Introduction
Non-small cell lung cancer (NSCLC) accounts for
about 85% of all lung cancer cases whereas adenocarcinoma (ADC) and
squamous cell carcinoma (SQCC) are the two major histological
subtypes (1). As a heterogeneous
disease, NSCLC is a challenge for treatment, and the 5-year
survival rate of patients with advanced tumor stages is still low
(2). Therefore, it is of critical
importance to uncover the molecular mechanism(s) behind NSCLC
progression.
We recently reported that expression levels of
serpin family A member 1 (SERPINA1), a gene-encoding
alpha1-antitrypsin (AAT), are significantly lower in tumor tissue
of both the SQCC and ADC subtypes of NSCLC compared to levels noted
in adjacent normal lung tissue (3). Remarkably, higher expression of the
SERPINA1 gene in tumor tissue, especially in smokers, was
associated with better overall and disease-free survival.
As mentioned above, the product of the
SERPINA1 gene is AAT, an acute-phase serum glycoprotein and
a broad-spectrum inhibitor of serine proteases. Several research
groups have demonstrated that AAT reduces acute lung injury by
suppressing inflammation and cell death. Interestingly, a recent
study identified AAT as an inhibitor of transmembrane serine
protease 2 (TMPRSS2) activity (4), a protease which plays a role in the
cellular entry of coronaviruses, including Severe Acute Respiratory
Syndrome Coronavirus Type 2 (SARS-CoV-2), SARS-CoV, and Middle East
Respiratory Syndrome (MERS)-CoV, as well as influenza viruses.
Recent research, based on pseudoparticles and replication-competent
viruses, are in line with the above observation as AAT prevents an
early step in the viral life cycle, and selectively inhibits
SARS-CoV-2 spike but not Vesicular stomatitis virus glycoprotein
(VSV-G)-mediated infection, which is independent from TMPRSS2
activation (5). Understanding how
TMPRSS2 protein expression varies in the lungs could reveal
important insights into differential susceptibility to influenza
and coronavirus infections but also into cancer development. In
general, patients with cancer seem to be more susceptible to
SARS-CoV (6). Specifically,
cancer type, staging and anti-cancer therapies seem to be
additional risk factors for severe COVID-19 (7).
Intriguingly, a recent study provided evidence that
tumor tissue from head and neck cancer patients have reduced
expression of TMPRSS2 when compared to non-tumorous tissues
(8). According to the
experimental data, SARS-CoV-infected TMPRSS2-knockdown (-/-) mice
show weakened inflammatory chemokine and/or cytokine responses,
lower virus spread within the airways and less severe
immunopathology (9). Hence,
hypothetically TMPRSS2 may be a cancer-suppressor gene or reflect
tumorigenic processes. Multiple studies have shown that in
prostate, lung, and other cancer cells, TMPRSS2 is regulated by the
androgen receptor, a nuclear receptor, and a member of the steroid
receptor family (10,11). TMPRSS2 is expressed as a 70-kDa
full-length form and has a 32-kDa protease domain, which is cleaved
and secreted after autocleavage (12). The increased activity of TMPRSS2
mediates signal transduction between cancer cells and the
extracellular environment, and thus determines different cellular
responses (13). Specifically,
TMPRSS2 can cleave and activate protease-activated receptor 2
(PAR2), which triggers the release of inflammatory mediators,
immune cell recruitment, tumor cell invasion, and apoptosis
(14). PAR2 receptor signaling in
the airway epithelium also causes calcium release (12) suggesting a role for TMPRSS2 in the
pathogenesis of cancer pain (13).
In the present study, we aimed to investigate RNA
and protein levels of TMPRSS2 in NSCLC patients and their
associations with survival prognosis. We also aimed to clarify
whether there is any relationship between TMPRSS2 and
SERPINA1 expression, and TMPRSS2 and AAT protein levels.
Materials and methods
Sample collection, characterization and
preparation
Tissue samples were provided by the Lung Biobank
Heidelberg, a member of the accredited Tissue Bank of the National
Center for Tumor Diseases (NCT) Heidelberg, the Biomaterial Bank
Heidelberg, and the Biobank Platform of the German Center for Lung
Research (DZL). The local ethics committees of the Medical Faculty
Heidelberg and Hannover Medical School [S-270/2001 (biobanking
vote) and 9155_BO_K_2020 (study-specific vote)] approved the use of
the biomaterial and data. All patients (for cohort overview see
Table I) included in the study
signed an informed consent, and the study was performed according
to the principles set out in the WMA Declaration of Helsinki.
 | Table IPatient data of the qPCR cohort. |
Table I
Patient data of the qPCR cohort.
Cohort description
|
|---|
| Parameters | n | (%) |
|---|
| Total | 347 | 100 |
| Median age
(years) | 65 (38-88) | |
| Sex | | |
| Male | 240 | 69 |
| Female | 107 | 31 |
| Histology | | |
|
Adenocarcinoma | 204 | 59 |
| Squamous | 143 | 41 |
| Therapy | | |
| OP | 203 | 59 |
| OP/RT | 11 | 3 |
| OP/ChT | 97 | 28 |
| OP/RT/ChT | 36 | 10 |
| Smoking status | | |
| Non-smoker | 36 | 10 |
| Ex-smoker | 183 | 53 |
| Smoker | 126 | 36 |
| No data | 2 | 1 |
| Pathological stage
(7th TNM edition) | | |
| IA | 34 | 10 |
| IB | 89 | 26 |
| IIA | 71 | 20 |
| IIB | 47 | 14 |
| IIIA | 94 | 27 |
| IIIB | 12 | 3 |
| ECOG | | |
| 0 | 297 | 86 |
| 1 | 46 | 13 |
| 2 | 4 | 1 |
Tumor and matched distant (>5 cm) tumor-free lung
tissue samples from NSCLC patients who underwent therapy-naïve
resection for primary lung cancer at Thoraxklinic at University
Hospital Heidelberg, Germany were collected between 2006 and 2011.
Tissues were snap-frozen within 30 min after resection and stored
at −80°C until the time of analysis. More detailed information is
provided elsewhere (15).
Total RNA isolation and cDNA
synthesis
RNA was isolated from tumor tissue and adjacent lung
tissue. For RNA isolation from patient tumor tissue, a tumor
content of ≥50% was the minimum prerequisite. A total of 10-15
tumor cryosections (10-15 µm) from each patient were sliced,
and the first as well as the last section of the series was stained
with hematoxylin and eosin (H&E). A lung pathologist determined
the proportion of viable tumor cells, stromal cells, healthy lung
cells, and necrotic areas. Total RNA was isolated from patient
tissue using an AllPrep DNA/RNA/miRNA Universal kit (Qiagen, Inc.)
according to the manufacturer's instructions. Afterwards, the
quality of total RNA was assessed by utilizing an Agilent 2100
Bioanalyzer and an Agilent RNA 6000 Nano kit (Agilent
Technologies). Using the Transcriptor First Strand cDNA Synthesis
kit (Roche), total RNA was transcribed to complementary DNA and
used for quantitative polymerase chain reaction (qPCR). A complete
description of the procedure is provided elsewhere (15).
Real-time polymerase chain reaction
(RT-PCR) analysis
For gene expression analyses of patient tissues,
volumes of 5 µl cDNA (corresponding to 5 ng of isolated
total RNA) were utilized for qPCR with the
LightCycler480® (Roche) in 384-well plates according to
the Minimum Information for Publication of qPCR Experiments (MIQE)
guidelines (16). Universal
ProbeLibrary (UPL) assay (Roche) was used as the amplification and
detection system. Gene-specific primers (TIB Molbiol were combined
with the primaQuant 2X qPCR Probe-MasterMix (Steinbrenner
Laborsysteme). Threshold cycle (Ct) values were evaluated with the
LightCycler480® software release 1.5 and the 2nd
derivative maximum method (Roche). For the comparison of gene
expression in tumor and non-malignant samples, the relative
expression of the genes was calculated (ΔCt values). A panel of
housekeeper genes was evaluated in tumor and lung samples in regard
to stable expression, and TMPRSS2 expression was normalized
to the housekeepers esterase D (ESD) and ribosomal protein S18
(RPS18). For the waterfall plots analyzing the fold-change
expression changes between tumor and normal tissues, the
2−ΔΔCt values were calculated. The following primers and
UPL were used for the detection of TMPRSS2: TMPRSS2
forward (UPL #71, 5′-ACC AGT GTG TCT GCC CAA C-3′) and
TMPRSS2 reverse (UPL #71, 5′-GCG TTC AGC ACT TCT GAG G-3′);
ESD forward (UPL#50, 5′-TCA GTC TGC TTC AGA ACA TGG-3′) and
ESD reverse (UPL#50 5′-CCT TTA ATA TTG CAG CCA CGA-3′);
RPS18 forward (UPL#46, 5′-CTT CCA CAG GAG GCC TAC AC-3′) and
RPS18 reverse (UPL#46, 5′-CGC AAA ATA TGC TGG AAC TTT-3′).
