Cancer is one of the leading causes of mortality,
accounting for ~13% of deaths worldwide (1). Carcinogenesis is characterized by the
enrichment of cells displaying phenotypes that result in excessive
proliferation, resistance to apoptosis, and metabolic dysregulation
(2). The cellular machineries that
determine these cellular behaviors remain unclear. However, it is
known that post-translational modification of proteins is a common
mechanism responsible for fine-tuning intracellular signaling and
metabolic pathways that either facilitate or impair malignant
transformation.
Lysine acetylation is a fundamental modification
that profoundly affects the activity of a protein, as evidenced by
proteomic studies revealing that >2,000 proteins involved in
diverse cellular processes are acetylated (3–5). The
lysine acetylation status of individual proteins is concurrently
determined by the balance between acetylation and deacetylation, a
process catalyzed by a cohort of acetyltransferases and
deacetylases, respectively. In contrast to the largely constitutive
and nonselective activities of several known acetyltransferases,
the mechanism of protein deacetylation has been studied
extensively, leading to the characterization of four classes of
deacetylase, all of which show variable catalytic characteristics
and substrate profiles. Sirtuins (SIRTs), a family comprising seven
members (SIRT 1–7), are class III deacetylases that deacetylate
diverse histone or non-histone proteins in a nicotinamide adenine
dinucleotide (NAD)-dependent manner. SIRTs are involved in
regulating diverse cellular activities such as cell proliferation,
survival, and metabolism (6–9).
Among this evolutionarily conserved deacetylase family, SIRT3 is a
primary mitochondrial deacetylase, whose dysregulation is involved
in the development of diseases such as diabetes (10), myocardial injury (11), and cancer (12,13).
In particular, SIRT3 plays a conflicting role in cancer initiation
and progression; the signaling networks involved in these processes
remain unclear (13–16).
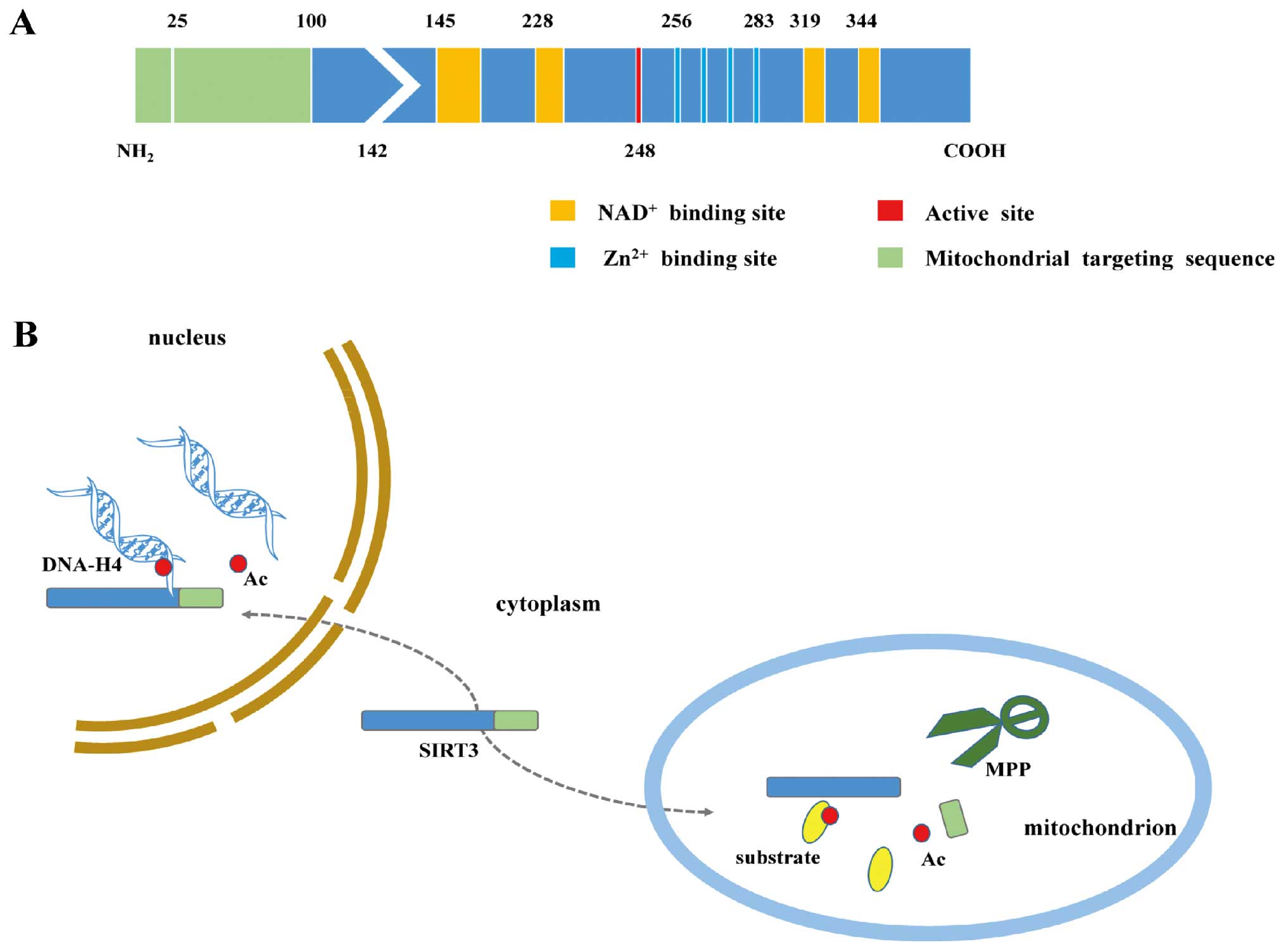
The SIRT3 protein comprises two main functional
domains. The larger Rossmann domain provides sites for
NAD+ binding, whereas the smaller domain binds zinc
atoms. The cleft between the two domains serves as a docking site
for acetylation substrates (17,18).
SIRT3 is first expressed in the cytoplasm as a 399-amino acid (44
kDa) inactive precursor. This precursor is subsequently targeted to
the mitochondrion, where it is cleaved at the N-terminus by the
mitochondrial matrix processing peptidase protein (MPP) to generate
a mature 28-kDa protein comprising 257 amino acid residues
(19–21). Mitochondrial trafficking depends on
the N-terminal 100 amino acids, of which residues 1–25 play an
important role both in mitochondrial targeting and proteolytic
processing (19). Intriguingly,
full-length SIRT3 also exists in the nucleus, where it normally
exhibits histone deacetylation activity and undergoes cytoplasmic
translocation under stress conditions (22) (Fig.
1).
