Introduction
Human and animal studies have demonstrated a strong
association between intrauterine growth retardation (IUGR) and
increased incidence of insulin resistance, obesity and type 2
diabetes during adult life (1,2).
This association has been conceptualized by a developmental
programming hypothesis, which proposes that disease risk begins
during fetal life as a result of ‘programming’ or long-term
alterations in gene expression and function resulting from a
suboptimal intrauterine milieu (3). Maternal undernutrition or abnormal
utero-placental function are capable of limiting availability of
substrates to the fetus and may induce secondary adaptations in the
metabolism and gene expression that may be beneficial during
intrauterine life but contribute to disease risk in later life.
Several IUGR rat models have been established in
order to investigate the mechanisms underlying the intrauterine
events and eventual adult phenotype. These animal models include
maternal protein-restricted (PR) diets (4), maternal semi-nutrient restriction
(5), maternal anemia (6), maternal hypoxia (7) and bilateral uterine artery ligation
(8) in rats. Among these IUGR
models, maternal PR diet is one of the most extensively studied and
the outcomes for offspring bear striking similarities to human
diabetes, both at the whole body and molecular level (9).
In order to understand the mechanisms responsible
for glucose intolerance that develop in later life, several in
vivo and in vitro studies in animals have focused on
skeletal muscle as an important target tissue of glucose disposal.
These studies suggest that major changes in the genes that regulate
glucose metabolism are associated with the development of type 2
diabetes mellitus. These changes have been shown to affect the
insulin receptor and its signaling system (10,11),
the insulin-responsive glucose uptake and transporter system
(12,13) and oxidative phosphorylation and ATP
production (14). The roles played
by other elements of the signaling system in IUGR rats are more
controversial. These include the insulin-responsive glucose
transporter 4 (GLUT4) (5,10–12),
transcriptional coactivator peroxisome proliferator-activated
receptor γ coactivator-1α (PGC-1α), which regulates the expression
of genes for oxidative phosphorylation and ATP production (15), and the insulin receptor (IR) and
insulin receptor substrate 1 (IRS-1), which are involved in insulin
receptor signaling.
It has recently been proposed that epigenetic
regulation of genes, particularly the methylation of clusters of
CpG dinucleotides (islands) in promoter regions of certain genes,
may contribute to metabolic reprogramming (16). Lillycrop et al demonstrated
that feeding a PR diet to pregnant rats increased glucocorticoid
receptor (GR) and peroxisome proliferator-activated receptor α
(PPARα) expression in the liver of the offspring by inducing
hypomethylation of respective promoters (17). These findings suggest that an
epigenetic mechanism induced by prenatal nutrition may produce an
altered phenotype in the offspring.
Given the metabolic phenotype in IUGR humans and the
importance of IUGR as a risk factor for type 2 diabetes, we
developed an IUGR experimental model using a maternal PR diet
during gestation. To avoid the confounding factors of gender and
hormones, only 18-month-old female offspring were selected for
investigation in the present study. Our objectives were to evaluate
the metabolic phenotype and insulin resistance status and to
determine expression changes in genes involved in key insulin
signaling, glucose metabolism and oxidative phosphorylation in the
skeletal muscles. We also investigated DNA methylation of candidate
genes that may contribute to metabolic phenotypes in IUGR
offspring.
Materials and methods
Animal procedures
Virgin, 7- to 8-week-old Sprague-Dawley (SD) rats
weighing 180±20 g were purchased from Shanghai Laboratory Animal
Center (Chinese Academy of Science, Shanghai, China). All the
animals were housed at 21–23°C, 65–69% humidity with a 12-h
light/dark cycle and had free access to food and tap water.
Following 10 days of habituation, female rats were mated overnight
with a male and copulation was verified the next morning by the
presence of spermatozoa in vaginal smears.
At conception, pregnant dams were housed
individually and fed isocaloric diets containing either normal
(20%) protein (control) or a PR diet containing 8% protein until
delivery. The composition of the diets has been described
previously (4). After delivery,
each mother rat fed eight pups (any extra pups were removed at
random). All mother rats were fed with normal rat chow during the
21-day lactation period. Following weaning, three or four rats from
the same group were housed in one cage. In order to avoid gender
and hormonal influence, only female offspring were selected.
