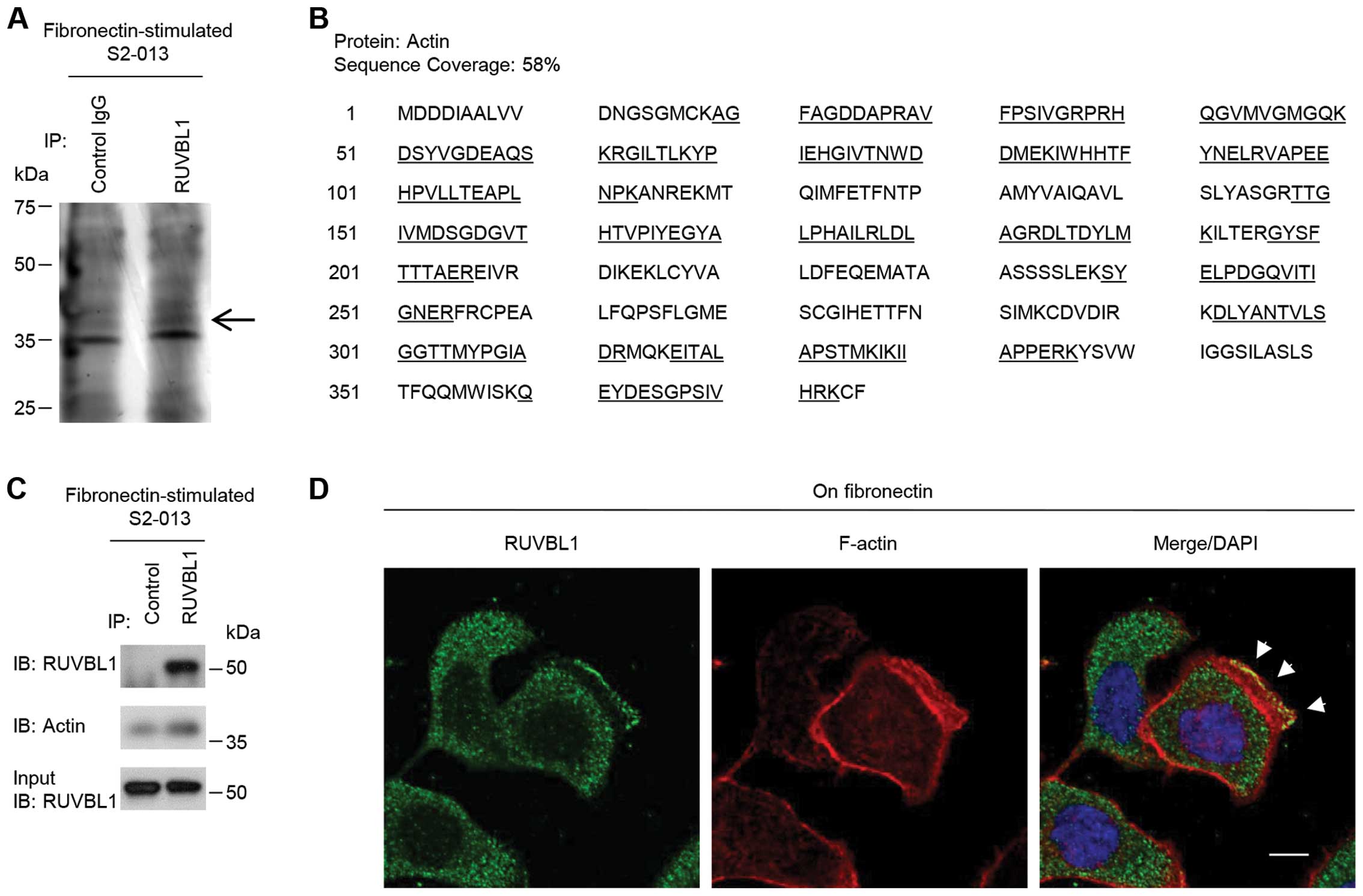
To investigate the mechanism by which RUVBL1
promoted cell motility and invasiveness, immunoprecipitation (IP)
experiments were performed with lysates from fibronectin-stimulated
S2-013 cells; a specific anti-RUVBL1 antibody was used to detect
multiprotein complexes that contained RUVBL1. Control and
anti-RUVBL1 immunoprecipitates were subject to SDS-PAGE; the
separated proteins were silver stained. A 40-kDa band was evident
in the anti-RUVBL1 sample that was very weak in the isotype control
sample (Fig. 3A). The band was
excised, and LC-MS/MS was used to identify the constituent protein
after in-gel trypsin digestion; the protein was actin. The peptide
sequence coverage was 58% (Fig.