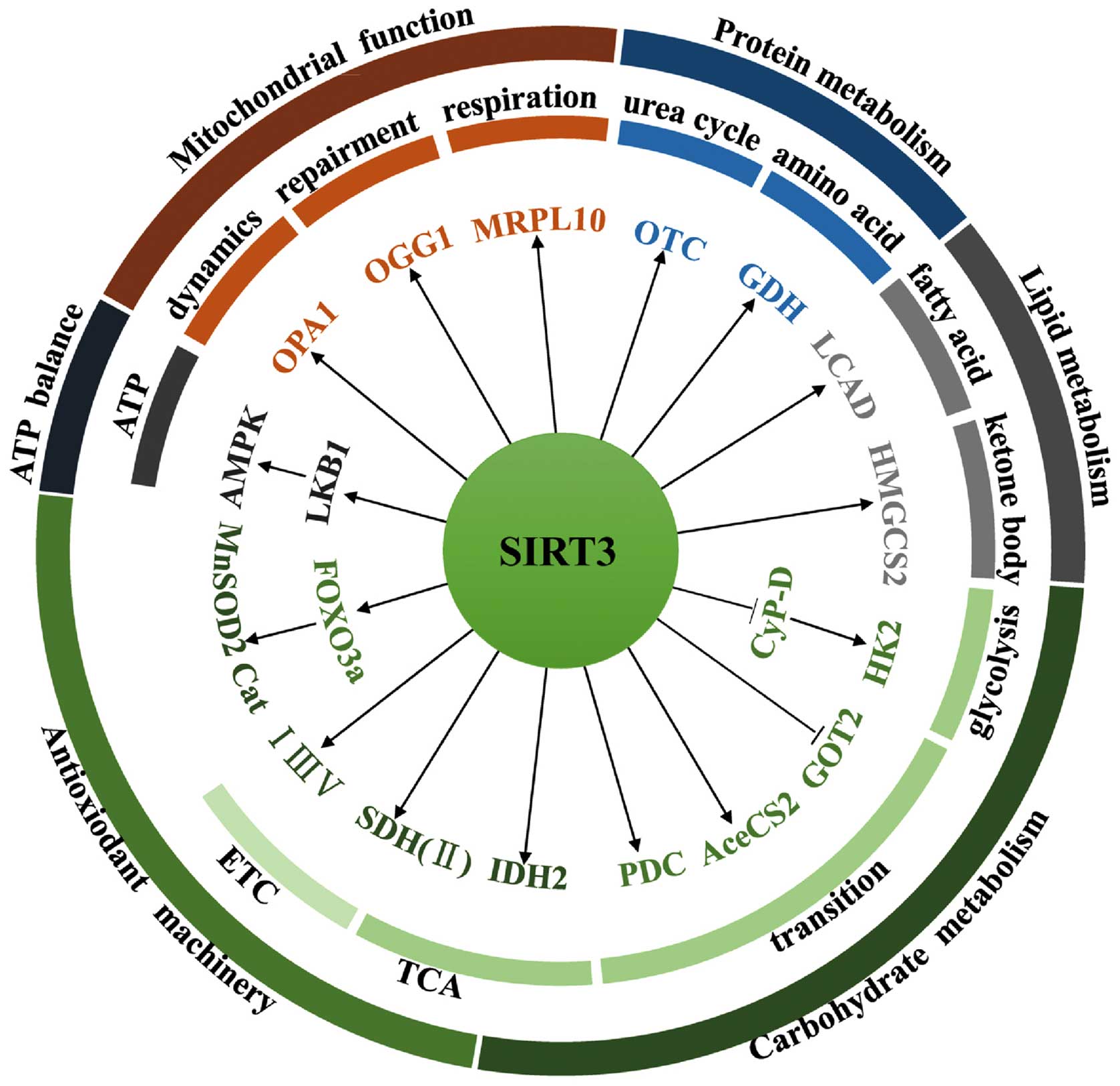
SIRT3 is the primary mitochondrial deacetylase that
determines the acetylation level of many mitochondrial proteins
(23). As a result, SIRT3 plays a
pivotal role in varied cellular events such as nutrient metabolism,
ATP balance, antioxidant machinery regulation, and some fundamental
mitochondrial functions (Fig.
2).
SIRT3 orchestrates global shifts in nutrient
metabolism by regulating the activity of diverse substrates
involved in multiple metabolic pathways. In carbohydrate
metabolism, for example, SIRT3 deacetylates and inactivates
cyclophilin D (CyP-D), resulting in the dissociation of hexokinase
2 (HK2) from the mitochondrial outer membrane and the subsequent
inhibition of glycolysis (24).
SIRT3 can also block the malate-aspartate shuttle by inhibiting
glutamate oxaloacetate transaminase 2 (GOT2) (25). Acetyl-CoA is a primary entry point
in the tricarboxylic acid cycle (TCA). To this end, SIRT3 can
deacetylate and activate acetyl-CoA synthetase 2 (AceCS2), thereby
providing increased acetyl-CoA to the TCA (26,27).
SIRT3 also increases acetyl-CoA levels by deacetylating the
upstream enzymes responsible for activating the pyruvate
dehydrogenase complex (PDC) (28).
In addition, SIRT3 accelerates the oxidation of isocitrate by
targeting isocitrate dehydrogenase 2 (IDH2), which plays a vital
role in glutathione antioxidant systems by generating NADPH
(29). Furthermore, SIRT3
coordinates the TCA cycle and the electron transport chain (ETC) by
activating complex II, e.g., the flavoprotein subunit of succinate
dehydrogenase (SDH) that plays a critical role in both of these
processes (30,31). Finally, SIRT3 interacts with
complexes I, III, and IV, thereby improving the efficiency of
electron transport (10,23).
SIRT3 participates in amino acid metabolism and in
the urea cycle. SIRT3 deacetylates and increases the enzymatic
activity of glutamate dehydrogenase (GDH), a key enzyme involved in
amino acid oxidation (32).
Moreover, SIRT3 deacetylates ornithine transcarbamylase (OTC),
thereby increasing urea cycle flux during energy restriction
(33).
During lipid metabolism, SIRT3 accelerates the
process of fatty acid oxidation by modulating the activity of long
chain acyl-CoA dehydrogenase (LCAD) (34). SIRT3 also enhances the enzymatic
activity of 3-hydroxy-3-methylglutaryl CoA synthase 2 (HMGCS2) to
promote synthesis of the ketone body, β-hydroxybutyrate (35).
SIRT3 acts as a watchdog that maintains redox
homeostasis by regulating the antioxidant machinery and ATP
balance. For instance, SIRT3 deacetylates and activates the
transcriptional factor forkhead box O3 (FOXO3a), leading to
upregulation of manganese-dependent superoxide dismutase 2 (MnSOD2)
and catalase (Cat), two important antioxidant proteins that deplete
intracellular reactive oxygen species (ROS) (36). As mentioned above, SIRT3 also
licenses glutathione antioxidant systems by targeting IDH2
(29), and facilitates electron
transportation by preventing slow ETC flux-triggered electron
leaking and ROS production (23).
SIRT3 also interacts with and deacetylates liver kinase B1 (LKB1).
The deacetylation of LKB1 activates 5′ AMP-activated protein kinase
(AMPK), leading to high levels of ATP (37). AMPK in turn increases SIRT3
activity by increasing cellular NAD+ levels via feedback
regulation (38). Furthermore, the
activity of SIRT3 depends on a high ratio of cellular
NAD+ to NADH, an indication of energy-deficient state
that increases AMPK activity (38,39).
SIRT3 plays a fundamental role in mitochondrial
functions such as repair, respiration, and dynamics. For example,
SIRT3 deacetylates and prevents the degradation of 8-oxoguanine
glycosylase 1 (OGG1), a DNA glycosylase enzyme responsible for
repair of mitochondrial DNA (40).
SIRT3 also regulates mitochondrial respiration by targeting
mitochondrial ribosomal protein 10 (MRPL10) (41). In addition, SIRT3 deacetylates and
activates optic atrophy 1 (OPA1), thereby modulating mitochondrial
dynamics (42). SIRT3 also plays a
role in maintaining mitochondrial permeability transition and
preventing accumulation of misfolded proteins within mitochondria
[the so-called mitochondrial unfolded protein response (UPRmt)]
(43,44).