Ten female pups born from mothers who received the
PR diet formed the IUGR group and 10 female pups from mothers fed a
normal diet formed the control group. The rats were weighed weekly.
All experiments were approved by the Animal Care and Use Committee
of Southeast University (Nanjing, China).
Intraperitoneal glucose tolerance test
(IGTT)
The rats were subjected to an IGTT as described
previously (8). Briefly,
18-month-old awake female control and IUGR rats received an
intraperitoneal injection of 2 g/kg glucose after fasting for 12 h.
Blood was collected from the tail veins 0, 15, 30, 60 and 120 min
after glucose administration. The EDTA tubes containing the blood
were gently mixed 10 times and centrifuged at 1500 × g for 10 min
at 4°C. The plasma was immediately transferred to a new tube and
stored at −20°C until assay. Plasma glucose was measured using the
glucose oxidase method (Sigma Diagnostics, St. Louis, MO, USA).
Insulin was quantified using a commercially available enzyme linked
immunoabsorbent assay (ELISA) kit (Cayman Chemical Co., Ann Arbor,
MI, USA). All measurements were performed in duplicate using a
microplate reader (Bio-Tek Instruments, Inc., Winooski, VT,
USA).
Skeletal muscle collection
At the end of the experimental period, 18-month-old
female rats were sacrificed by decapitation. The gastrocnemius
muscles of the right posterior limb were rapidly removed, frozen in
liquid nitrogen and stored at −80°C.
RNA isolation and quantitative real-time
PCR
Total RNA was extracted from the skeletal muscles
using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to
the manufacturer’s instructions. DNA contamination was removed
using an Amibion DNA-free kit (Applied Biosystems, Foster City, CA,
USA). Aliquots (2 μg) of total RNA were reverse-transcribed using
an iScript™ cDNA synthesis kit (Bio-Rad, Hercules, CA, USA) at a
final volume of 40 μl according to the manufacturer’s instructions.
The reaction was terminated by heating for 5 min at 25°C, for 30
min at 42°C and for 5 min at 85°C and quickly cooling on ice.
The expression of IR, IRS-1, GLUT4, PGC-1α and the
housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH)
were assessed simultaneously in individual samples. Quantitative
real-time PCR analysis was performed using SYBR-Green Master mix
(Bio-Rad) and a CFX96™ Real-Time PCR Detection System instrument
(Bio-Rad). The cycling consisted of 2 min at 50°C and 2 min at
95°C, followed by 40 cycles of 15 sec at 95°C and 45 sec at 60°C.
Following completion of the final cycle, a melting curve analysis
was performed to monitor the purity of the PCR products. Each
sample was analyzed in duplicate.
RNA levels in the IUGR group were calculated
relative to the control group, for which values were arbitrary set
to 1 to obtain estimates of relative abundance. All primers were
synthesized by Shengneng Bicolor Biotech (Shanghai, China) and were
designed according to published sequences in GenBank as listed in
Table I.
 | Table ISequence of DNA oligonucleotide
primers used in quantitative real-time RT-PCR (qRT-PCR) and
bisulphate sequencing PCR (BSP-PCR) experiments. |
Table I
Sequence of DNA oligonucleotide
primers used in quantitative real-time RT-PCR (qRT-PCR) and
bisulphate sequencing PCR (BSP-PCR) experiments.