3B). Immunoblot analysis showed that a faint band of actin was
detected in control-immunoprecipitates from fibronectin-stimulated
S2-013 cells (Fig. 3C), suggesting
the presence of low levels of non-specific actin in all IP samples.
Strong actin band was detected in the
anti-RUVBL1-immunoprecipitates (Fig.
3C), indicating that actin was enriched in RUVBL1-IP materials
compared to control IgG-IPs.
Immunocytochemical signal from RUVBL1 and
fluorescent signal from peripheral F-actin structures (labeled by
phalloidin) were colocalized in cell protrusions of
fibronectin-stimulated S2-013 cells (arrowheads in Fig. 3D). Thus, we hypothesized that the
RUVBL1-actin complexes localized in cell protrusions could function
in promotion of the motility and invasiveness.
Effects of RUVBL1 on in vitro actin
polymerization
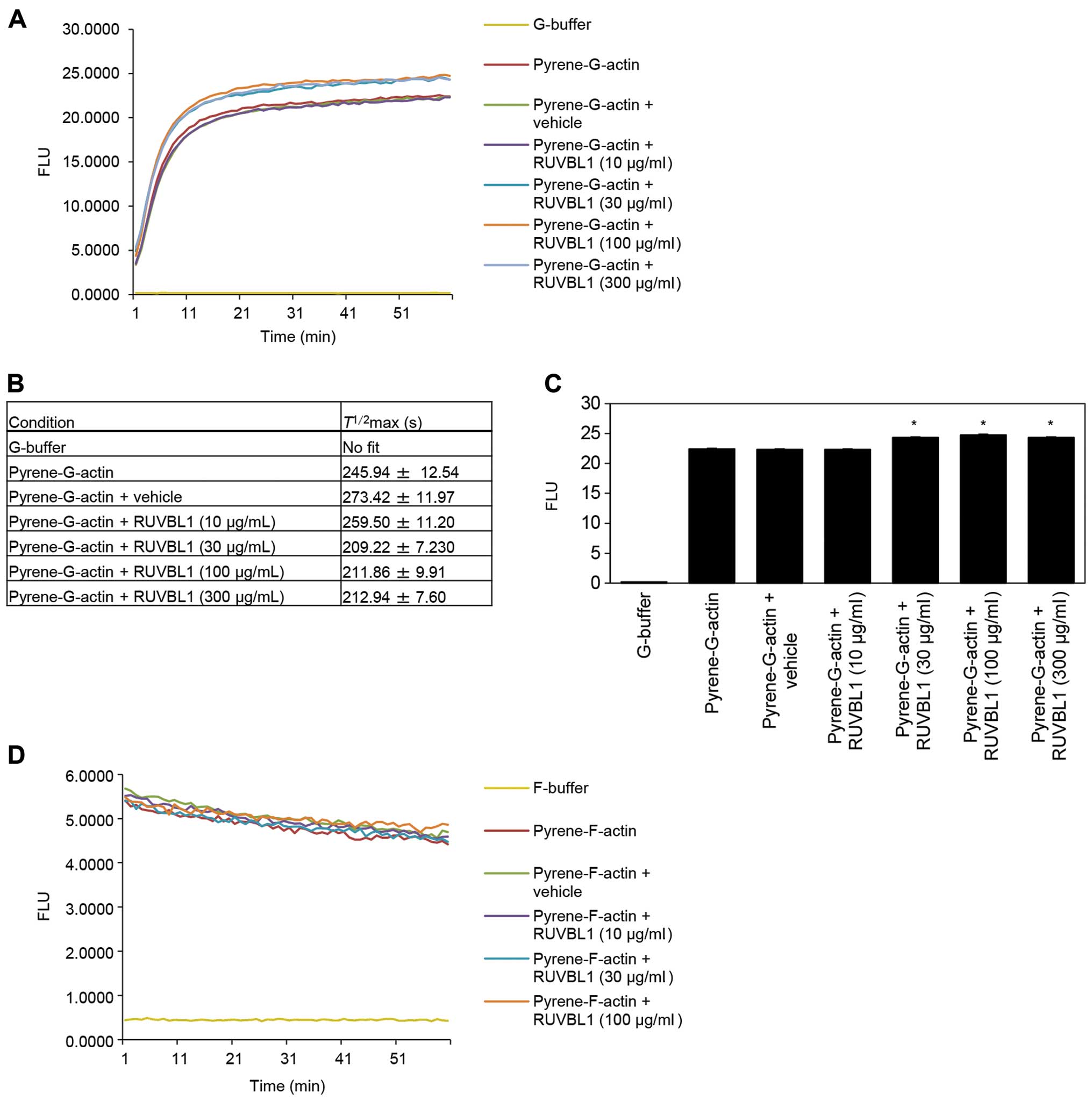
To investigate whether RUVBL1 influenced the
structural organization of F-actin, we assessed whether RUVBL1 had
an effect on the apparent rate of actin polymerization. Actin
polymerization was monitored using an in vitro
pyrene-labeled G-actin polymerization assay. The kinetics of actin
polymerization in the presence of 30, 100 or 300 μg/ml
RUVBL1 was measured by increase in pyrene fluorescence (Fig. 4A). Half-maximal saturated
polymerization values [T1/2max (s)] calculated
from Boltzmann sigmoidal curve fits are summarized in Fig. 4B. It is likely that
T1/2max values for actin reactions in the