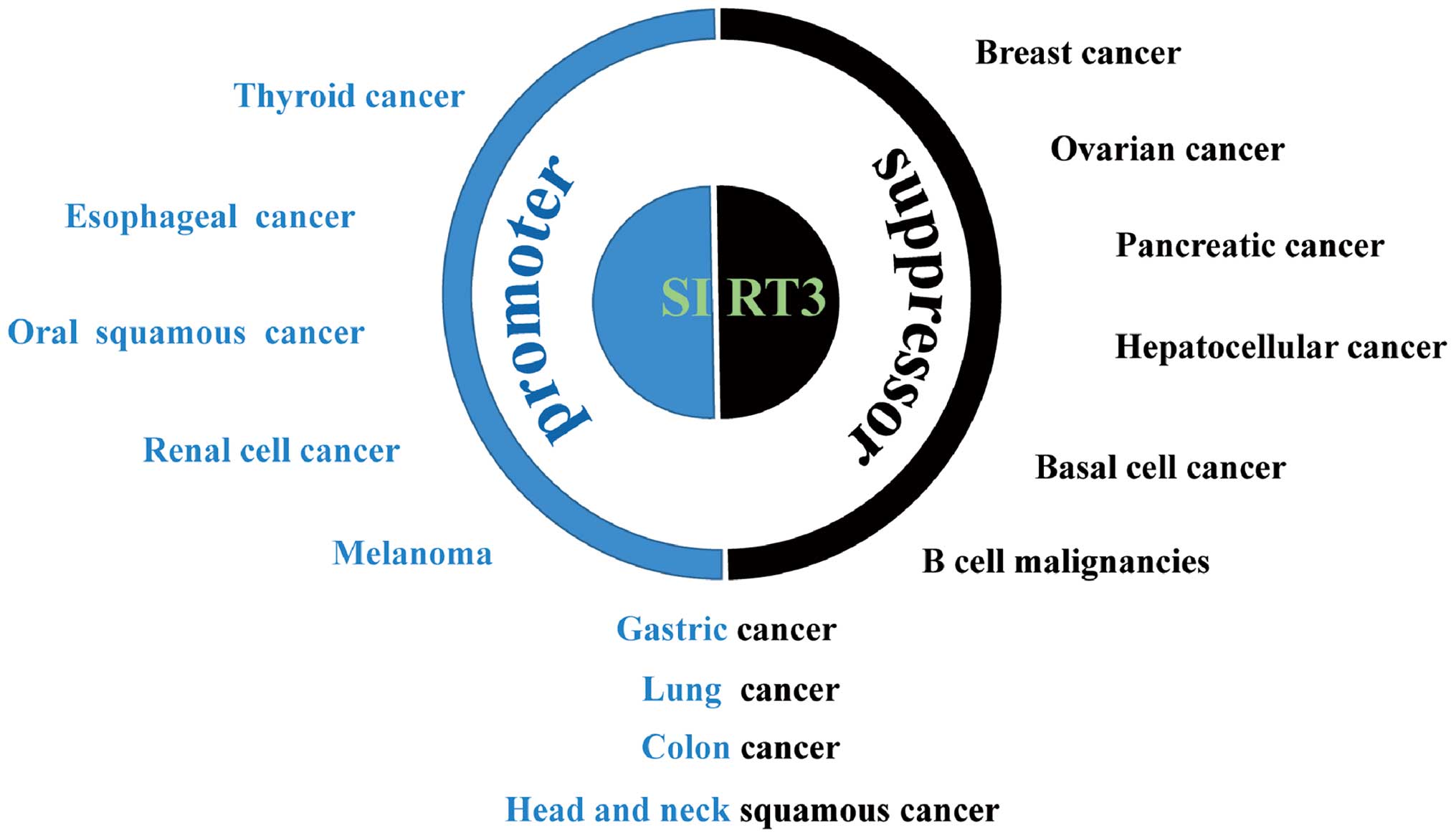
The causal relationship between SIRT3 deregulation
and cancer is well documented. However, SIRT3 may exhibit
tumor-promoting or -suppressive capacity depending upon the type of
cancer and, probably, the context of intracellular signal pathways
(Fig. 3). SIRT3 also plays
conflicting roles in malignancies originating from the same types
of tissue, e.g., gastric cancer (45,46),
lung cancer (47,48), colon cancer (49,50),
and head and neck squamous cell cancer (51–53).
SIRT3 is an oncogenic factor for many types of
carcinoma: high expression correlates with hallmarks of malignancy
and a poor clinical prognosis. This is exemplified by esophageal
cancer, in which high expression of SIRT3 is associated with a poor
outcome; indeed, the level of SIRT3 expression is an independent
predictor for cancer prognosis (54). High SIRT3 expression is also
associated with poor prognosis in patients with grade 3 breast
cancer; SIRT3 knockdown increases the efficacy of cisplatin and
tamoxifen, which are used to treat this disease (55). SIRT3 is also overexpressed in oral
squamous cell carcinoma cells. Silencing of SIRT3 inhibits cancer
cell proliferation, reduces tumor burden, and sensitizes these
cells to radiation or chemotherapy (56). Furthermore, SIRT3 expression in
markedly elevated in melanoma compared with melanocytic nevi
tissues; the same is true in cell lines derived from these tissues.
Knockdown of SIRT3 reduces proliferation, colony formation, and
cellular migration, and induces senescence and G1-phase arrest in
human melanoma cells, with a corresponding increase in p16 and p21
expression and a decrease in expression of cyclin D1 and E1 and
cyclin-dependent kinases (CDKs) 2, 4, and 6. SIRT3 knockdown
significantly inhibits tumorigenesis in a xenograft model of
melanoma. Conversely, forced overexpression of SIRT3 increases the
proliferative potential of melanoma cell lines (57). SIRT3 is also overexpressed in renal
cancer, in which SIRT3 depletion or an inactive mutant inhibits
cell proliferation and xenograft tumor growth (58). Immunohistochemical analysis of
thyroid cancer demonstrated that SIRT3 expression is associated
with tumors showing low SIRT3 levels in benign thyroid tissues,
moderately increased levels in follicular carcinomas, and high
levels in papillary thyroid carcinomas and well-differentiated
thyroid carcinomas. Such a distribution is in accordance with the
expression pattern of nicotinamide phosphoribosyltransferase
(NAMPT), a rate-limiting enzyme in NAD+ biosynthesis,
supporting the critical involvement of the
NAD+-dependent deacetylase, SIRT3, in thyroid gland
(59).
SIRT3 inhibits proliferation, metabolic
reprogramming, and other malignant phenotypes of cells, and
correlates with a good outcome for many types of cancer. For
instance, SIRT3 expression is markedly lower in breast cancer cells
than in paired normal breast epithelium, and lower SIRT3 expression
is associated with shorter locoregional relapse-free survival
(60). SIRT3 expression also
correlates with clinical characteristics such as the distribution
of estrogen receptors and levels of oxidative stress (61). SIRT3 induces inhibition of
glycolysis and reverses metabolic reprogramming in breast cell
lines (24). The tumor-suppressive
role of SIRT3 is also manifest in hepatocellular cancer (HCC), in
which SIRT3 expression in cancerous tissues is much lower than that
in adjacent non-cancerous tissue. Survival analyses indicate that
high SIRT3 expression correlates with increased overall survival
and recurrence-free survival. In addition, lower SIRT3 expression
is associated with unfavorable clinicopathological parameters such
as differentiation, clinical stage, and tumor multiplicity
(62–64). Overexpression of SIRT3 inhibits
growth and induces apoptosis in HCC cells and increases their
sensitivity to chemotherapeutic agents (65–67).
Furthermore, lower SIRT3 expression correlates with increased
aggressiveness of pancreatic cancer and a shorter time to relapse
and patient survival in the absence of chemotherapeutic
intervention, suggesting that SIRT3 is tumor-suppressive in the
context of pancreatic cancer and may also represent a novel
predictive biomarker for chemotherapeutic response (68). Mechanistically, SIRT3 inhibits
pancreatic cell metabolism and growth by impeding malate-aspartate
shuttle activity (25) or
preventing iron metabolism (69).
SIRT3 expression in samples from patients with B cell malignancies
is lower than that in primary B cells from healthy donors, and low
SIRT3 expression predicts worse overall survival. Overexpression of
SIRT3 decreases the proliferation and diminishes the Warburg-like
phenotype of SIRT3-deficient B cell lymphoma cells (70). As a potential prognostic biomarker
for basal cell cancer, SIRT3 expression is lower in cancerous
tissues than in normal tissues (71). In addition, SIRT3 expression is
significantly downregulated in metastatic ovarian cancer tissues
and in highly metastatic ovarian cancer cell lines. Knockdown of
SIRT3 increases ovarian cancer cell migration and invasion in
vitro and liver metastasis in vivo by downregulating
Twist, thereby suppressing epithelial-to-mesenchymal transition
(EMT) (72).