| Method | Target gene | Forward primer | Reverse primer |
|---|
| qRT-PCR | IR |
5′-TTCGAGGAGAGACCTTGGAA-3′ |
5′-TCGTGAGGTTGTGCTTGTTC-3′ |
| IRS-1 |
5′-TGGATGCAAGTGGATGACTC-3′ |
5′-CGGAGGATTGTTGAGATGGT-3′ |
| GLUT4 |
5′-ACAATGTCTTGGCTGTGCTG-3′ |
5′-TCCCACATACATAGGCACCA-3′ |
| PGC1α |
5′-TCTGGAACTGCAGGCCTAACTC-3′ |
5′-GCAAGAGGGCTTCAGCTTTG-3′ |
| GAPDH |
5′-CATGACAACTTTGGCATCGT-3′ |
5′-GGATGCAGGGATGATGTTCT-3′ |
| BSP-PCR | PGC1α |
5′-TTAGAGATTTAGGGGTGAAGTAA-3′ |
5′-CTAATCTTCAAAACCCCAAAAT-3′ |
| GLUT4 |
5′-TTTAGGAATTAATGTAGAGAAATG-3′ |
5′-AATAACTATTTTTAACTCCCAC-3′ |
DNA methylation detection
DNA methylation in promoters was detected using
bisulphate sequencing PCR (BSP-PCR). Briefly, genomic DNA from rat
skeletal muscle was extracted using DNeasy Mini kits (Qiagen)
according to the manufacturer’s instructions. The genomic DNA (1
μg) was subjected to bisulphate modification using a CpGenome DNA
Modification kit (Chemicon, Temecula, CA, USA) according to the
manufacturer’s instructions. The chemically modified DNA was then
used as a template for methylation-specific PCR in 2 target genes
(PGC-1α and GLUT4) in skeletal muscle. All primers (Table I) were designed according to the
NCBI genome database using Methyl Primer Express v1.0 (ABI) and
were synthesized by Shengneng Bicolor Biotech.
The PCR products were separated on 1% agarose gel,
and the bands were purified with an agarose gel DNA purification
kit (Promega,, Madison, WI, USA). The purified DNA was subcloned
onto the pGEM-T Easy Vector (Promega). Positive clones were
sequenced using M13 primer from Shanghai Invitrogen Biotech Co.,
Ltd. (Shanghai, China). The final sequence results were processed
using an online computer program: http://biq-analyzer.bioinf.mpi-sb.mpg.de/ (18).
Statistical analysis
Statistical analyses were performed using SPSS
version 15.0 statistical software. The data are presented as the
means ± standard error (SEM). The differences between control and
IUGR groups were determined by two-tailed Student’s t-tests or
χ2 tests. Values of P<0.05 were considered to
indicate a statistically significant difference.
Results
Body weight at birth and postnatally
The gestation period of pregnant rats fed both
normal protein and PR diet was between 21 and 22 days. There were
no significant differences in litter sizes or litter gender
distribution (Table II) between
normal and PR diet dams. The average birth weight of pups from
normal diet pregnant dams was calculated. Pups whose birth weight
was below the 10th percentile for the average birth weight were
defined as having IUGR. The incidence of IUGR in pregnant rats on
the PR diet (66.4%) was significantly higher than that of rats on
the normal diet (4.48%, P<0.001) (Table II). These results confirmed that
administration of an isocaloric PR diet to gestating rats did not
affect fertility and provided a convincing IUGR model (4).
 | Table IICharacteristics at birth in normal and
protein restriction diet pregnant rats. |
Table II
Characteristics at birth in normal and
protein restriction diet pregnant rats.
| Control rats
(n=7) | PR rats (n=12) | P-value |
|---|
| Litter size | 9.57±0.53 | 9.67±0.43 | 0.89 |
| Litter gender
distribution (M/F) | 1.11±0.07 | 1.15±0.04 | 0.60 |
| Incidence of | 4.48 | 66.38 | <0.001 |
| IUGR (%) | | | |
The average body weight at birth and at different
periods of postnatal life in control and IUGR female rats is shown
in Table III. Birth weights of
IUGR rats (4.93±0.16 g) were markedly lower than those of control
rats (6.65±0.20 g; P<0.05). At 4 weeks of age, the weights of
IUGR rats began to approach those of rats in the control group. At
4–8 weeks of age, the growth of IUGR rats accelerated and surpassed
that of control rats. The difference at this time point was not
statistically significant. However, at 12 weeks of age, IUGR rats
were significantly obese (244.14±8.31 g) compared with control rats
(214.18±7.94 g; P<0.05). This difference persisted until the end
of the experiment.
 | Table IIIBody weights of the female rats at
different times in the control and IUGR group. |
Table III
Body weights of the female rats at
different times in the control and IUGR group.