presence of 30, 100 or 300 μg/ml RUVBL1 were faster than
that in vehicle control. A significant difference in synergistic
polymerization rates between vehicle control and added RUVBL1
became evident at a time point of 60 min (Fig. 4C). In contrast, RUVBL1 had no
discernible effect on the kinetics of in vitro
pyrene-labeled F-actin depolymerisation assays that involved
measurement of F-actin fluorescence (Fig. 4D). These results indicated that
binding of RUVBL1 to F-actin enhanced elongation of existing actin
filaments.
Effects of RUVBL1 on G-actin
concentration in cell protrusions
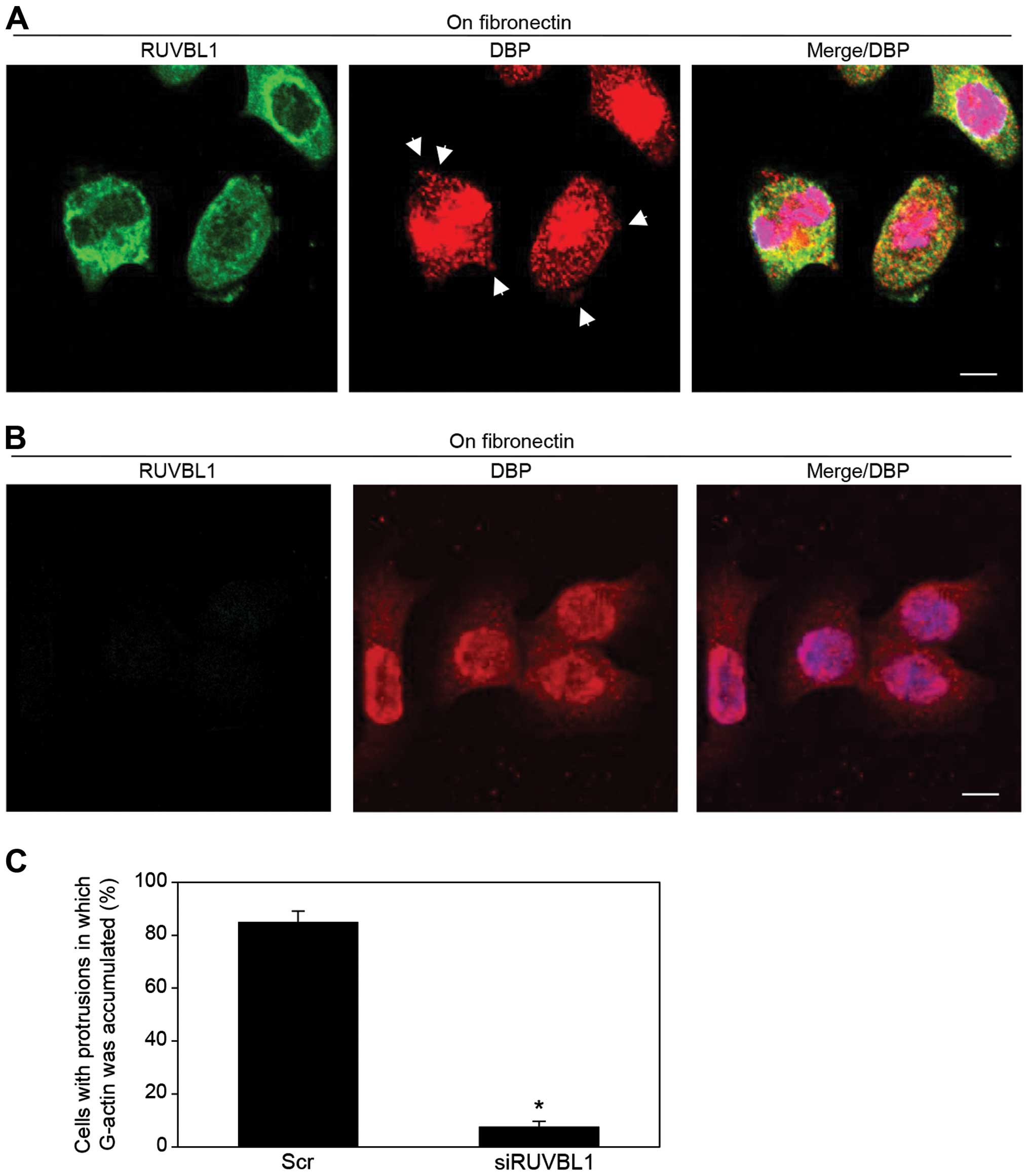
Immunocytochemistry and an antibody against vitamin
D-binding protein [DBP (27)], a
protein that specifically binds G-actin, was used to indirectly
assess the presence and localization of G-actin in
fibronectin-stimulated S2-013 cells; the cells had transfected with
either scrambled control-siRNA or RUVBL1-siRNA. In control cells,
G-actin accumulated in cell protrusions of motile cells (arrows in
Fig. 5A); notably, RUVBL1 and
peripheral F-actin filaments were colocalized in these cell
protrusions (Fig. 3D), but RUVBL1
did not bind to G-actin in these control protrusions (Fig. 5A). In contrast, siRNA-mediated
suppression of RUVBL1 inhibited the accumulation of G-actin near
the cell membranes (Fig. 5B). A
significant difference in rates of G-actin concentration in cell
protrusion between scrambled control and knockdown of RUVBL is
shown in Fig. 5C. Suppression of