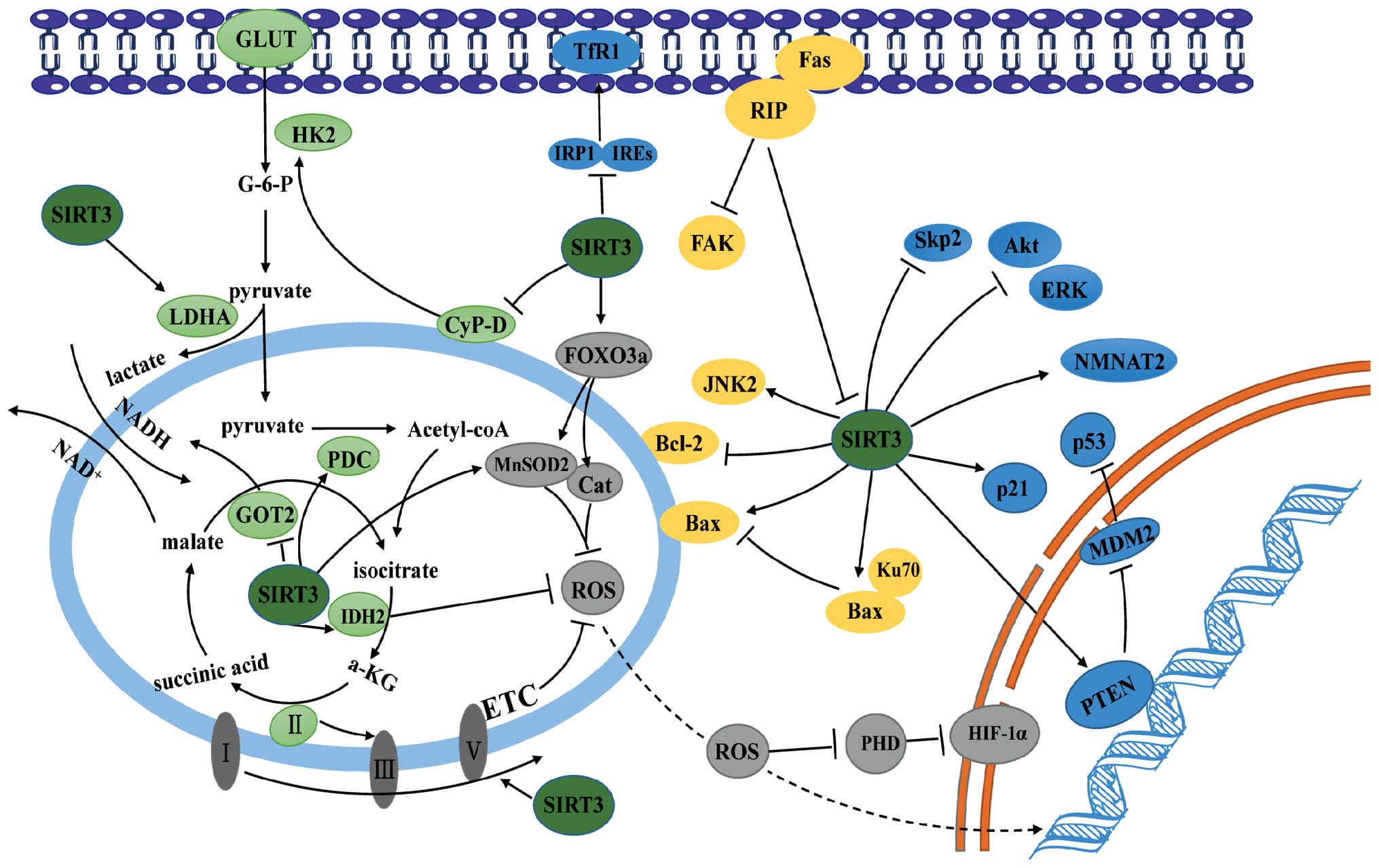
The precise mechanism by which SIRT3 regulates the
pathogenesis of cancer remains a matter for debate. Nevertheless,
depletion of ROS, modulation of metabolism, and regulation of
proliferative or apoptotic pathways provide a biochemical rationale
for its regulatory role in cancer (Fig. 4).
ROS either expedite or inhibit malignancy. These
controversial findings are attributed to the diverse roles played
by ROS during cancer development (73). ROS can damage macromolecules like
DNA by oxidizing specific intracellular chemical moieties, leading
to genetic mutations and the activation of biochemical pathways
that stimulate cell proliferation and neoplastic transformation
(74). As shown above, SIRT3
reduces ROS levels by activating the antioxidant defense system.
Loss of SIRT3 triggers oxidative damage, activates ROS-mediated
signaling, and causes carcinogenesis in various cell types
(14). For example, mouse
embryonic fibroblasts (MEFs) lacking SIRT3 exhibit increased ROS
levels and high genomic instability, and can be transformed by
ectopic expression of a single oncogene. By contrast, wild-type
MEFs require both Myc and Ras to acquire a malignant phenotype
(75). SIRT3 knockdown facilitates
tumorigenesis in xenograft models, which is inhibited by treatment
with the antioxidant, N-acetyl cysteine, whereas overexpression of
SIRT3 decreases tumorigenesis in xenografts (76). Loss of SIRT3 causes MnSOD2
acetylation and elevates ROS levels, resulting in endocrine therapy
resistance in human luminal B breast cancer (77). SIRT3 deficiency promotes the growth
of B cell lymphoma via a ROS-dependent mechanism. Consistent with
this, low SIRT3 levels in clinical B lymphomas correlated with
hyperacetylation and reduced activities of the mitochondrial
proteins IDH2 and MnSOD2 and with high levels of ROS (70). Therefore, SIRT3 functions as a
tumor suppressor by scavenging ROS.
Consistent with the role of ROS in tumor formation
and progression, removal of ROS may also underlie the oncogenic
role of SIRT3. For example, overexpression of SIRT3 in gastric
cancer promotes cell proliferation and glycolysis, which is
ascribed to the ability of SIRT3 to maintain the low levels of
intracellular ROS (46).
Interestingly, SIRT3 activates hypoxia-induced mitophagy in human
glioma cells by increasing the interaction between VDAC1 and
Parkin, while inhibiting SIRT3-mediated mitophagy further decreases
the mitochondrial membrane potential, and increases the
accumulation of ROS, which in turn triggers degradation of the
antiapoptotic proteins Mcl-1 and survivin via the proteasomal
pathway; this suggests that SIRT3 may maintain ROS at levels that
contribute to the malignant phenotype of cancer cells (78).
Cancer cells prefer to utilize glycolysis for energy
production, even in the presence of sufficient oxygen: this
phenomenon is called ‘aerobic glycolysis’ or ‘the Warburg effect’
(79). The resulting metabolic
reprogramming, in addition to enabling rapid energy supply,
provides enough glycolytic intermediates for biosynthetic pathways
and facilitates the synthesis of the macromolecules and organelles
needed for generating new cancer cells (80–82).
Accumulating studies support a suppressive role for SIRT3 in
cancer-related metabolic reprogramming via two different pathways
(13). First, SIRT3-mediated
reductions in ROS levels induce activation of oxygen-dependent
prolyl hydroxylases (PHD), followed by degradation of
hypoxia-inducible factor 1-α (HIF-1α), a key mediator of metabolic
reprogramming. Consistent with this, loss of SIRT3 from breast
cancer cells provides an impetus for metabolic reprogramming via
the ROS/HIF-1α pathway (83). In
this regard, the anticancer role of profilin1 (Pfn1) in pancreatic
cancer was ascribed to its capacity to upregulate SIRT3, resulting
in destabilization of HIF-1α and suppressed expression of
glycolytic genes (84). Second,
SIRT3 stimulates the TCA or inhibits glycolysis by directly
targeting metabolic enzymes. As described above, SIRT3 blocks a
critical step of glycolysis in both breast cancer and gastric
cancer by inhibiting HK2 (24,45).
Another key enzyme modified by SIRT3 is PDC, which links the
glycolysis metabolic pathway to the TCA by converting pyruvate into
acetyl-CoA. The activity of PDC is either inhibited by pyruvate
dehydrogenase kinase or restored by pyruvate dehydrogenase
phosphatase. In cancer cells, mitochondrial ACAT1 acetylates and
inhibits PDC by recruiting PDK1, while PDC is deacetylated by
SIRT3. ACAT1 and SIRT3 also acetylate and deacetylate PDP1,
resulting in the dissociation from, or association with, PDC,
respectively (28,85). In pancreatic cells, SIRT3
deacetylates and inhibits the activity of GOT2, the limiting enzyme
in the malate-aspartate shuttle, thereby impairing the transport of
cytosolic NADH into the mitochondria, a process required for
glycolysis (25).