| Body weight
(g) |
|---|
|
|
|---|
| Age | CON (n=12) | IUGR (n=12) | P-value |
|---|
| At birth | 6.65±0.20 | 4.93±0.16b | <0.001 |
| 1 week | 12.19±0.44 | 9.67±0.51b | <0.01 |
| 4 weeks | 88.81±4.66 | 85.72±4.76 | 0.65 |
| 8 weeks | 176.72±6.91 | 189.90±7.96 | 0.22 |
| 12 weeks | 214.18±7.94 | 244.14±8.31a | 0.02 |
| 12 months | 232.84±8.05 | 259.98±7.52a | 0.02 |
| 18 months | 234.70±8.15 | 261.81±9.32a | 0.04 |
Plasma glucose and insulin
concentrations
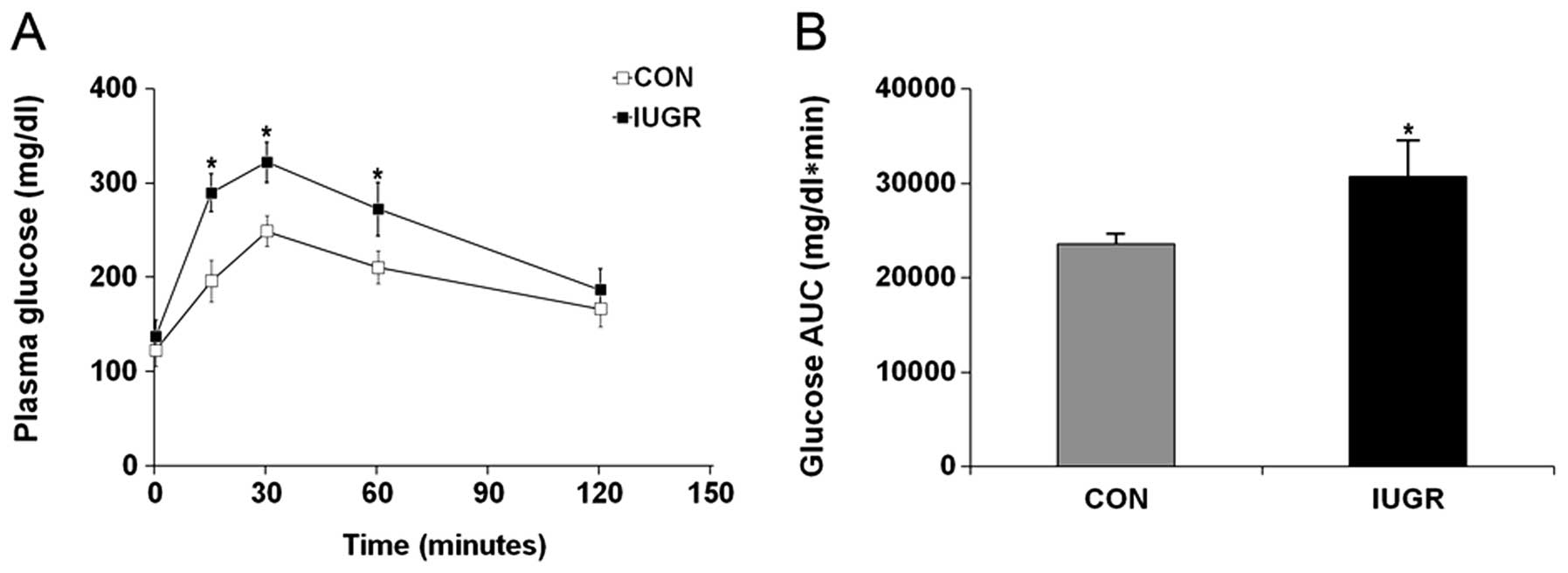
IGTT was performed in 18-month-old female IUGR and
control rats to investigate whether older female IUGR rats develop
insulin resistance. The analysis revealed that the fasting glucose
in the IUGR rats was slightly higher than in control rats, but the
difference was not statistically significant (Fig. 1A; P=0.09). However, plasma glucose
concentrations at 15, 30 and 60 min were significantly higher in
IUGR rats than in control rats (P<0.001 at all three time
points) resulting in a significantly higher area under the curve
(AUC) (Fig. 1B; P<0.001).