RUVBL1 decreased G-actin concentration in the protrusion of S2-013.
These results indicated that RUVBL1 played a role in induction of
G-actin concentration in cell protrusions.
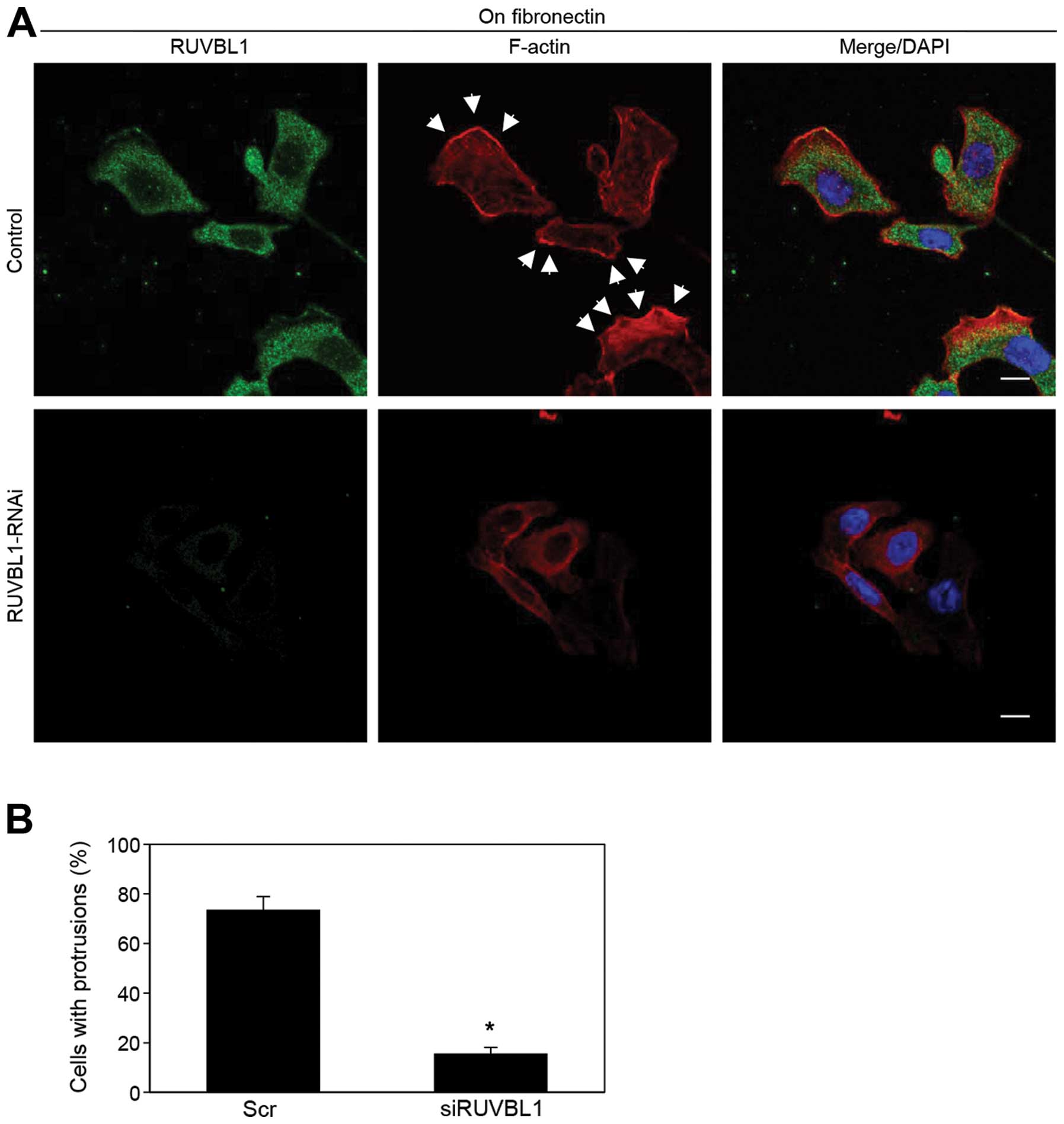
RUVBL1 induces the formation of membrane
protrusions
We analyzed peripheral F-actin structures in
membrane ruffles of S2-013 cells transfected with scrambled control
or RUVBL1 siRNA; all cells were cultured on fibronectin. Knockdown
of RUVBL1 inhibited the increase in peripheral F-actin structures
compared to control cells (Fig.
6A). To determine whether RUVBL1 that increased F-actin
structures in cell protrusions has a role in inducing membrane
protrusions, immunofluorescence was carried out; all cells were
treated with either scrambled control or RUVBL1 RNAi and cultured
on fibronectin. siRNA-mediated suppression of RUVBL1 significantly
inhibited the formation of fibronectin-mediated membrane
protrusions in which peripheral actin structures were abundant,
compared to scrambled control (Fig.
6B). These results indicated that RUVBL1 induced peripheral
F-actin polymerization in cell protrusions that promoted formation
of cell protrusions, and that these RUVBL1-mediated actin
rearrangements promote the motility and invasiveness of PDAC
cells.
Discussion
RUVBL1 is overexpressed in a variety of human solid
tumors including, colorectal (28), gastric (29), bladder (30) and non-small cell lung (31) cancers. As noted in the