SIRT3 also shows a dark side when regulating
metabolic reprogramming. For example, in gastric cancer cells,
SIRT3 can deacetylate and activate lactate dehydrogenase A, thereby
promoting anaerobic glycolysis and carcinogenesis (46). In addition, a cluster of
glycolysis-associated proteins (HK2 and glucose transporters) is
upregulated in SIRT3-expressing gastric carcinoma cells (46). These findings suggest that SIRT3
may expedite malignant transformation by promoting metabolic
reprogramming.
Cell division is driven by intracellular signals
normally initiated by growth factor engagement of cognate
receptors, culminating in activation of transcriptional factors and
expression of proteins required for cell cycle progression. The
canonical signaling pathways that contribute to cell proliferation
are tailored and orchestrated by numerous regulators. To this end,
SIRT3 deacetylates and modifies the activity of components or
master regulators of proliferative signaling and is, therefore,
essential for tumorigenesis.
First, SIRT3 plays a regulatory role in cellular
levels of P53, the well-characterized tumor suppressor. SIRT3
attenuates p53 degradation by downregulating the mouse double
minute 2 homolog (MDM2) in HCC cells (65). SIRT3-mediated deacetylation also
upregulates the activity of phosphatase and tension homolog,
leading to a reduction in MDM2 transcription and p53 degradation in
various cancer cells (48,86).
Second, SIRT3 dictates cell proliferation by
modulating the classical mitogen-activated protein
kinases/extracellular signal-regulated kinases (MAPK/ERK) and
protein kinase B (PKB/Akt) signaling pathways. In HCC and prostate
cancer cells, ectopic expression of SIRT3 reduces phosphorylation
of ERK1/2 and Akt, thereby suppressing cell survival and
proliferation (65). While reduced
mitochondrial ROS production may underlie SIRT3 suppression of the
PI3K/Akt pathway, the ubiquitination and degradation of the
oncoprotein c-MYC is responsible for inhibiting cell proliferation
downstream of SIRT3/Akt signaling (87,88).
Third, SIRT3 can suppress mitosis by directly
targeting cell cycle proteins. S-phase kinase-associated protein 2
(Skp2) enables cell cycle progression through the G1/S checkpoint
by promoting destruction of p27 (89). Acetylation of Skp2 by p300
increases its stability and cytoplasmic translocation, whereas
SIRT3 deacetylates Skp2 and antagonizes p300-mediated Skp2
activation (90).
Finally, SIRT3 suppresses the growth of pancreatic
cancer by modulating cellular iron metabolism. Overexpression of
SIRT3 impairs enrichment of iron regulatory protein 1 (IRP1) on the
iron-responsive elements, thereby downregulating the transferrin
receptor 1 (TfR1) and suppressing TfR1-mediated iron uptake and
cell growth (69).
It is noteworthy that SIRT3 also facilitates
carcinogenesis by reinforcing proliferative signal pathways. In
bladder tumor-derived EJ-P53 cells, SIRT3 appears to rescue
p53-induced growth arrest by abrogating p53 activity (91). In non-small cell lung cancer cells,
SIRT3 promotes growth and proliferation by interacting with
nicotinamide mononucleotide adenylyltransferase 2 (NMNAT2), which
catalyzes an essential step in NAD (and NADP) biosynthesis
(47).
Programmed cell death or apoptosis plays an
important role in ontogenesis and the development of various
disorders. Resistance to apoptosis is a hallmark of cancer cells
(2). Recent studies show that
SIRT3 extensively participates in the regulation of the apoptotic
machinery.
The B cell lymphoma 2 (Bcl-2) family proteins are
key regulators of the apoptotic pathway. Its proapoptotic members,
such as Bcl-2-associated X protein (Bax), damage the integrity of
the outer mitochondrial membrane, leading to cytochrome c
release into the cytosol and a subsequent cascade of caspase
activation that triggers apoptosis. The mitochondrial poreforming
activity of these proapoptotic proteins is inhibited by their
association with antiapoptotic members. SIRT3 regulates both
proapoptotic and antiapoptotic members of the Bcl-2 family. For
example, SIRT3 mediates Bcl-2- and JNK2-regulated apoptosis in
colorectal carcinoma cells under basal conditions (92). Moreover, SIRT3 overexpression in
HCC cells promotes chemotherapeutic agent- and sorafenib-induced
apoptosis, while SIRT3 silencing confers resistance to
chemotherapy. Mechanistically, SIRT3 decreases the amount of
glutathione S-transferase P1 (GSTP1), causing sequential activation
of JNK and c-Jun and upregulating the c-Jun transcriptional target,
Bim (67). The proapoptotic role
of SIRT3 was also verified in lung adenocarcinoma cells, in which
SIRT3 overexpression induces apoptosis by increasing the Bax/Bcl-2
ratio (48). Kaempferol, a
flavonoid compound, induces apoptosis by upregulating Bax and SIRT3
and downregulating Bcl-2 (93).
Interestingly, the BH3-only protein mimetic S1, a novel pan Bcl-2
inhibitor, simultaneously interrupts glucose metabolism and induces
apoptosis of ovarian cancer cells by upregulating and inducing the
mitochondrial translocation of nuclear SIRT3, suggesting reciprocal
regulation between SIRT3 and Bcl-2 family proteins in cancer cells
(94). Finally, given the
regulatory role of SIRT3 on p53, SIRT3 may also induce apoptosis
through transcriptional activation of Bax and downregulation of
Bcl-2.
Conversely, SIRT3 could also function as an
antiapoptotic protein in various types of cell. SIRT3 blocks
apoptosis in cervical carcinoma HeLa cells by deacetylating Ku70,
thereby facilitating its association with Bax and preventing Bax
translocation to the mitochondrion (11). Cancer cells have evolved the
ability to escape anoikis, a specific form of apoptosis triggered
by loss of contact with the extracellular matrix (95,96).
In oral squamous cell carcinoma, dissociation of focal adhesion
kinase from receptor interacting protein (RIP) promotes RIP binding
to Fas and the formation of the death-inducing signaling complex
that triggers anoikis; however, SIRT3 functions as a negative
regulator of RIP downstream signaling, thereby rendering neoplastic
cells resistant to anoikis (97,98).
As a crucial mitochondrial deacetylase, SIRT3
modulates the activity of an increasing list of substrate proteins.
Mounting evidence supports causal involvement of SIRT3 in various
disorders, including carcinogenesis. However, studies using
different carcinoma models have reached diverse conclusions
concerning the precise role of SIRT3 in tumorigenesis. These
studies may have reached different conclusions because SIRT3 acts
on a wide array of substrates in different cellular contexts. Also,
SIRT3 catalyzes deacetylation of both histone and non-histone
proteins in the mitochondrion and nucleus, which has different
effects on individual proteins. These are further complicated by
the non-physiological manipulation of genes in vitro.
Therefore, further studies are required to discriminate
SIRT3-regulated key events that drive carcinogenesis from those
that do not (or even suppress it). In this respect, while
utilization of SIRT3-deficient cells or animal models allows
systemic study of its global contributions to intracellular
signaling or metabolic pattern shifts, studies based on clinical
cancers will help define a bona fide correlation between
SIRT3 and the pathogenesis of different malignancies. Nonetheless,
a clearer understanding of the role of SIRT3 in carcinogenesis will
eventually open new avenues for cancer treatment by targeting this
mitochondrial deacetylase.
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Choudhary C, Kumar C, Gnad F, Nielsen ML,
Rehman M, Walther TC, Olsen JV and Mann M: Lysine acetylation
targets protein complexes and co-regulates major cellular
functions. Science. 325:834–840. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kim SC, Sprung R, Chen Y, Xu Y, Ball H,
Pei J, Cheng T, Kho Y, Xiao H, Xiao L, et al: Substrate and
functional diversity of lysine acetylation revealed by a proteomics
survey. Mol Cell. 23:607–618. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Guan KL and Xiong Y: Regulation of
intermediary metabolism by protein acetylation. Trends Biochem Sci.