The complete procedure including housekeeper selection is described
elsewhere (15).
Western blot analyses of TMPRSS2 and
AAT
Cryo-conserved tumor and corresponding lung tissues
from patients were evaluated by a pathologist as described in the
'Total RNA isolation and cDNA synthesis' section and cut in
100-µm pieces with a HM 500 OM microm (Thermo Fisher
Scientific, Inc.). Tissues were disrupted using 500 µl PBS
(#14190-094, Thermo Fisher Scientific, Inc.) per 100 mg of tissue,
Halt™ Protease Inhibitor Cocktail (#78430, Thermo Fisher
Scientific, Inc.) and a TissueLyser Mixer-Mill Disruptor (Qiagen)
for 2 min at 25 Hz. Afterwards, the cell lysates were centrifuged
for 10 min at 13.000 × g and 4°C. The supernatants were transferred
to a new tube, and the protein concentration was determined using a
BCA protein assay (#23225, Thermo Fisher Scientific, Inc.). A total
of 24 randomly selected patients were used as representative
examples. Total protein (100 µg) was used for the immunoblot
analysis of TMPRSS2 and AAT and heated for 5 min at 95°C in an SDS
(sodium dodecyl sulfate)-sample buffer containing
ß-mercaptoethanol, glycine and pyronin. Samples were separated
using 10% SDS-PAGE and blotted on a nitrocellulose membrane
(#GE10600002 Merck KGaA) that was blocked with nonfat dried milk
powder (5%, #A0830, AppliChem) in PBS/Tween 20 (0.1%, #A4974,
AppliChem) for 1 h at room temperature. For the detection of
TMPRSS2, a rabbit monoclonal antibody (#MA5-35756, Thermo Fisher
Scientific, Inc.) was applied at a dilution of 1:750 overnight at
4°C. For the detection of AAT, a rabbit polyclonal antibody
(#A0012, Dako) was used at a dilution of 1:10,000 overnight at 4°C.
A β-actin antibody (#A5441 Merck KGaA) was applied at a dilution of
1:10,000 for 1 h at room temperature as a loading control. A
peroxidase system was used for signal development. A secondary
anti-mouse IgG antibody (#A4416, Merck KGaA) was applied at a
dilution of 1:10,000 for 1 h at room temperature, and a secondary
anti-rabbit IgG antibody (#A6154, Merck KGaA) was applied at a
dilution of 1:5,000. Signals were visualized with Chemiluminate-HRP
Picodetect (#A3417, Applichem). Quantification of the signals was
evaluated with Image Studio Lite V. 5.2 (LI-COR Biosciences, GmbH)
and adapted to actin levels (Figs.
S3-S6).
Immunohistochemical analyses
For detection of TMPRSS2, a monoclonal TMPRSS2
antibody (MABF2158, Merck KGaA) was used. Before tissue microarray
(TMA) construction, a H&E-stained slide of each block was
analyzed to select tumor-containing regions. A TMA machine
(AlphaMetrix Biotech) was used to extract tandem 1.0-mm cylindrical
core sample from each tissue donor block (for cohort overview see
Table II). Paraffin-embedded
tissue sections were deparaffinized with the following steps: 2×10
min in xylol, 2×5 min in 100% ethanol, 1×3 min in 98% ethanol and
1×3 min in 70% ethanol. Antigen retrieval was performed in a
steamer with sodium-citrate-buffer (10 mM sodium citrate, 0.05%
Tween 20, pH 6.0) for 15 min. Peroxidases were blocked for 10 min
at room temperature (RT) using 3% H2O2
(Applichem). Slides were incubated with normal goat serum for 1 h
at RT to avoid unspecific background staining (Cell Signaling
Technology, Inc.). The primary antibody was incubated at a dilution
of 1:200 overnight at 4°C in a humid chamber. The staining