Introduction, RUVBL1 is an ATPase protein that is associated with
several chromatin-remodelling complexes in the nucleus. RUVBL1
localized in the nucleus promotes histone H3K9 trimethylation and
negatively regulates p53 expression in colon cancer cells (26). Substantial evidence clearly
demonstrates that RUVBL1 localized in the nucleus is required for
cell growth and viability (32,33);
however, the role of RUVBL1 in motility and invasion of cancer
cells has not been fully examined. Here, we found that RUVBL1
localized mainly to the cytoplasm and some population of
cytoplasmic RUVBL1 accumulated in cell protrusions of PDAC cells.
We describe a newly discovered function for RUVBL1 localized at
cell protrusions in cell motility and invasion in PDAC. RUVBL1
played a role as an F-actin-binding protein in mediating actin
polymerization, but it did not interact with G-actin. Notably,
RUVBL1 enhanced elongation of existing actin filaments via the
direct binding to F-actin in cell protrusions of spreading PDAC
cells. PDAC cells depend on actin-based motility to invade nearby
organs such as the duodenum, stomach or liver (34). Knockdown of RUVBL1 inhibited the
formation of cell protrusions via decrease in peripheral actin
rearrangements. Suppression of RUVBL1 did not affect cell growth in
an in vitro MTT assay using S2-013 and PANC-1 (data not
shown); therefore, it is likely that cytoplasmic RUVBL1 was
associated with actin-based motility and invasiveness of PDAC
cells.