36:108–116. 2011. View Article : Google Scholar :
|
|
6
|
Frye RA: Phylogenetic classification of
prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res
Commun. 273:793–798. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Haigis MC and Guarente LP: Mammalian
sirtuins - emerging roles in physiology, aging, and calorie
restriction. Genes Dev. 20:2913–2921. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Weir HJ, Lane JD and Balthasar N: SIRT3: A
central regulator of mitochondrial adaptation in health and
disease. Genes Cancer. 4:118–124. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hallows WC, Albaugh BN and Denu JM: Where
in the cell is SIRT3? - functional localization of an
NAD+-dependent protein deacetylase. Biochem J.
411:e11–e13. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jing E, Emanuelli B, Hirschey MD, Boucher
J, Lee KY, Lombard D, Verdin EM and Kahn CR: Sirtuin-3 (Sirt3)
regulates skeletal muscle metabolism and insulin signaling via
altered mitochondrial oxidation and reactive oxygen species
production. Proc Natl Acad Sci USA. 108:14608–14613. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sundaresan NR, Samant SA, Pillai VB,
Rajamohan SB and Gupta MP: SIRT3 is a stress-responsive deacetylase
in cardiomyocytes that protects cells from stress-mediated cell
death by deacetylation of Ku70. Mol Cell Biol. 28:6384–6401. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kumar S and Lombard DB: Mitochondrial
sirtuins and their relationships with metabolic disease and cancer.
Antioxid Redox Signal. 22:1060–1077. 2015. View Article : Google Scholar :
|
|
13
|
Haigis MC, Deng C-X, Finley LWS, Kim H-S
and Gius D: SIRT3 is a mitochondrial tumor suppressor: A scientific
tale that connects aberrant cellular ROS, the Warburg effect, and
carcinogenesis. Cancer Res. 72:2468–2472. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Finley LWS and Haigis MC: Metabolic
regulation by SIRT3: Implications for tumorigenesis. Trends Mol
Med. 18:516–523. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Alhazzazi TY, Kamarajan P, Verdin E and
Kapila YL: Sirtuin-3 (SIRT3) and the hallmarks of cancer. Genes
Cancer. 4:164–171. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen Y, Fu LL, Wen X, Wang XY, Liu J,
Cheng Y and Huang J: Sirtuin-3 (SIRT3), a therapeutic target with
oncogenic and tumor-suppressive function in cancer. Cell Death Dis.
5:e10472014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nguyen GT, Schaefer S, Gertz M, Weyand M
and Steegborn C: Structures of human sirtuin 3 complexes with
ADP-ribose and with carba-NAD+ and SRT1720: Binding
details and inhibition mechanism. Acta Crystallogr D Biol
Crystallogr. 69:1423–1432. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jin L, Wei W, Jiang Y, Peng H, Cai J, Mao
C, Dai H, Choy W, Bemis JE, Jirousek MR, et al: Crystal structures
of human SIRT3 displaying substrate-induced conformational changes.
J Biol Chem. 284:24394–24405. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Schwer B, North BJ, Frye RA, Ott M and
Verdin E: The human silent information regulator (Sir)2 homologue
hSIRT3 is a mitochondrial nicotinamide adenine
dinucleotide-dependent deacetylase. J Cell Biol. 158:647–657. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cooper HM, Huang JY, Verdin E and
Spelbrink JN: A new splice variant of the mouse SIRT3 gene encodes
the mitochondrial precursor protein. PLoS One. 4:e49862009.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Onyango P, Celic I, McCaffery JM, Boeke JD
and Feinberg AP: SIRT3, a human SIR2 homologue, is an NAD-dependent
deacetylase localized to mitochondria. Proc Natl Acad Sci USA.
99:13653–13658. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Scher MB, Vaquero A and Reinberg D: SirT3
is a nuclear NAD+-dependent histone deacetylase that
translocates to the mitochondria upon cellular stress. Genes Dev.
21:920–928. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ahn B-H, Kim H-S, Song S, Lee IH, Liu J,
Vassilopoulos A, Deng CX and Finkel T: A role for the mitochondrial
deacetylase Sirt3 in regulating energy homeostasis. Proc Natl Acad
Sci USA. 105:14447–14452. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wei L, Zhou Y, Dai Q, Qiao C, Zhao L, Hui
H, Lu N and Guo QL: Oroxylin A induces dissociation of hexokinase
II from the mitochondria and inhibits glycolysis by SIRT3-mediated
deacetylation of cyclophilin D in breast carcinoma. Cell Death Dis.
4:e6012013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang H, Zhou L, Shi Q, Zhao Y, Lin H,
Zhang M, Zhao S, Yang Y, Ling ZQ, Guan KL, et al: SIRT3-dependent
GOT2 acetylation status affects the malate-aspartate NADH shuttle
activity and pancreatic tumor growth. EMBO J. 34:1110–1125. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hallows WC, Lee S and Denu JM: Sirtuins
deacetylate and activate mammalian acetyl-CoA synthetases. Proc
Natl Acad Sci USA. 103:10230–10235. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Schwer B, Bunkenborg J, Verdin RO,
Andersen JS and Verdin E: Reversible lysine acetylation controls
the activity of the mitochondrial enzyme acetyl-CoA synthetase 2.
Proc Natl Acad Sci USA. 103:10224–10229. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shan C, Kang H-B, Elf S, Xie J, Gu TL,
Aguiar M, Lonning S, Hitosugi T, Chung TW, Arellano M, et al:
Tyr-94 phosphorylation inhibits pyruvate dehydrogenase phosphatase
1 and promotes tumor growth. J Biol Chem. 289:21413–21422. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Someya S, Yu W, Hallows WC, Xu J, Vann JM,
Leeuwenburgh C, Tanokura M, Denu JM and Prolla TA: Sirt3 mediates
reduction of oxidative damage and prevention of age-related hearing
loss under caloric restriction. Cell. 143:802–812. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cimen H, Han MJ, Yang Y, Tong Q, Koc H and
Koc EC: Regulation of succinate dehydrogenase activity by SIRT3 in
mammalian mitochondria. Biochemistry. 49:304–311. 2010. View Article : Google Scholar :
|
|
31
|
Finley LW, Haas W, Desquiret-Dumas V,
Wallace DC, Procaccio V, Gygi SP and Haigis MC: Succinate
dehydrogenase is a direct target of sirtuin 3 deacetylase activity.
PLoS One. 6:e232952011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Schlicker C, Gertz M, Papatheodorou P,
Kachholz B, Becker CF and Steegborn C: Substrates and regulation
mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. J
Mol Biol. 382:790–801. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hallows WC, Yu W, Smith BC, Devries MK,
Ellinger JJ, Someya S, Shortreed MR, Prolla T, Markley JL, Smith
LM, et al: Sirt3 promotes the urea cycle and fatty acid oxidation
during dietary restriction. Mol Cell. 41:139–149. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hirschey MD, Shimazu T, Goetzman E, Jing
E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S,
Ilkayeva OR, et al: SIRT3 regulates mitochondrial fatty-acid
oxidation by reversible enzyme deacetylation. Nature. 464:121–125.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Shimazu T, Hirschey MD, Hua L,
Dittenhafer-Reed KE, Schwer B, Lombard DB, Li Y, Bunkenborg J, Alt
FW, Denu JM, et al: SIRT3 deacetylates mitochondrial
3-hydroxy-3-methylglutaryl CoA synthase 2 and regulates ketone body
production. Cell Metab. 12:654–661. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sundaresan NR, Gupta M, Kim G, Rajamohan
SB, Isbatan A and Gupta MP: Sirt3 blocks the cardiac hypertrophic
response by augmenting Foxo3a-dependent antioxidant defense
mechanisms in mice. J Clin Invest. 119:2758–2771. 2009.PubMed/NCBI
|
|
37
|
Pillai VB, Sundaresan NR, Kim G, Gupta M,
Rajamohan SB, Pillai JB, Samant S, Ravindra PV, Isbatan A and Gupta
MP: Exogenous NAD blocks cardiac hypertrophic response via
activation of the SIRT3-LKB1-AMP-activated kinase pathway. J Biol
Chem. 285:3133–3144. 2010. View Article : Google Scholar :
|
|
38
|
Cantó C, Gerhart-Hines Z, Feige JN,
Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P and Auwerx
J: AMPK regulates energy expenditure by modulating NAD+
metabolism and SIRT1 activity. Nature. 458:1056–1060. 2009.