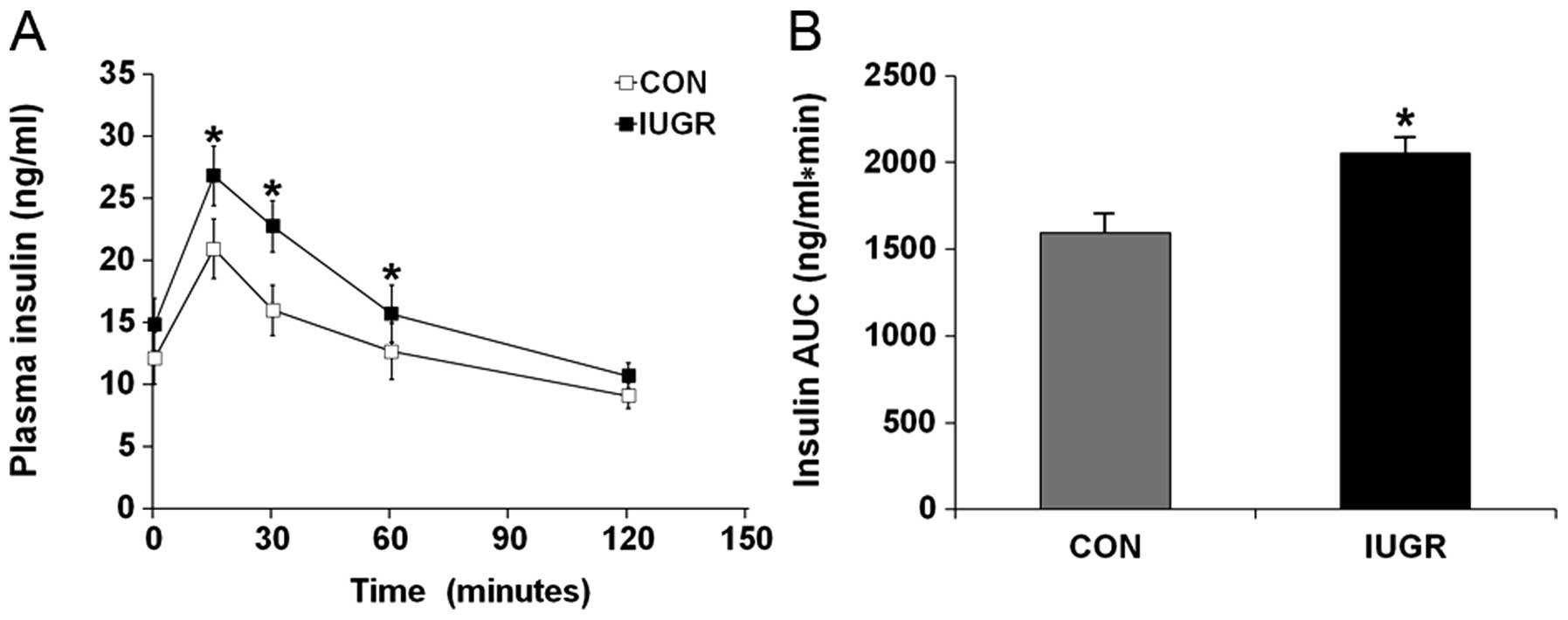
Hyperglycemia during IGTT in the IUGR group was
associated with a significant increase in the insulin response.
Plasma insulin concentrations in the IUGR group 15, 30 and 60 min
after the glucose challenge were significantly higher than in the
control group (Fig. 2A;
P<0.001). This resulted in a higher insulin AUC than in the
control group (Fig. 2B;
P<0.001).