G-actin is the building block for F-actin, and local
concentrations of G-actin directly affect the rate of filament
assembly (35). Cell protrusions
produced in motile cells contain G-actin (36), but whether spatio-temporal
regulation of G-actin is important to cancer cell motility and
invasion is unknown. We examined the intracellular distribution of
G-actin and of F-actin in RNAi-treated S2-013 cells; all cells were
treated with either scrambled control or RUVBL1 RNAi and cultured
on fibronectin. G-actin was abundantly localized to membrane
protrusions in scrambled control cells, whereas the peripheral
concentration of G-actin was inhibited by suppression of RUVBL1
(Fig. 5A–C). RUVBL1 was associated
with polymerization of G-actin, but it failed to depolymerize
F-actin in in vitro assays (Fig. 4A–D); therefore, RUVBL1 was probably
involved in the spatial arrangement of G-actin and its localization
to membrane protrusions, resulted in increased F-actin structures
in the protrusions. These results indicate that reductions in
G-actin concentration were tightly associated with cessation and
retraction of actin polymerization (Fig. 6A) and membrane protrusions
(Fig. 6B) in RUVBL1 RNAi cells.
Consistent with our findings, the concentration of G-actin in cell
protrusions is sufficiently high to support actin polymerization at
the tip of the cell protrusions in breast cancer cells (37).
Crawling cells typically move over substrates by the
combined effects of i) actin-based protrusions at leading cell
edges; ii) adhesion to the substrate; and iii) myosin-based
contraction at the cell rear (38). Dynamic, actin-based plasma membrane
protrusions that control growth cone path-finding include i)
lamellipodia in which the actin cytoskeleton assumes a crosslinked
and branched meshwork; and ii) filo-podia, which consist of
parallel bundles of actin filaments protruding from the growth cone
or lamellipodial margin (39).
Migratory competence of tumor cells requires activation of the
motile cycle, the first step of which is actin remodeling; this
remodeling drives the formation of cell protrusions, defines the
direction of migration, and initiates the growth of the
lamellipodium (40). In this
study, fibronectin-stimulated peripheral actin rearrangements and
subsequent formation of membrane protrusions were inhibited when
RUVBL1 RNAi S2-013 cells were plated on fibronectin. Thus, RUVBL1,
which associates with F-actin filaments, is probably a
physiological activator that induces peripheral actin
rearrangements, which themselves promote formation of membrane
protrusions.
The findings presented in this report are consistent
with the hypothesis that RUVBL1 that localized in cell protrusions
has pivotal roles in the coordinated regulation of cortical actin
changes via the direct binding to F-actin. We have established the
functional significance of RUVBL1 and that RUVBL1-mediated actin
polymerization promoted i) the spatio-temporal localization of
G-actin to protrusions, and ii) the formation of additional
membrane protrusions in motile PDAC cells; the RUVBL1-mediated
actin polymerization may play an important role in PDAC motility
and invasiveness. Inhibition of binding between RUVBL1 and actin
filaments may be a rational approach to a targeted molecular
therapy for PDAC because such a therapy would inhibit the formation
of cell protrusions and consequently limit the motility and
invasiveness of PDAC cells.
Acknowledgements
We thank Aki Tanouchi and Chiaki Okura
for their excellent technical assistance. This study was supported
by the Grants-in-Aid for Scientific Research (KAKENHI) (to K.T. and
S.I.), by the Pancreas Research Foundation of Japan (to K.T.), and
by the Japanese Foundation for Multidisciplinary Treatment of
Cancer (to K.T.).
References
|
1.
|
Ammelburg M, Frickey T and Lupas AN:
Classification of AAA+ proteins. J Struct. 156:2–11. 2006.
|
|
2.
|
Kanemaki M, Kurokawa Y, Matsuura T, Makino
Y, Masani A, Okazaki K, Morishita T and Tamura TA: TIP49b, a new
RuvB-like DNA helicase, is included in a complex together with
another RuvB-like DNA helicase, TIP49a. J Biol Chem.
274:22437–22444. 1999. View Article : Google Scholar
|
|
3.
|
Qiu XB, Lin YL, Thome KC, Pian P, Schlegel
BP, Weremowicz S, Parvin JD and Dutta A: An eukaryotic RuvB-like
protein (RUVBL1) essential for growth. J Biol Chem.
273:27786–27793. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Wood MA, McMahon SB and Cole MD: An
ATPase/helicase complex is an essential cofactor for oncogenic
transformation by c-Myc. Mol Cell. 5:321–330. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Bauer A, Chauvet S, Huber O, Usseglio F,
Rothbacher U, Aragnol D, Kemler R and Pradel J: Pontin52 and
reptin52 function as antagonistic regulators of beta-catenin
signalling activity. EMBO J. 19:6121–6130. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Hoverter NP and Waterman ML: A Wnt-fall
for gene regulation: repression. Sci Signal. 1:e432008. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Ni L, Saeki M, Xu L, Nakahara H, Saijo M,
Tanaka K and Kamisaki Y: RPAP3 interacts with Reptin to regulate
UV-induced phosphorylation of H2AX and DNA damage. J Cell Biochem.