View Article : Google Scholar
|
|
39
|
Yang H, Yang T, Baur JA, Perez E, Matsui
T, Carmona JJ, Lamming DW, Souza-Pinto NC, Bohr VA, Rosenzweig A,
et al: Nutrient-sensitive mitochondrial NAD+ levels
dictate cell survival. Cell. 130:1095–1107. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Cheng Y, Ren X, Gowda ASP, Shan Y, Zhang
L, Yuan YS, Patel R, Wu H, Huber-Keener K, Yang JW, et al:
Interaction of Sirt3 with OGG1 contributes to repair of
mitochondrial DNA and protects from apoptotic cell death under
oxidative stress. Cell Death Dis. 4:e7312013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yang Y, Cimen H, Han MJ, Shi T, Deng JH,
Koc H, Palacios OM, Montier L, Bai Y, Tong Q, et al:
NAD+-dependent deacetylase SIRT3 regulates mitochondrial
protein synthesis by deacetylation of the ribosomal protein MRPL10.
J Biol Chem. 285:7417–7429. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Samant SA, Zhang HJ, Hong Z, Pillai VB,
Sundaresan NR, Wolfgeher D, Archer SL, Chan DC and Gupta MP: SIRT3
deacetylates and activates OPA1 to regulate mitochondrial dynamics
during stress. Mol Cell Biol. 34:807–819. 2014. View Article : Google Scholar :
|
|
43
|
Shulga N and Pastorino JG: Ethanol
sensitizes mitochondria to the permeability transition by
inhibiting deacetylation of cyclophilin-D mediated by sirtuin-3. J
Cell Sci. 123:4117–4127. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Papa L and Germain D: SirT3 regulates the
mitochondrial unfolded protein response. Mol Cell Biol. 34:699–710.
2014. View Article : Google Scholar :
|
|
45
|
Yang B, Fu X, Shao L, Ding Y and Zeng D:
Aberrant expression of SIRT3 is conversely correlated with the
progression and prognosis of human gastric cancer. Biochem Biophys
Res Commun. 443:156–160. 2014. View Article : Google Scholar
|
|
46
|
Cui Y, Qin L, Wu J, Qu X, Hou C, Sun W, Li
S, Vaughan AT, Li JJ and Liu J: SIRT3 enhances glycolysis and
proliferation in SIRT3-expressing gastric cancer cells. PLoS One.
10:e01298342015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Li H, Feng Z, Wu W, Li J, Zhang J and Xia
T: SIRT3 regulates cell proliferation and apoptosis related to
energy metabolism in non-small cell lung cancer cells through
deacetylation of NMNAT2. Int J Oncol. 43:1420–1430. 2013.PubMed/NCBI
|
|
48
|
Xiao K, Jiang J, Wang W, Cao S, Zhu L,
Zeng H, Ouyang R, Zhou R and Chen P: Sirt3 is a tumor suppressor in
lung adenocarcinoma cells. Oncol Rep. 30:1323–1328. 2013.PubMed/NCBI
|
|
49
|
Liang L, Li Q, Huang L, Li D and Li X:
Sirt3 binds to and deacetylates mitochondrial pyruvate carrier 1 to
enhance its activity. Biochem Biophys Res Commun. 468:807–812.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Liu C, Huang Z, Jiang H and Shi F: The
sirtuin 3 expression profile is associated with pathological and
clinical outcomes in colon cancer patients. Biomed Res Int.
2014:871263. 2014.PubMed/NCBI
|
|
51
|
Lai C-C, Lin P-M, Lin S-F, Hsu CH, Lin HC,
Hu ML, Hsu CM and Yang MY: Altered expression of SIRT gene family
in head and neck squamous cell carcinoma. Tumour Biol.
34:1847–1854. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Mahjabeen I and Kayani MA: Loss of
mitochondrial tumor suppressor genes expression is associated with
unfavorable clinical outcome in head and neck squamous cell
carcinoma: Data from retrospective study. PLoS One.
11:e01469482016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Alhazzazi TY, Kamarajan P, Xu Y, Ai T,
Chen L, Verdin E and Kapila YL: A novel sirtuin-3 inhibitor,
LC-0296, inhibits cell survival and proliferation, and promotes
apoptosis of head and neck cancer cells. Anticancer Res. 36:49–60.
2016.PubMed/NCBI
|
|
54
|
Zhao Y, Yang H, Wang X, Zhang R, Wang C
and Guo Z: Sirtuin-3 (SIRT3) expression is associated with overall
survival in esophageal cancer. Ann Diagn Pathol. 17:483–485. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Torrens-Mas M, Pons DG, Sastre-Serra J,
Oliver J and Roca P: SIRT3 silencing sensitizes breast cancer cells
to cytotoxic treatments through an increment in ROS production. J
Cell Biochem. Jul 15–2016.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Alhazzazi TY, Kamarajan P, Joo N, Huang
JY, Verdin E, D’Silva NJ and Kapila YL: Sirtuin-3 (SIRT3), a novel
potential therapeutic target for oral cancer. Cancer.
117:1670–1678. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
George J, Nihal M, Singh CK, Zhong W, Liu
X and Ahmad N: Pro-proliferative function of mitochondrial sirtuin
deacetylase SIRT3 in human melanoma. J Invest Dermatol.
136:809–818. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Choi J, Koh E, Lee YS, Lee HW, Kang HG,
Yoon YE, Han WK, Choi KH and Kim KS: Mitochondrial Sirt3 supports
cell proliferation by regulating glutamine-dependent oxidation in
renal cell carcinoma. Biochem Biophys Res Commun. 474:547–553.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Shackelford R, Hirsh S, Henry K,
Abdel-Mageed A, Kandil E and Coppola D: Nicotinamide
phosphoribosyltransferase and SIRT3 expression are increased in
well-differentiated thyroid carcinomas. Anticancer Res.
33:3047–3052. 2013.PubMed/NCBI
|
|
60
|
Desouki MM, Doubinskaia I, Gius D and
Abdulkadir SA: Decreased mitochondrial SIRT3 expression is a
potential molecular biomarker associated with poor outcome in
breast cancer. Hum Pathol. 45:1071–1077. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Sastre-Serra J, Nadal-Serrano M, Pons DG,
Valle A, Garau I, García-Bonafé M, Oliver J and Roca P: The
oxidative stress in breast tumors of postmenopausal women is
ERα/ERβ ratio dependent. Free Radic Biol Med. 61:11–17. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zhang CZ, Liu L, Cai M, Pan Y, Fu J, Cao Y
and Yun J: Low SIRT3 expression correlates with poor
differentiation and unfavorable prognosis in primary hepatocellular
carcinoma. PLoS One. 7:e517032012. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Zhang B, Qin L, Zhou C-J, Liu Y-L, Qian
H-X and He S-B: SIRT3 expression in hepatocellular carcinoma and
its impact on proliferation and invasion of hepatoma cells. Asian
Pac J Trop Med. 6:649–652. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wang J-X, Yi Y, Li Y-W, Cai XY, He HW, Ni
XC, Zhou J, Cheng YF, Jin JJ, Fan J, et al: Down-regulation of
sirtuin 3 is associated with poor prognosis in hepatocellular
carcinoma after resection. BMC Cancer. 14:2972014. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Zhang Y-Y and Zhou L-M: Sirt3 inhibits
hepatocellular carcinoma cell growth through reducing Mdm2-mediated
p53 degradation. Biochem Biophys Res Commun. 423:26–31. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Li Y, Wang W, Xu X, Sun S and Qu XJ:
{2-[1-
(3-methoxycarbonylmethyl-1H-indol-2-yl)-1-methyl-ethyl]-1H-indol-3-yl}-acetic
acid methyl ester (MIAM) inhibited human hepatocellular carcinoma
growth through upregulation of Sirtuin-3 (SIRT3). Biomed
Pharmacother. 69:125–132. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Tao NN, Zhou HZ, Tang H, Cai XF, Zhang WL,
Ren JH, Zhou L, Chen X, Chen K, Li WY, et al: Sirtuin 3 enhanced
drug sensitivity of human hepatoma cells through glutathione
S-transferase pi 1/JNK signaling pathway. Oncotarget.