procedure was performed with SignalStain® DAB Substrate
Kit (#8059, Cell Signaling Technology, Inc.) according to
manufacturer's instructions. The last developing step was performed
for 2 min. Cell nuclei were stained using Mayer's Hematoxylin
Solution (Sigma-Aldrich/Merck KGaA). Slides were mounted using
ImmunoHistoMount™ (Sigma-Aldrich/Merck KGaA). Staining was observed
with an Olympus IX-71 inverted microscope. Images were captured
with an Olympus Color View II digital camera and Olympus Cell-F
software (cellSense dimension, V1.11, Olympus). Tiffs were
assembled into figures using Photoshop CS6 (Adobe Systems, Inc.).
Only changes in brightness and contrast were applied. TMA slides
were scanned and analyzed using Aperio ImageScope (v12.4.3.5008,
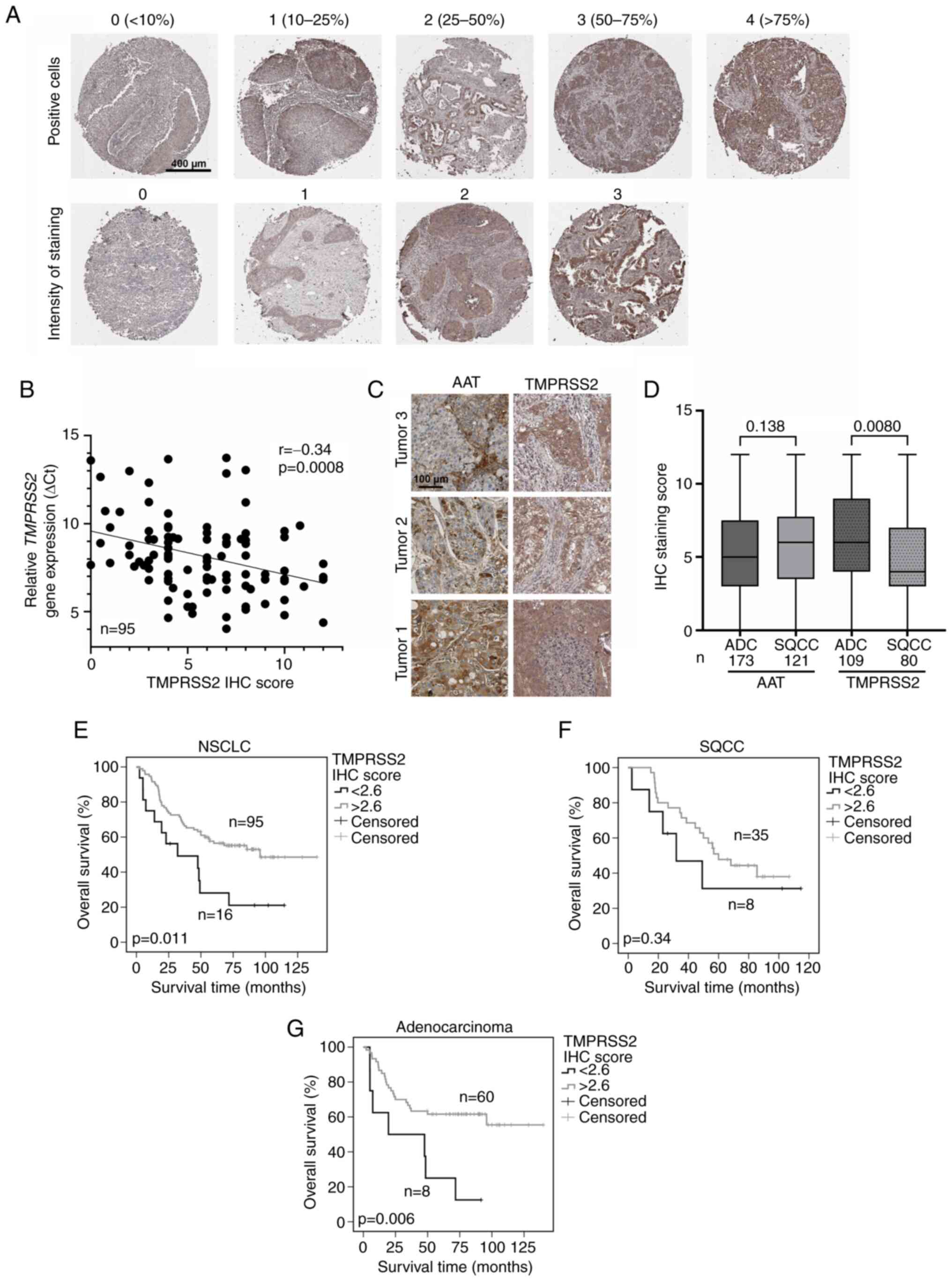
Leica Biosystems). Scoring was performed by multiplication of
staining intensity (0-3) with the proportion of positive cells
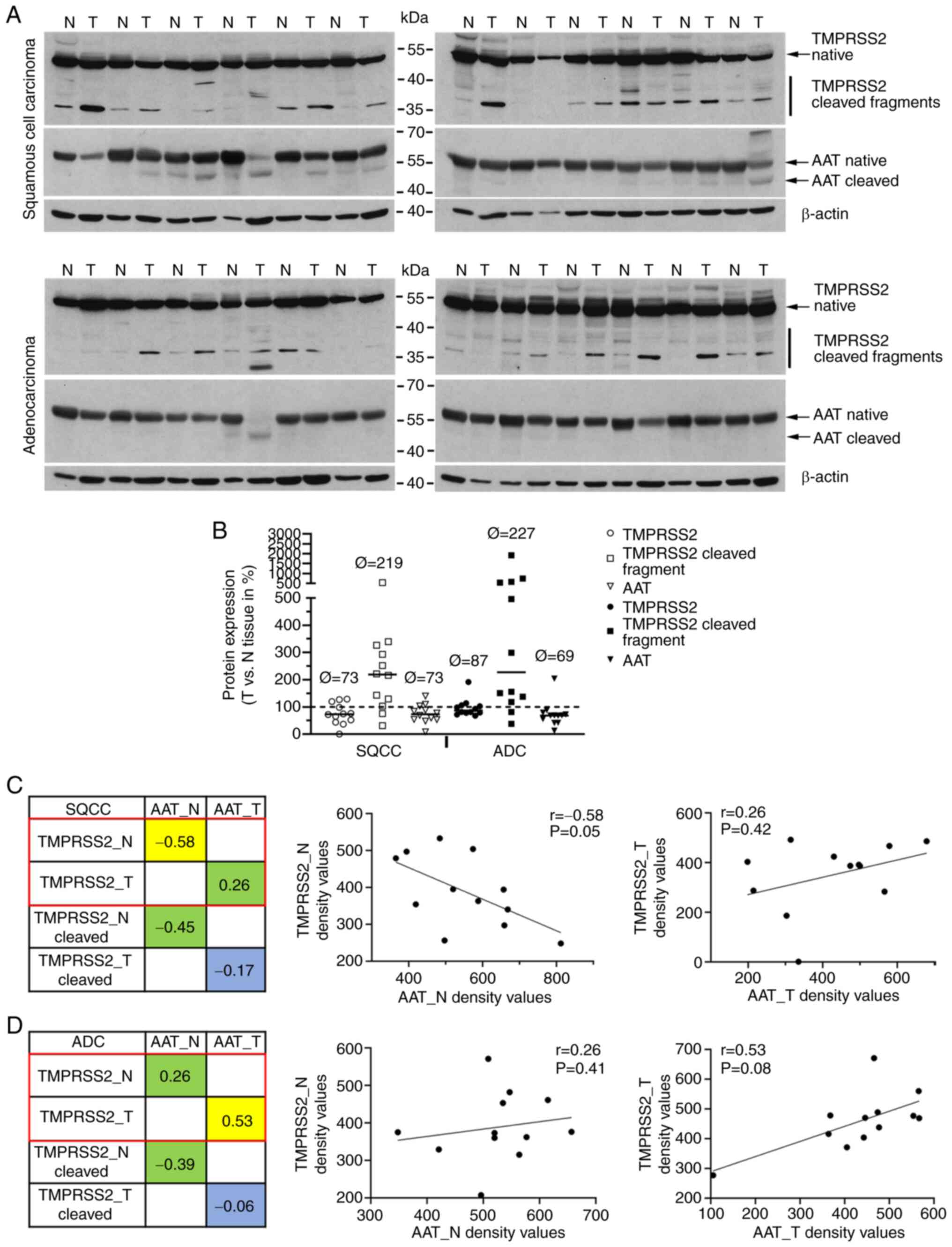
(0-4) (Fig. 4A).
 | Table IIPatient data of the IHC cohort. |
Table II
Patient data of the IHC cohort.
Cohort description
|
|---|
| Parameter | n | (%) |
|---|
| Total | 189 | 100 |
| Median age (range)
in years | 63 (39-81) | |
| Sex | | |
| Male | 115 | 61 |
| Female | 74 | 39 |
| Histology | | |
|
Adenocarcinoma | 109 | 58 |
| Squamous | 80 | 42 |
| Therapy | | |
| OP | 106 | 56 |
| OP/RT | 8 | 4 |
| OP/ChT | 53 | 28 |
| OP/RT/ChT | 22 | 12 |
| Smoking status | | |
| Non-smoker | 16 | 8 |
| Ex-smoker | 116 | 61 |
| Smoker | 49 | 26 |
| No data | 8 | 4 |
| Pathological stage
(7th TNM edition) | | |
| IA | 24 | 13 |
| IB | 41 | 22 |
| IIA | 27 | 14 |
| IIB | 18 | 10 |
| IIIA | 64 | 34 |
| IIIB | 6 | 3 |
| IVA | 4 | 2 |
| IVB | 3 | 2 |
| ECOG | | |
| 0 | 126 | 67 |
| 1 | 55 | 29 |
| 2 | 7 | 4 |
| 4 | 1 | 1 |
Statistical analyses
Data of qPCR and IHC analyses were statistically
analyzed under REMARK criteria (17) with SPSS 25.0 for Windows (IBM
Corp.). The endpoint of the study was overall survival. Overall
survival time was calculated from the date of diagnosis until the
last date of contact or death. The cut-offs used for survival
analyses were selected using the software tool 'Cutoff-Finder'
(http://molpath.charite.de/cutoff/index.jsp).
Multivariate survival analyses were performed using the Cox
proportional hazards model (Table
III). Univariate analysis of survival data was performed
according to Kaplan and Meier (Figs.
1C-F, 2 and 4). The log-rank test was used to test
the significance between the groups. The non-parametric, Wilcoxon
matched-pairs signed rank test and the Mann-Whitney test were used
to investigate significant differences between the patient groups
(Figs. 1A, 4D and S1). The Bonferroni's correction for
multiple testing was considered for Fig. 1A. Correlation analyses were
performed using the nonparametric Spearman's rank correlation
analysis (Figs. 3, 4B, 5C and
D and S2). A P-value
<0.05 was considered significant. Data were visualized with
GraphPad Prism 9 (GraphPad Software, Inc.) and SPSS 25.0 (IBM
Corp.).
 | Table IIIMultivariate analysis. |
Table III
Multivariate analysis.
All patients
|
|---|
| Variable | Significance
(P-value) | Hazard ratio (95%
CI) |
|---|
| Age (years) | 0.001 | 1.036
(1.016-1.056) |
| Sex (male vs.
female) | 0.041 | 1.502
(1.018-2.216) |
| Histology (ADC vs.