106:920–928. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Ikura T, Ogryzko VV, Grigoriev M, Groisman
R, Wang J, Horikoshi M, Scully R, Qin J and Nakatani Y: Involvement
of the TIP60 histone acetylase complex in DNA repair and apoptosis.
Cell. 102:463–473. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Venteicher AS, Meng Z, Mason PJ, Veenstra
TD and Artandi SE: Identification of ATPases pontin and reptin as
telomerase components essential for holoenzyme assembly. Cell.
132:945–957. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Rousseau B, Ménard L, Haurie V, Taras D,
Blanc JF, Moreau-Gaudry F, Metzler P, Hugues M, Boyault S, Lemière
S, Canron X, Costet P, Cole M, Balabaud C, Bioulac-Sage P,
Zucman-Rossi J and Rosenbaum J: Overexpression and role of the
ATPase and putative DNA helicase RuvB-like 2 in human
hepatocellular carcinoma. Hepatology. 46:1108–1118. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Ménard L, Taras D, Grigoletto A, Haurie V,
Nicou A, Dugot-Senant N, Costet P, Rousseau B and Rosenbaum J: In
vivo silencing of Reptin blocks the progression of human
hepatocellular carcinoma in xenografts and is associated with
replicative senescence. J Hepatol. 52:681–689. 2010.PubMed/NCBI
|
|
12.
|
Grigoletto A, Lestienne P and Rosenbaum J:
The multifaceted proteins reptin and pontin as major players in
cancer. Biochim Biophys Acta. 1815:147–157. 2011.PubMed/NCBI
|
|
13.
|
Izumi N, Yamashita A, Iwamatsu A, Kurata
R, Nakamura H, Saari B, Hirano H, Anderson P and Ohno S: AAA+
proteins RUVBL1 and RUVBL2 coordinate PIKK activity and function in
nonsense-mediated mRNA decay. Sci Signal. 23:ra272010.
|
|
14.
|
Hawley SB, Tamura T and Miles LA:
Purification, cloning, and characterization of a profibrinolytic
plasminogen-binding protein, TIP49a. J Biol Chem. 276:179–186.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Ridley AJ: Rho GTPases and cell migration.
J Cell Sci. 114:2713–2722. 2001.PubMed/NCBI
|
|
16.
|
Totsukawa G, Wu Y, Sasaki Y, Hartshorne
DJ, Yamakita Y, Yamashiro S and Matsumura F: Distinct roles of MLCK
and ROCK in the regulation of membrane protrusions and focal
adhesion dynamics during cell migration of fibroblasts. J Cell
Biol. 164:427–439. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Pollard TD, Blanchoin L and Mullins RD:
Molecular mechanisms controlling actin filament dynamics in
nonmuscle cells. Annu Rev Biophys Biomol Struct. 29:545–576. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Wang W, Goswami S, Sahai E, Wyckoff JB,
Segall JE and Condeelis JS: Tumor cells caught in the act of
invading: their strategy for enhanced cell motility. Trends Cell
Biol. 15:138–145. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Pollard TD and Borisy GB: Cellular
motility driven by assembly and disassembly of actin filaments.
Cell. 112:453–465. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Ponti A, Matov A, Adams M, Gupton S,
Waterman-Storer CM and Danuser G: Periodic patterns of actin
turnover in lamellipodia and lamellae of migrating epithelial cells
analyzed by quantitative Fluorescent Speckle Microscopy. Biophys J.
89:3456–3469. 2005. View Article : Google Scholar
|
|
21.
|
Baumgart M, Heinmöller E, Horstmann O,
Becker H and Ghadimi BM: The genetic basis of sporadic pancreatic
cancer. Cell Oncol. 27:3–13. 2005.
|
|
22.
|
Taniuchi K, Yokotani K and Saibara T: BART
inhibits pancreatic cancer cell invasion by Rac1 inactivation
through direct binding to active Rac1. Neoplasia. 14:440–450.
2012.PubMed/NCBI
|
|
23.
|
Taniuchi K, Iwasaki S and Saibara T: BART
inhibits pancreatic cancer cell invasion by inhibiting
ARL2-mediated RhoA inactivation. Int J Oncol. 39:1243–1252.