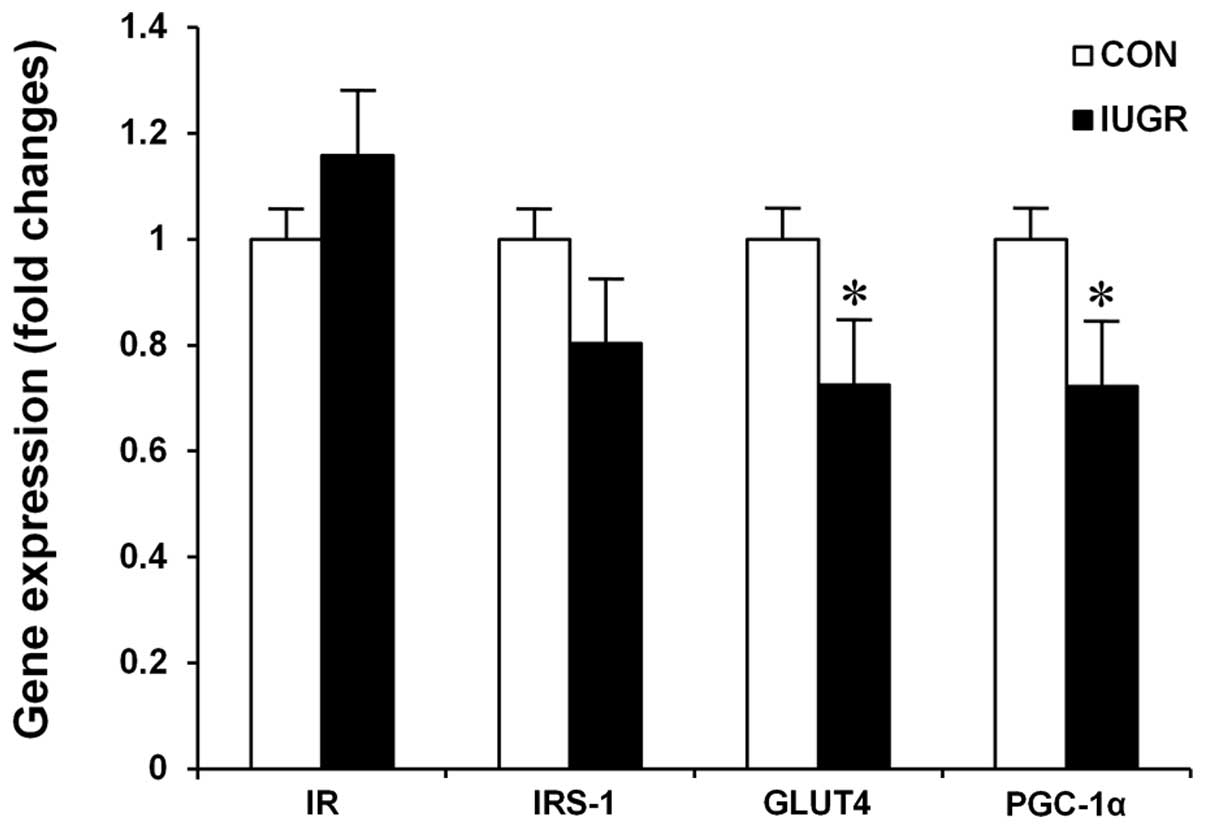
Quantitative real-time PCR
In comparison with age-matched control rats,
18-month-old IUGR rats exhibited a significant decrease in
expression of GLUT4 (P=0.0308) and PGC-1α mRNA (P=0.0416) (Fig. 3). No statistically significant
between-group differences were found for IR (P=0.2589) or IRS-1
(P=0.2265) genes.
DNA methylation
As shown in Fig. 4,
the 11 CpG sites in the examined promoter sequence of GLUT4 were
rarely methylated but did include some sporadic methylated sites.
Despite this, methylation of the PGC-1α gene was significantly
higher in the IUGR group (average of 16.18% of all the 17 CpG
sites) than in the control group (9.31%; P<0.05) (Fig. 5).
Discussion
The nutritional environment at the fetal and
neonatal stages has been suggested to be a critical factor in
development (1). In the current
study, we focused primarily on the female offspring in order to
avoid gender and hormone effects on metabolism. Our data showed
that a maternal PR diet during gestation had a profound and
long-term impact on the offspring. The offspring initially
exhibited low birth weight and IUGR but went on to develop obesity
and peripheral insulin resistance in older age. We also showed that
GLUT4 and PGC-1α mRNA expression were reduced in skeletal muscles
from the older female IUGR rats, and demonstrated that epigenetic
mechanisms are likely to be operative in the pathogenesis of
insulin resistance and metabolic phenotype, since DNA methylation
of the PGC-1α promoter was found to be increased.
Data from IUGR animal models support the opinion
that poor fetal growth has permanent consequences in adulthood.