SQCC) | 0.450 | 0.873
(0.614-1.242) |
| ECOG | 0.062 | 1.430
(0.983-2.080) |
| Pathological
stage |
<0.001 | 1.032
(1.020-1.043) |
| Smoking status | 0.166 | 1.149
(0.944-1.399) |
| TMPRSS2 expression
(low vs. high) | 0.033 | 1.536
(1.035-2.279) |
|
Squamous cell
carcinoma
|
| Variable | Significance
(P-value) | Hazard ratio (95%
CI) |
|
| Age | 0.031 | 1.033
(1.003-1.065) |
| Sex (male vs.
female) | 0.035 | 2.483
(1.067-5.776) |
| ECOG | 0.17 | 1.462
(0.850-2.515) |
| Pathological
stage | 0.003 | 1.027
(1.009-1.046) |
| Smoking status | 0.315 | 1.281
(0.791-2.075) |
| TMPRSS2 expression
(low vs. high) | 0.299 | 0.692
(0.346-1.386) |
Squamous cell
carcinoma
|
Adenocarcinoma
|
| Variable | Significance
(P-value) | Hazard ratio (95%
CI) |
|
| Age (years) | 0.003 | 1.040
(1.013-1.067) |
| Sex (male vs.
female) | 0.081 | 1.591
(0.945-2.681) |
| ECOG | 0.332 | 1.119
(0.892-1.045) |
| Pathological
stage |
<0.001 | 1.037
(1.022-1.053) |
| Smoking status | 0.305 | 1.266
(0.807-1.987) |
| TMPRSS2 expression
(low vs. high) | 0.002 | 2.065
(1.308-3.260) |
Results
Lower expression of the TMPRSS2 gene in
tumor tissues of NSCLC patients is a prognostic factor for overall
survival
First, we analyzed TMPRSS2 gene expression in
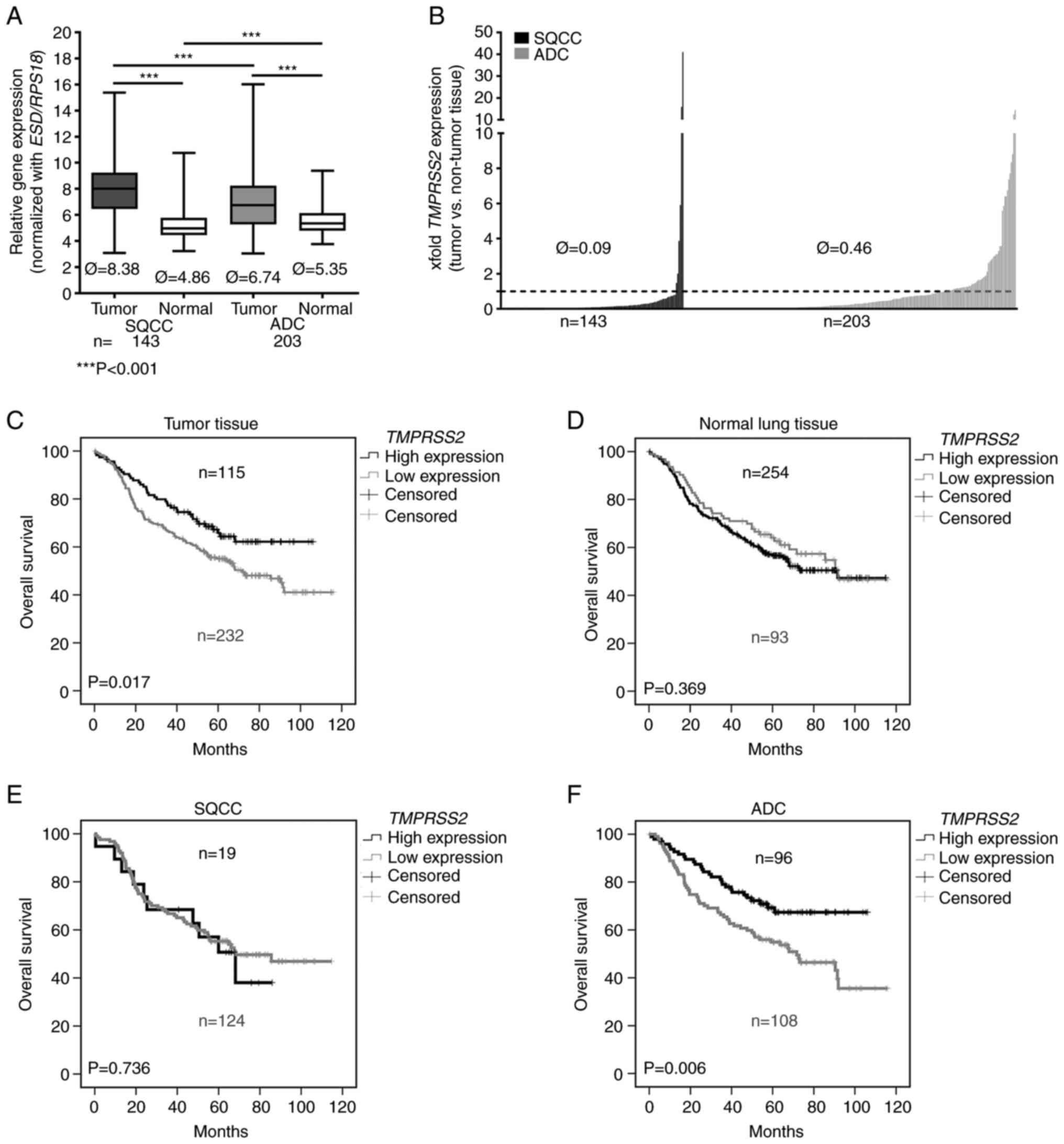
a large NSCLC patient cohort (n=347, Table I). We observed a significantly
lower expression of TMPRSS2 in the tumor tissues of patients
with SQCC and ADC compared to that noted in the adjacent non-tumor
lung tissues (Fig. 1A; Please
note that higher values indicate a lower expression). TMPRSS2 was
significantly upregulated in pathological stage I ADC compared to
stage II, but not in SQCC (Fig.
S1; Please note that higher values indicate a lower
expression). Comparing tumor and normal lung tissues, we found an
approximately 10-fold downregulation of TMPRSS2 in the tumor
tissues of patients with SQCC and an approximately 2-fold
downregulation in the tumor tissues of patients with ADC (Fig. 1B). In the SQCC group,
TMPRSS2 was downregulated in 137/143 patients (~96%,
Fig. 1B).
In a further approach, we focused on the influence
of TMPRSS2 on patient overall survival. We used the software
Cut-off Finder to separate patients into two groups: one with a
higher and one with a lower TMPRSS2 expression. A
multivariate survival analysis (Table III) including the most important
clinical parameters of the entire NSCLC cohort revealed that age
(HR=1.036, P=0.001), sex (male vs. female, HR=1.502, P=0.041)
pathological tumor stage (HR=1.032, P<0.001) and a low
expression of TMPRSS2 (HR=1.536, P=0.033) are independent
prognostic indicators of a dismal prognosis (Table III). A more detailed analyses of
the two subgroups of NSCLC revealed that age and pathological tumor
stage are prognostic indicators for both, SQCC and ADC, patients.
However, the lower expression of TMPRSS2 was found as a
significant independent prognostic factor only for ADC (HR=2.065,
P=0.002) (Table III). A low
expression of TMPRSS2 resulted in a more than 2-fold hazard
ratio. Kaplan-Meier analyses including all patients (Fig. 1C) confirmed that low
TMPRSS2 expression in tumor tissue is prognostic for poor
overall survival (P=0.017). In contrast, the expression of
TMPRSS2 in adjacent normal lung tissue was not correlated
with survival prognosis (Fig.
1D). Remarkably, we found that low TMPRSS2 expression
had a prognostic value only for patients harboring an ADC (P=0.006)
(Fig. 1F) but not for patients
with SQCC (P=0.736 (Fig. 1E).