2011.PubMed/NCBI
|
|
24.
|
Taniuchi K, Yokotani K and Saibara T: BART
inhibits pancreatic cancer cell invasion by PKCα inactivation
through binding to ANX7. PLoS One. 7:e356742012.PubMed/NCBI
|
|
25.
|
Iwamura T, Katsuki T and Ide K:
Establishment and characterization of a human pancreatic cancer
cell line (SUIT-2) producing carcinoembryonic antigen and
carbohydrate antigen 19–9. Jpn J Cancer Res. 78:54–62.
1987.PubMed/NCBI
|
|
26.
|
Taniue K, Oda T, Hayashi T, Okuno M and
Akiyama T: A member of the ETS family, EHF, and the ATPase RUVBL1
inhibit p53-mediated apoptosis. EMBO Rep. 12:682–689. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Meijerman I, Blom WM, de Bont HJ, Mulder
GJ and Nagelkerke JF: Changes of G-actin localisation in the
mitotic spindle region or nucleus during mitosis and after heat
shock: a histochemical study of G-actin in various cell lines with
fluorescent labelled vitamin D-binding protein. Biochim Biophys
Acta. 1452:12–24. 1999. View Article : Google Scholar
|
|
28.
|
Ki DH, Jeung HC, Park CH, Kang SH, Lee GY,
Lee WS, Kim NK, Chung HC and Rha SY: Whole genome analysis for
liver metastasis gene signatures in colorectal cancer. Int J
Cancer. 121:2005–2012. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Li W, Zeng J, Li Q, Zhao L, Liu T,
Björkholm M, Jia J and Xu D: Reptin is required for the
transcription of telomerase reverse transcriptase and
over-expressed in gastric cancer. Mol Cancer. 9:1322010. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Dyrskjøt L, Kruhøffer M, Thykjaer T,
Marcussen N, Jensen JL, Møller K and Ørntoft TF: Gene expression in
the urinary bladder: a common carcinoma in situ gene expression
signature exists disregarding histopathological classification.
Cancer Res. 64:4040–4048. 2004.PubMed/NCBI
|
|
31.
|
Dehan E, Ben-Dor A, Liao W, Lipson D,
Frimer H, Rienstein S, Simansky D, Krupsky M, Yaron P, Friedman E,
Rechavi G, Perlman M, Aviram-Goldring A, Izraeli S, Bittner M,
Yakhini Z and Kaminski N: Chromosomal aberrations and gene
expression profiles in non-small cell lung cancer. Lung Cancer.
56:175–184. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Ducat D, Kawaguchi S, Liu H, Yates JR III
and Zheng Y: Regulation of microtubule assembly and organization in
mitosis by the AAA+ ATPase pontin. Mol Biol Cell. 19:3097–3110.
2008.
|
|
33.
|
Schlabach MR, Luo J, Solimini NL, Hu G, Xu
Q, Li MZ, Zhao Z, Smogorzewska A, Sowa ME, Ang XL, Westbrook TF,
Liang AC, Chang K, Hackett JA, Harper JW, Hannon GJ and Elledge SJ:
Cancer proliferation gene discovery through functional genomics.
Science. 319:620–624. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Kedrin D, van Rheenen J and Segall JE:
Cell motility and cytoskeletal regulation in invasion and
metastasis. J Mammary Gland Biol. 12:143–152. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Lee CW, Vitriol EA, Shim S, Wise AL,
Velayutham RP and Zheng JQ: Dynamic localization of G-actin during
membrane protrusion in neuronal motility. Current Biology.
23:1046–1056. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Van Goor D, Hyland C, Schaefer AW and
Forscher P: The role of actin turnover in retrograde actin network
flow in neuronal growth cones. PLoS One. 7:e309592012.PubMed/NCBI
|
|
37.
|
Kiuchi T, Nagai T, Ohashi K and Mizuno K:
Measurements of spatiotemporal changes in G-actin concentration
reveal its effect on stimulus-induced actin assembly and
lamellipodium extension. J Cell Biol. 193:365–380. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Mogilner A and Keren K: The shape of
motile cells. Curr Biol. 19:R762–R771. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Gallo G and Letourneau PC: Regulation of
growth cone actin filaments by guidance cues. J Neurobiol.
58:92–102. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Eiseler T, Döppler H, Yan IK, Kitatani K,
Mizuno K and Storz P: Protein kinase D1 regulates cofilin-mediated
F-actin reorganization and cell motility through slingshot. Nat
Cell Biol. 11:545–556. 2009. View Article : Google Scholar : PubMed/NCBI
|