7:50117–50130. 2016.PubMed/NCBI
|
|
68
|
McGlynn LM, McCluney S, Jamieson NB,
Thomson J, MacDonald AI, Oien K, Dickson EJ, Carter CR, McKay CJ
and Shiels PG: SIRT3 & SIRT7: Potential novel biomarkers for
determining outcome in pancreatic cancer patients. PLoS One.
10:e01313442015. View Article : Google Scholar :
|
|
69
|
Jeong SM, Lee J, Finley LWS, Schmidt PJ,
Fleming MD and Haigis MC: SIRT3 regulates cellular iron metabolism
and cancer growth by repressing iron regulatory protein 1.
Oncogene. 34:2115–2124. 2015. View Article : Google Scholar
|
|
70
|
Yu W, Denu RA, Krautkramer KA, Grindle KM,
Yang DT, Asimakopoulos F, Hematti P and Denu JM: Loss of SIRT3
provides growth advantage for B cell malignancies. J Biol Chem.
291:3268–3279. 2016. View Article : Google Scholar
|
|
71
|
Temel M, Koc MN, Ulutas S and Gogebakan B:
The expression levels of the sirtuins in patients with BCC. Tumour
Biol. 37:6429–6435. 2016. View Article : Google Scholar
|
|
72
|
Dong XC, Jing LM, Wang WX and Gao YX:
Down-regulation of SIRT3 promotes ovarian carcinoma metastasis.
Biochem Biophys Res Commun. 475:245–250. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Chandel NS and Tuveson DA: The promise and
perils of antioxidants for cancer patients. N Engl J Med.
371:177–178. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Finkel T: Signal transduction by reactive
oxygen species. J Cell Biol. 194:7–15. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Kim H-S, Patel K, Muldoon-Jacobs K, Bisht
KS, Aykin-Burns N, Pennington JD, van der Meer R, Nguyen P, Savage
J, Owens KM, et al: SIRT3 is a mitochondria-localized tumor
suppressor required for maintenance of mitochondrial integrity and
metabolism during stress. Cancer Cell. 17:41–52. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Bell EL, Emerling BM, Ricoult SJH and
Guarente L: SirT3 suppresses hypoxia inducible factor 1α and tumor
growth by inhibiting mitochondrial ROS production. Oncogene.
30:2986–2996. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Zou X, Santa-Maria CA, O’Brien J, Gius D
and Zhu Y: Manganese superoxide dismutase acetylation and
dysregulation, due to loss of SIRT3 activity, promote a luminal
B-like breast carcinogenic-permissive phenotype. Antioxid Redox
Signal. 25:326–336. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Qiao A, Wang K, Yuan Y, Guan Y, Ren X, Li
L, Chen X, Li F, Chen AF, Zhou J, et al: Sirt3-mediated mitophagy
protects tumor cells against apoptosis under hypoxia. Oncotarget.
May 30–2016.(Epub ahead of print). View Article : Google Scholar
|
|
79
|
Warburg O: On the origin of cancer cells.
Science. 123:309–314. 1956. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Vander Heiden MG, Cantley LC and Thompson
CB: Understanding the Warburg effect: The metabolic requirements of
cell proliferation. Science. 324:1029–1033. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
DeBerardinis RJ, Lum JJ, Hatzivassiliou G
and Thompson CB: The biology of cancer: Metabolic reprogramming
fuels cell growth and proliferation. Cell Metab. 7:11–20. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Christofk HR, Vander Heiden MG, Harris MH,
Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL and
Cantley LC: The M2 splice isoform of pyruvate kinase is important
for cancer metabolism and tumour growth. Nature. 452:230–233. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Wei L, Zhou Y, Qiao C, Ni T, Li Z, You Q,
Guo Q and Lu N: Oroxylin A inhibits glycolysis-dependent
proliferation of human breast cancer via promoting SIRT3-mediated
SOD2 transcription and HIF1α destabilization. Cell Death Dis.
6:e17142015. View Article : Google Scholar
|
|
84
|
Yao W, Ji S, Qin Y, Yang J, Xu J, Zhang B,
Xu W, Liu J, Shi S, Liu L, et al: Profilin-1 suppresses
tumorigenicity in pancreatic cancer through regulation of the
SIRT3-HIF1α axis. Mol Cancer. 13:1872014. View Article : Google Scholar
|
|
85
|
Fan J, Shan C, Kang HB, Elf S, Xie J,
Tucker M, Gu TL, Aguiar M, Lonning S, Chen H, et al: Tyr
phosphorylation of PDP1 toggles recruitment between ACAT1 and SIRT3
to regulate the pyruvate dehydrogenase complex. Mol Cell.
53:534–548. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Zhao K, Zhou Y, Qiao C, Ni T, Li Z, Wang
X, Guo Q, Lu N and Wei L: Oroxylin A promotes PTEN-mediated
negative regulation of MDM2 transcription via SIRT3-mediated
deacetylation to stabilize p53 and inhibit glycolysis in wt-p53
cancer cells. J Hematol Oncol. 8:412015. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Quan Y, Wang N, Chen Q, Xu J, Cheng W, Di
M, Xia W and Gao WQ: SIRT3 inhibits prostate cancer by
destabilizing oncoprotein c-MYC through regulation of the PI3K/Akt
pathway. Oncotarget. 6:26494–26507. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Pillai VB, Sundaresan NR and Gupta MP:
Regulation of Akt signaling by sirtuins: Its implication in cardiac
hypertrophy and aging. Circ Res. 114:368–378. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Carrano AC, Eytan E, Hershko A and Pagano
M: SKP2 is required for ubiquitin-mediated degradation of the CDK
inhibitor p27. Nat Cell Biol. 1:193–199. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Wang Z, Inuzuka H, Zhong J, Liu P, Sarkar
FH, Sun Y and Wei W: Identification of acetylation-dependent
regulatory mechanisms that govern the oncogenic functions of Skp2.
Oncotarget. 3:1294–1300. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Li S, Banck M, Mujtaba S, Zhou M-M, Sugrue
MM and Walsh MJ: p53-induced growth arrest is regulated by the
mitochondrial SirT3 deacetylase. PLoS One. 5:e104862010. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Allison SJ and Milner J: SIRT3 is
pro-apoptotic and participates in distinct basal apoptotic
pathways. Cell Cycle. 6:2669–2677. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Marfe G, Tafani M, Indelicato M,
Sinibaldi-Salimei P, Reali V, Pucci B, Fini M and Russo MA:
Kaempferol induces apoptosis in two different cell lines via Akt
inactivation, Bax and SIRT3 activation, and mitochondrial
dysfunction. J Cell Biochem. 106:643–650. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Xiang XY, Kang JS, Yang XC, Su J, Wu Y,
Yan XY, Xue YN, Xu Y, Liu YH, Yu CY, et al: SIRT3 participates in
glucose metabolism interruption and apoptosis induced by BH3
mimetic S1 in ovarian cancer cells. Int J Oncol. 49:773–784.
2016.PubMed/NCBI
|
|
95
|
Shin SI, Freedman VH, Risser R and Pollack
R: Tumorigenicity of virus-transformed cells in nude mice is
correlated specifically with anchorage independent growth in vitro.
Proc Natl Acad Sci USA. 72:4435–4439. 1975. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Kantak SS and Kramer RH: E-cadherin
regulates anchorage-independent growth and survival in oral
squamous cell carcinoma cells. J Biol Chem. 273:16953–16961. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Kamarajan P, Bunek J, Lin Y, Nunez G and
Kapila YL: Receptor-interacting protein shuttles between cell death
and survival signaling pathways. Mol Biol Cell. 21:481–488. 2010.
View Article : Google Scholar :
|
|
98
|
Kamarajan P, Alhazzazi TY, Danciu T,
D’silva NJ, Verdin E and Kapila YL: Receptor-interacting protein
(RIP) and Sirtuin-3 (SIRT3) are on opposite sides of anoikis and
tumorigenesis. Cancer. 118:5800–5810. 2012. View Article : Google Scholar : PubMed/NCBI
|