Prognostic value of TMPRSS2 depends on
smoking status and sex
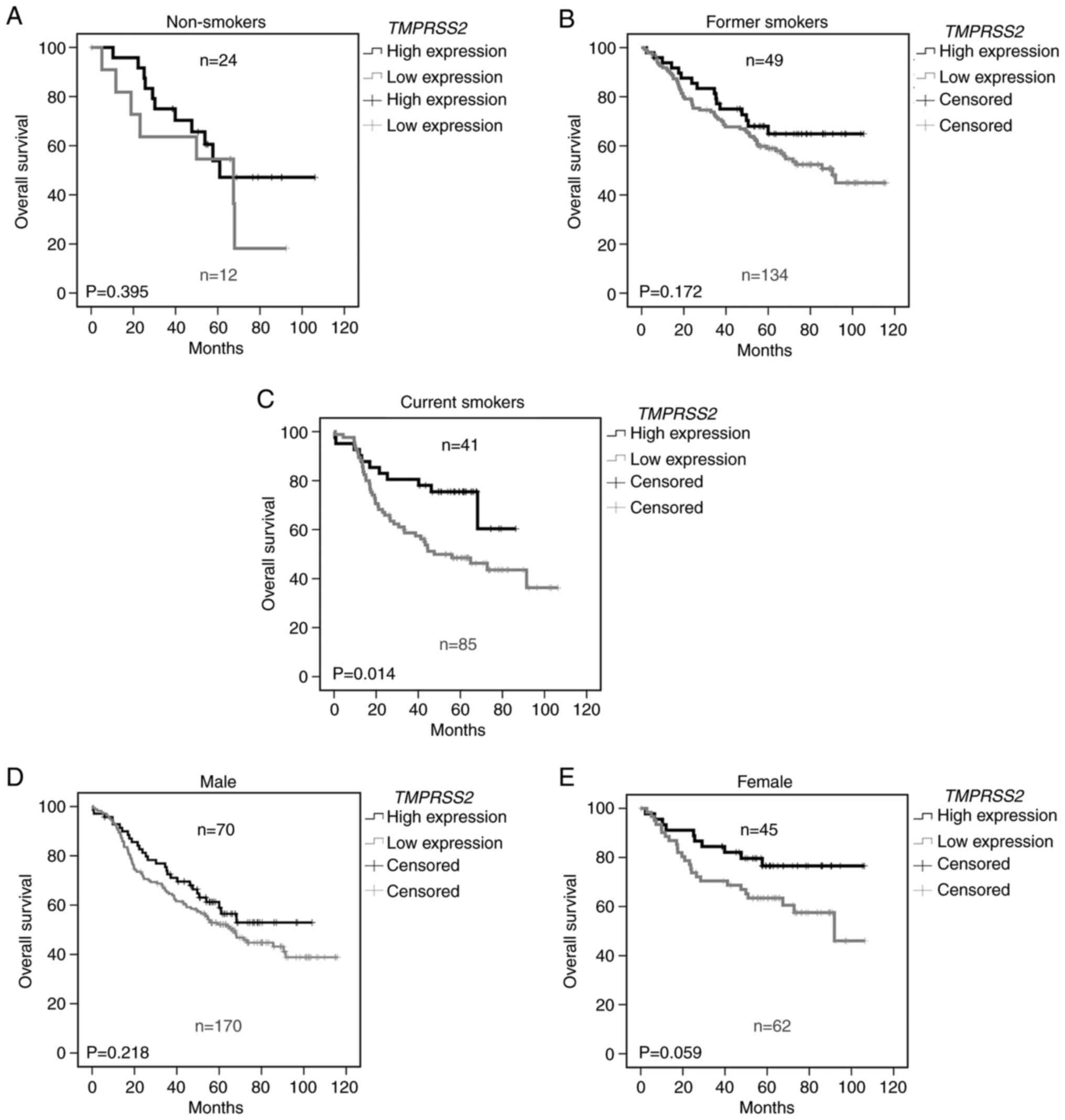
A recent publication demonstrated an increased
expression of TMPRSS2 in lung airway of smokers compared to
non-smokers(18). Therefore, we
further analyzed TMPRSS2 in relationship to the smoking
status (Fig. 2). Indeed, we found
that the prognostic effect of TMPRSS2 expression depends on
the smoking history. While TMPRSS2 expression had no
influence on overall survival in non-smokers (Fig. 2A, P=0.395), a trend was observed
for former smokers (Fig. 2B,
P=0.172) and a significantly worse prognosis was noted in current
smokers (Fig. 2C, P=0.014) with
lower TMPRSS2 expression in tumor tissue. Since
TMPRSS2 is an androgen-regulated gene (19), we analyzed its relationship to
sex. We observed a tendency for male patients (Fig. 2D) and a nearly significant
survival benefit in female patients expressing higher levels of the
TMPRSS2 gene in the lungs (P=0.059, Fig. 2E).
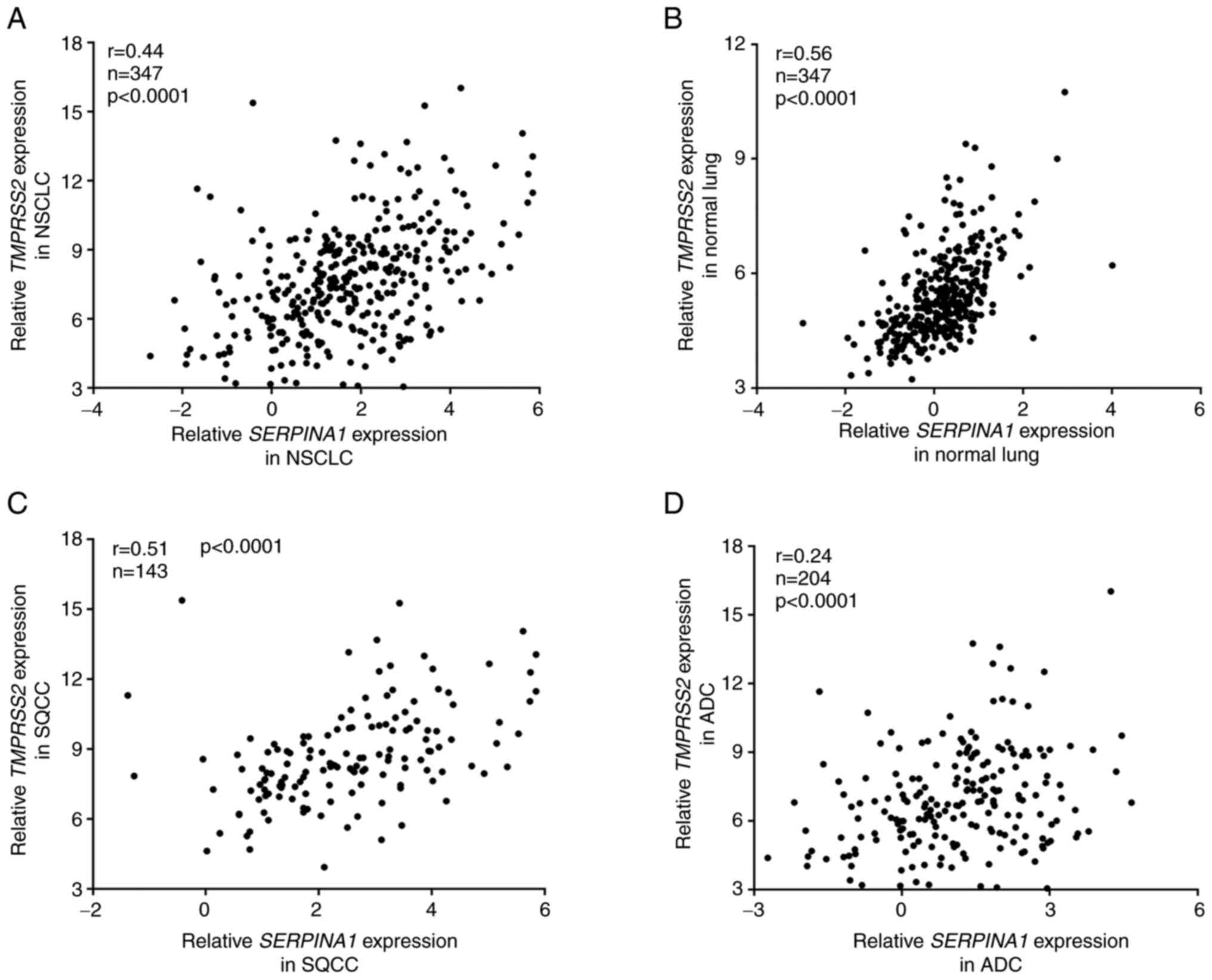
Expression of TMPRSS2 and SERPINA1 genes
is correlated in normal lung and SQCC, but not in ADC tissues
AAT has been shown to inhibit TMPRSS2 activity
(4). Moreover, TMPRSS2 and
SERPINA1 are co-expressed in human lungs (GTEx Multi Gene
Query. GTEx Portal 2020. https://gtexportal.org/home/multiGeneQueryPage/TMPRSS2,SERPINA1).
These facts prompted us to perform correlation analyses between
TMPRSS2 and SERPINA1 expression in our NSCLC patient
cohort (Fig. 3). Overall, we
found a weak correlation (r=0.44) between TMPRSS1 and
SERPINA1 expression in NSCLC tumor tissues (Fig. 3A). However, in normal lung
tissues, the correlation between these two genes was higher
(r=0.56, Fig. 3B).
When we further stratified NSCLC cases into SQCC and
ADC, we found that TMPRSS2 and SERPINA1 genes were
correlated in SQCC (r=0.51, Fig.
3C) but not in the ADC subgroup (r=0.24, Fig. 3D).
TMPRSS2 protein level has prognostic
value for overall survival but does not correlate with AAT
staining
We next performed IHC staining for TMPRSS2 using
tissue microarrays (TMA) to include protein expression data in our
analyses. In total, 104 patients with ADC and 84 patients with SQCC
were evaluated (Fig. 4A). Each
patient was evaluated in duplicates for the proportion of positive
tumor cells (upper panel) and intensity of staining (lower panel).
The total IHC score for each patient was calculated by
multiplication of both values. Out of all the evaluated IHC
patients, 95 were from the qPCR cohort presented in Fig. 1. As shown in Fig. 4B, protein and RNA levels of
TMPRSS2 showed a slight correlation. The same TMAs were evaluated
for AAT expression in our previous study (3). While AAT was detected
heterogeneously and found in both tumor and stroma (Fig. 4C, left panels), TMPRSS2 protein
mainly occurred in tumor cells (Fig.
4C, right panels). While AAT protein levels were similar
between ADC and SQCC (Fig. 4D),
TMPRSS2 levels were significantly higher in ADC than that noted in
SQCC. Furthermore, we found that high protein levels of TMPRSS2 are
associated with better patient overall survival (Fig. 4E) and have prognostic value for
ADC (P=0.006, Fig. 4G), but not
for SQCC (Fig. 4F). This latter
finding is in line with our gene expression data (Fig. 1E and F). However, we did not find
any correlation between TMPRSS2 and AAT protein levels in the
tissue samples analyzed (Fig.
S2).
Since AAT has been described as an inhibitor of
TMPRSS2 activity (4), we further
investigated TMPRSS2 and AAT protein profiles in NSCLC patients.
Therefore, we homogenized normal and tumor tissues (tumor content
>50%) from 24 randomly selected patients (12 SQCC and 12 ADC).
Equal amounts of proteins were separated electrophoretically
followed by western blot analyses (Fig. 5A). By using specific anti-TMPRSS2
and anti-AAT antibodies we were able to detect various molecular
forms of TMPRSS2 (upper panels) and AAT (lower panel) proteins.
Using semi-quantitative densitometric analyses, we compared the
protein profiles of TMPRSS2 in tumor and normal samples (Figs. 5B and S3-S6). We observed an increased amount
of TMPRSS2 cleaved form in tumor tissues of both SQCC and ADC (with
a median increase of 238 and 359%, respectively). The levels of
full-length TMPRSS2 and AAT protein were lower in SQCC (73% for
TMPRSS2 and AAT) and ADC (87% for TMPRSS2 and 69% for AAT) as
compared to normal lung tissues. We further investigated the
relationship between TMPRSS2 and AAT protein levels and did not
find any correlation in patients with SQCC (Fig. 5C). Interestingly, a slight
negative correlation between the two proteins was found in normal
lung tissues of patients with SQCC and ADC (Fig. 5D).
Discussion
Lung cancer is a leading cause of cancer-related
death worldwide and is linked to tobacco smoking and persistent
inflammation (20). To date, the
molecular pathogenesis of lung cancer remains incompletely defined,
and the differences in molecular signatures between adenocarcinoma
(ADC) and squamous cell carcinoma (SQCC), two predominant subtypes
of non-small cell lung cancer (NSCLC), are not well characterized.
SQCC mostly develops in smokers while ADC is common in people who
have never smoked. Moreover, ADC and SQCC exhibit different
mutation spectra and hence patients are treated with different
strategies (1). Recently, Cai
et al highlighted several gene sets that showed different
behavioral patterns in ADC and SQCC. For example, the higher
expression of genes encoding blood coagulation factors were
associated with worse survival predominantly in SQCC whereas
specific changes in cell cycle gene expression were associated with
worse survival in ADC (21). The
investigations of the genes are of clinical importance as they can
provide new insights into mechanisms contributing to ADC and SQCC,
and help to discover diagnostic and prognostic biomarkers for
NSCLC.
Based on our previous findings that low expression
of SERPINA1 (AAT encoding gene) in tumor tissues is
associated with worse NSCLC patient prognosis (3), and that TMPRSS2 and
SERPINA1 genes are co-expressed in human lungs (GTEx Multi
Gene Query, GTEx Portal 2020, https://gtexportal.org/home/multiGeneQueryPage/TMPRSS2,SERPINA1),
we aimed to investigate the relationship between SERPINA1
and TMPRSS2 expression in NSCLC patients. According to RNA
and protein expression data available at Human Protein Atlas
database, the lung, among all organs, has the highest
TMPRSS2 gene expression. Here we showed that patients with
NSCLC have significantly lower TMPRSS2 expression in tumor
than in adjacent non-tumor lung tissue. The most intriguing finding
of our current study is that although a strongly reduced expression
of TMPRSS2 was observed in both SQCC and ADC, only in patients
harboring an ADC was TMPRSS2 gene and protein expression correlated
with a poor overall survival. Our results are in concordance with
recent studies showing lower levels of TMPRSS2 in lung
cancer, and in head and neck cancer (8,22).
The fact that TMPRSS2 expression is higher in the very early
stage of the disease provides a hint that tumors benefit from
reduced levels of TMPRSS2 especially when they start to
metastasize. However, the biological function of TMPRSS2 remains
largely unknown and available data are inconsistent. For instance,
based on publicly available gene expression datasets, Asselta et
al reported that TMPRSS2 expression is higher in
bronchial epithelial cells of males than females, whereas the
expression in the lung is similar (23). Based on three microarray datasets
(GSE40419, GSE19804 and GSE10072) from NCBI GEO, Piva et al
found no statistical differences in TMPRSS2 expression in
lung tissues by stratifying cases for sex or smoking habits, but
found a decrease in TMPRSS2 with increasing age (24). Another study, based on a different
dataset analysis, reported that patients with lung cancer showed
only minor changes in expression of TMPRSS2 when compared to
healthy controls with identical smoking status (25). By contrast, in our large patient
cohort (n=347), we found significantly worse prognosis in current
smokers with NSCLC having low tumor tissue expression of
TMPRSS2 relative to non-smokers or ex-smokers. These
discrepancies may reflect specificities of the clinical cohorts, as
the disease definition in different datasets is not always
consistent. On the other hand, specific microRNAs might target
TMPRSS2 post-transcriptionally, thereby leading to its reduced
expression levels in tumor tissues. All the above postulations
warrant further investigations.
The TMPRSS2 protein contains a 32-kDa extracellular
serine protease domain (26). The
activation of the serine protease requires its cleavage, which is
autocatalytic suggesting that TMPRSS2 may be its own substrate
(27). This active serine
protease with trypsin-like specificity is then shed into the
extracellular space, where it is predicted to interact with other
proteins on the cell surface, matrix components and proteins on
adjacent cells (12).
We further found that in ADC tumor tissues TMPRSS2
protein is correlated with levels of AAT protein. These results
show that there is a direct relationship in ADC tissue. The lower
AAT protein levels within tumor tissues are correlated with a
higher cleavage of TMPRSS2 protein. In general, this scenario seems
to be associated with a worse survival among ADC patients.
Among others, TMPRSS2 may activate the protease
activated receptor PAR2 (12) and
certain metalloproteinases (MMPs) (28). Indeed, it has been reported that
the expression of PAR2 is significantly upregulated in NSCLC cells
(29), and that expression of
MMPs is higher in ADC than in SQCC (30). It is suggested that PAR2 is
involved in epithelial-mesenchymal transition in lung ADC cells and
can serve as a therapeutic target for metastatic lung ADC and a
potential biomarker for predicting the prognosis of lung ADC. These
latter findings in part explain why we found that lower TMPRSS2
expression in tumor tissue is linked to a poor overall survival in
patient with ADC. PAR2 is a G protein-coupled receptor for trypsin
as well as for TMPRRS2 (12),
which after proteolytic activation contributes to growth,
anti-apoptosis, and migration in lung cancer (31). Hypothetically, the inhibition of
TMPRSS2 by AAT may indirectly suppress the activation of PAR2 and
subsequently prevent cancer progression. Therefore, higher
expression of the SERPINA1 gene and higher levels of AAT
protein in tumor tissues, and lower expression of the
TMPRSS2 gene or lower cleavage of TMPRSS2 might suggest
better prognosis. Consequently, we thought that correlations
between SERPINA1 and TMPRSS2 genes in tumor tissues
and occurrence of cleaved forms of TMPRSS2 protein may reflect a
pro-tumorigenic feature of lung cancer. Unfortunately, according to
IHC staining, TMPRSS2 did not correlate with AAT protein. Based on
the western blot analyses, we found only a slight negative
correlation between full-length TMPRSS2 and AAT proteins in
non-tumor tissues of SQCC cases, which seems to be related to
smoking status. Since SQCC patients are often heavy smokers, we
believe that the observed negative correlation in normal lung
tissues of SQCC may be related to smoking status (SQCC patients of
our qPCR cohort had a median of 45 packyears as compared to 30
packyears for ADC). Moreover, ADC often develop in the distal lung
while SQCC typically grows proximal to the bronchus. Consequently,
normal tissue may be more distal or more proximal which might also
influence the expression/correlation of both proteins.
The SERPINA1 gene has been proposed as a
biomarker for a variety of cancer entities, such as cutaneous
squamous cell carcinoma (32),
insulinomas (33), breast cancer
(34) and NSCLC (35). In a large cohort of NSCLC
patients, we previously reported that lower SERPINA1
expression in tumor but higher in adjacent non-tumor lung tissues
as well as higher serum levels of AAT protein were associated with
worse survival rates (3).
Moreover, according to our previous findings, higher serum levels
of AAT in NSCLC patients have prognostic value for a patient's
impaired outcome but do not correlate with SERPINA1
expression in tumor or non-tumor lung tissues or with staining
intensities for tumor-related AAT protein. We pointed out that
protein levels do not necessarily mirror gene expression, and that
protein and protein-coding gene can reflect different processes in
tumor biology. The circulating AAT is mainly produced and secreted
by the liver (36), and can be
taken up by tumor cells (3).
Hence, it is possible that higher tumor levels of AAT prevent
TMPRSS2 auto-cleavage or cleavage by other proteases and/or inhibit
its tumorigenic activity. Although lower TMPRSS2 levels seem to be
associated with a worse survival among lung cancer patients, the
expression of TMPRSS2 and SERPINA1 genes were
correlated only within the normal lung tissue of SQCC and tumor
tissue of ADC.
We demonstrated that TMPRSS2 is a new prognostic
marker for patients with lung ADC. Our data well support the recent
assumption that TMPRSS2 and SERPINA1 genes play a
role in NSCLC. However, the mechanisms regulating the associations
between the two genes remain to be defined and due to limitations
of sample sizes, the findings of our explorative study must be
validated using a larger cohort. We plan to further extend our
research focus on the interaction and correlation of TMPRSS2 and
AAT using additional biobank samples. Using qPCR, we will try to
distinguish between the full-length and the spliced form of TMPRSS2
to investigate which form is relevant for survival prognosis.
Moreover, patient-derived cell culture models will be used to
investigate the interaction of both proteins in the lung. A better
understanding of the mechanisms that regulate the network of
proteases and their inhibitors may identify novel approaches to
disrupt processes important in cancer progression.
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
Conceptualization of the research study was achieved
by MAS, SJ, and SW. Methodology was the responsibility of MAS and
SJ. Software was the responsibility of MAS. Validation of the data
was conducted by MAS, SJ, ARG, and TM. qPCR measurements and IHC
stainings were conducted by MAS and SR. Resources were acquired by
MAS, SJ, TM, HW, TW and MM. Data curation was the responsibility of
MAS, SR, MK, SJ, and SW. Writing-original draft preparation was
conducted by MAS and SJ. Writing-review and editing was conducted
by SJ, ARG, SR, MM, MK, HW, TW and TM. Visualization was the
responsibility of MAS. Supervision was performed by TM, MM, and SJ.
Project administration was the responsibility of MAS and SJ.
Funding acquisition was the responsibility of MM, TM, TW and SJ.
All authors have read and agreed to the final version of the
manuscript for publication.
Ethics approval and consent to
participate
Informed consent was obtained from all subjects
involved in the study. The local ethics committees of the Medical
Faculty Heidelberg and Hannover Medical School (S-270/2001 and
9155_BO_K_2020) approved the use of the biomaterial and data. All
patients signed a broad informed consent for the use of their
samples and data for research purposes (S-270/2001). The consent is
available from the corresponding author on reasonable request.
Patient consent for publication
Not applicable.
Competing interests
The authors declare no competing interests. The
funders had no role in the design of the study; in the collection,
analyses, or interpretation of data; in the writing of the
manuscript, or in the decision to publish the results.
Acknowledgments
The authors would like to thank, Martin Fallenbüchel
and Christa Stolp for the technical support.
Funding
This research was funded by the German Center for Lung Research
(DZL), grant nos. 82DZL00402 and 82DZL002A1.
References
|
1
|
Herbst RS, Morgensztern D and Boshoff C:
The biology and management of non-small cell lung cancer. Nature.
553:446–454. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Miller KD, Nogueira L, Mariotto AB,
Rowland JH, Yabroff KR, Alfano CM, Jemal A, Kramer JL and Siegel
RL: Cancer treatment and survivorship statistics, 2019. CA Cancer J
Clin. 69:363–385. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ercetin E, Richtmann S, Delgado BM,
Gomez-Mariano G, Wrenger S, Korenbaum E, Liu B, DeLuca D, Kuhnel
MP, Jonigk D, et al: Clinical significance of SERPINA1 gene and its
encoded Alpha1-antitrypsin protein in NSCLC. Cancers (Basel).
11:13062019. View Article : Google Scholar
|
|
4
|
Azouz NP, Klingler AM, Callahan V,
Akhrymuk IV, Elez K, Raich L, Henry BM, Benoit JL, Benoit SW, Noé
F, et al: Alpha 1 antitrypsin is an inhibitor of the
SARS-CoV-2-priming protease TMPRSS2. Pathog Immun. 6:55–74. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wettstein L, Conzelmann C, Müller JA, Weil
T, Groß R, Hirschenberger M, Seidel A, Klute S, Zech F, Bozzo CP,
et al: Alpha-1 antitrypsin inhibits SARS-CoV-2 infection. bioRxiv.
2020.
|
|
6
|
Gupta S, Hayek SS, Wang W, Chan L, Mathews
KS, Melamed ML, Brenner SK, Leonberg-Yoo A, Schenck EJ, Radbel J,
et al: Factors associated with death in critically Ill patients
with coronavirus disease 2019 in the US. JAMA Intern Med.
180:1436–1447. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Derosa L, Melenotte C, Griscelli F, Gachot
B, Marabelle A, Kroemer G and Zitvogel L: The immuno-oncological
challenge of COVID-19. Nat Cancer. 1:946–964. 2020. View Article : Google Scholar
|
|
8
|
Sacconi A, Donzelli S, Pulito C, Ferrero
S, Spinella F, Morrone A, Rigoni M, Pimpinelli F, Ensoli F,
Sanguineti G, et al: TMPRSS2, a SARS-CoV-2 internalization protease
is downregulated in head and neck cancer patients. J Exp Clin
Cancer Res. 39:2002020. View Article : Google Scholar
|
|
9
|
Iwata-Yoshikawa N, Okamura T, Shimizu Y,
Hasegawa H, Takeda M and Nagata N: TMPRSS2 contributes to virus
spread and immunopathology in the airways of murine models after
coronavirus infection. J Virol. 93:e01815–e01818. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Clinckemalie L, Spans L, Dubois V, Laurent
M, Helsen C, Joniau S and Claessens F: Androgen regulation of the
TMPRSS2 gene and the effect of a SNP in an androgen response
element. Mol Endocrinol. 27:2028–2040. 2013. View Article : Google Scholar
|
|
11
|
Leach DA, Mohr A, Giotis ES, Cil E, Isac
AM, Yates LL, Barclay WS, Zwacka RM, Bevan CL and Brooke GN: The
antiandrogen enzalutamide downregulates TMPRSS2 and reduces
cellular entry of SARS-CoV-2 in human lung cells. Nat Commun.
12:40682021. View Article : Google Scholar
|
|
12
|
Wilson S, Greer B, Hooper J, Zijlstra A,
Walker B, Quigley J and Hawthorne S: The membrane-anchored serine
protease, TMPRSS2, activates PAR-2 in prostate cancer cells.
Biochem J. 388:967–972. 2005. View Article : Google Scholar :
|
|
13
|
Lam DK, Dang D, Flynn AN, Hardt M and
Schmidt BL: TMPRSS2, a novel membrane-anchored mediator in cancer
pain. Pain. 156:923–930. 2015. View Article : Google Scholar
|
|
14
|
Jairaman A, Yamashita M, Schleimer RP and
Prakriya M: Store-operated Ca2+ release-activated
Ca2+ channels regulate PAR2-activated Ca2+
signaling and cytokine production in airway epithelial cells. J
Immunol. 195:2122–2133. 2015. View Article : Google Scholar
|
|
15
|
Schneider MA, Granzow M, Warth A, Schnabel
PA, Thomas M, Herth FJ, Dienemann H, Muley T and Meister M:
Glycodelin: A new biomarker with immunomodulatory functions in
non-small cell lung cancer. Clin Cancer Res. 21:3529–3540. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bustin SA, Benes V, Garson JA, Hellemans
J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL,
et al: The MIQE guidelines: Minimum information for publication of
quantitative real-time PCR experiments. Clin Chem. 55:611–622.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
McShane LM, Altman DG, Sauerbrei W, Taube
SE, Gion M and Clark GM; Statistics Subcommittee of NCI-EORTC
Working Group on Cancer Diagnostics. REporting recommendations for
tumor MARKer prognostic studies (REMARK). Breast Cancer Res Treat.
100:229–235. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Saheb Sharif-Askari N, Saheb Sharif-Askari
F, Alabed M, Temsah MH, Al Heialy S, Hamid Q and Halwani R: Airways
expression of SARS-CoV-2 receptor, ACE2, and TMPRSS2 is lower in
children than adults and increases with smoking and COPD. Mol Ther
Methods Clin Dev. 18:1–6. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Qiao Y, Wang XM, Mannan R, Pitchiaya S,
Zhang Y, Wotring JW, Xiao L, Robinson DR, Wu YM, Tien JC, et al:
Targeting transcriptional regulation of SARS-CoV-2 entry factors
ACE2 and TMPRSS2. Proc Natl Acad Sci USA. Dec 11–2020.Epub ahead of
print.
|
|
20
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cai L, Luo D, Yao B, Yang DM, Lin S,
Girard L, DeBerardinis RJ, Minna JD, Xie Y and Xiao G: Systematic
analysis of gene expression in lung adenocarcinoma and squamous
cell carcinoma with a case study of FAM83A and FAM83B. Cancers
(Basel). 11:8862019. View Article : Google Scholar
|
|
22
|
Kong Q, Xiang Z, Wu Y, Gu Y, Guo J and
Geng F: Analysis of the susceptibility of lung cancer patients to
SARS-CoV-2 infection. Mol Cancer. 19:802020. View Article : Google Scholar
|
|
23
|
Asselta R, Paraboschi EM, Mantovani A and
Duga S: ACE2 and TMPRSS2 variants and expression as candidates to
sex and country differences in COVID-19 severity in Italy. Aging
(Albany NY). 12:10087–10098. 2020. View Article : Google Scholar
|
|
24
|
Piva F, Sabanovic B, Cecati M and
Giulietti M: Expression and co-expression analyses of TMPRSS2, a
key element in COVID-19. Eur J Clin Microbiol Infect Dis.
40:451–455. 2021. View Article : Google Scholar
|
|
25
|
Yin J, Kasper B, Petersen F and Yu X:
Association of cigarette smoking, COPD, and lung cancer with
expression of SARS-CoV-2 entry genes in human airway epithelial
cells. Front Med (Lausanne). 7:6194532020. View Article : Google Scholar
|
|
26
|
Paoloni-Giacobino A, Chen H, Peitsch MC,
Rossier C and Antonarakis SE: Cloning of the TMPRSS2 gene, which
encodes a novel serine protease with transmembrane, LDLRA, and SRCR
domains and maps to 21q22.3. Genomics. 44:309–320. 1997. View Article : Google Scholar
|
|
27
|
Afar DE, Vivanco I, Hubert RS, Kuo J, Chen
E, Saffran DC, Raitano AB and Jakobovits A: Catalytic cleavage of
the androgen-regulated TMPRSS2 protease results in its secretion by
prostate and prostate cancer epithelia. Cancer Res. 61:1686–1692.
2001.
|
|
28
|
Reid JC, Matsika A, Davies CM, He Y,
Broomfield A, Bennett NC, Magdolen V, Srinivasan B, Clements JA and
Hooper JD: Pericellular regulation of prostate cancer expressed
kallikrein-related peptidases and matrix metalloproteinases by cell
surface serine proteases. Am J Cancer Res. 7:2257–2274. 2017.
|
|
29
|
Jiang Y, Zhuo X, Fu X, Wu Y and Mao C:
Targeting PAR2 overcomes gefitinib resistance in non-small-cell
lung cancer cells through inhibition of EGFR transactivation. Front
Pharmacol. 12:6252892021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Thomas P, Khokha R, Shepherd FA, Feld R
and Tsao MS: Differential expression of matrix metalloproteinases
and their inhibitors in non-small cell lung cancer. J Pathol.
190:150–156. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tsai CC, Chou YT and Fu HW:
Protease-activated receptor 2 induces migration and promotes
slug-mediated epithelial-mesenchymal transition in lung
adenocarcinoma cells. Biochim Biophys Acta Mol Cell Res.
1866:486–503. 2019. View Article : Google Scholar
|
|
32
|
Farshchian M, Kivisaari A, Ala-Aho R,
Riihilä P, Kallajoki M, Grénman R, Peltonen J, Pihlajaniemi T,
Heljasvaara R and Kähäri VM: Serpin peptidase inhibitor clade A
member 1 (SerpinA1) is a novel biomarker for progression of
cutaneous squamous cell carcinoma. Am J Pathol. 179:1110–1119.
2011. View Article : Google Scholar
|
|
33
|
de Sa SV, Correa-Giannella ML, Machado MC,
Krogh K, de Almeida MQ, Albergaria Pereira MA, Coelho Siqueira SA,
Patzina RA, Ibuki FS, Sogayar MC, et al: Serpin peptidase inhibitor
clade A member 1 as a potential marker for malignancy in
insulinomas. Clin Cancer Res. 13:5322–5330. 2007. View Article : Google Scholar
|
|
34
|
Chan HJ, Li H, Liu Z, Yuan YC, Mortimer J
and Chen S: SERPINA1 is a direct estrogen receptor target gene and
a predictor of survival in breast cancer patients. Oncotarget.
6:25815–25827. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhao W, Yang Z, Liu X, Tian Q, Lv Y, Liang
Y, Li C, Gao X and Chen L: Identification of alpha1-antitrypsin as
a potential prognostic biomarker for advanced nonsmall cell lung
cancer treated with epidermal growth factor receptor tyrosine
kinase inhibitors by proteomic analysis. J Int Med Res. 41:573–583.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Janciauskiene SM, Bals R, Koczulla R,
Vogelmeier C, Kohnlein T and Welte T: The discovery of
alpha1-antitrypsin and its role in health and disease. Respir Med.
105:1129–1139. 2011. View Article : Google Scholar
|