Birth weights of IUGR induced by PR diet (19) and bilateral uterine artery ligation
(8) during gestation have been
reported by others to be significantly lower than those of
controls. Our finding that administration of a PR diet during
pregnancy also interfered with the general growth of the pups,
initially resulting in a lower birth weight but subsequently
resulting in obesity, also supports the previously reported
findings (8,19). We also found that IUGR rats
exhibited peripheral insulin resistance and displayed hyperglycemia
and hyperinsulinemia during IGTT. Such observations suggest that
animals with IUGR secrete more insulin than control rats, but are
unable to sustain normal glycemia. This finding agrees with earlier
studies in malnourished animals during gestation (20). The findings from our animal model
support the hypothesis that intrauterine protein restriction
results in a phenotype that mirrors the epidemiological association
between low birth weight and subsequent development of impaired
glucose tolerance and type 2 diabetes in humans (21).
Skeletal muscle is the major tissue presenting
insulin-responsive glucose uptake. In our study, hyperinsulinemia
was associated with a decrease in skeletal muscle GLUT4 expression.
Previous in vitro investigations have demonstrated variable
results regarding IUGR-induced changes in skeletal muscle GLUT4
mRNA expression. Different rat models of IUGR adult offspring have
also shown conflicting results. No change in expression was
reported with a utero-placental insufficiency model (22), whereas a total calorie restriction
model resulted in a significant decrease in expression (23). Our investigation using PR
diet-induced IUGR also demonstrated significantly decreased
skeletal muscle GLUT4 mRNA concentration in mature animals. This
observation is consistent with the decline in total GLUT4
concentration reported previously in IUGR (24) and replicated in the young adult
IUGR human skeletal muscle (25).
Other investigators have demonstrated that insulin resistance is
associated with an impaired regulation of insulin-induced GLUT4
gene expression in skeletal muscle and adipose tissue in human IUGR
subjects (26).
Transcriptional coactivator PGC-1α is a key
metabolic factor regulating the expression of genes for oxidative
phosphorylation in several tissues including skeletal muscle, liver
and adipose tissue and is an important factor in the development of
type 2 diabetes (15). Previous
studies suggest that reduced expression of PGC-1α in the islets
(27) and skeletal muscle
(28) is associated with insulin
resistance in patients with type 2 diabetes. However, the level of
PGC-1α in skeletal muscles from mature IUGR offspring has been
unknown to date. Our results indicate that the expression of the
PGC-1α gene is reduced in skeletal muscle from 18-month-old female
IUGR offspring. However, we found no difference in IR or IRS-1 gene
expression between the IUGR and control groups. The lack of
statistical significance in IR and IRS-1 mRNA expression may
suggest that the molecular defect lies downstream of the insulin
receptor. Together, these data suggest that whole body glucose
intolerance in our model may be due to dysregulation of GLUT4 and
PGC-1α expression.
There is growing evidence that gene
promoter-specific DNA methylation changes (17) are involved in nutritional
aberrations. Since decreased GLUT4 and PGC-1α gene expression both
progress in old age, we hypothesized that such reductions may be
mediated in part by altered DNA methylation. However, our study did
not find any changes in DNA methylation in the GLUT4 promoter. In
other studies, genes such as the insulin-like growth factor 2 were
shown to be differentially methylated in regions far upstream of
the entire gene and were found to modify downstream gene
transcription (29). Whether a
similar situation exists in the case of GLUT4 expression cannot be
ruled out by our studies, as we primarily focused on the gene
promoter region.
Other workers have reported that histone code
modifications repress skeletal muscle GLUT4 transcription in the
postnatal period and that these changes persist in adult female
IUGR offspring (23). Upstream of
the GLUT4 promoter, there are several binding sites for various
transcriptional factors that could potentially regulate GLUT4
expression under different situations (30). Thus, we speculate that reduced
expression of GLUT4 in skeletal muscle from 18-month-old female
IUGR rats may be due to altered methylation of other genomic
region(s), altered histone modification, or changes in
binding/expression of other transcription factors regulating
GLUT4.
It has also been reported that DNA methylation of
PGC-1α increased in human diabetic islets from T2D patients
(27) and the umbilical cord of
newborns from mothers with high pregestational BMI (31). Furthermore, PGC-1α promoters were
found to be methylated to a higher extent in skeletal muscle
biopsies from young and lean low birth weight (LBW) offspring
compared with normal birth weight (NBW) subjects subjected to an
isocaloric control diet (32). Our
finding that 18-month-old female IUGR rats exhibited increased DNA
methylation of PGC-1α in muscle tissues is in accordance with these
reports, and suggests that IUGR may be involved in the reduced
PGC-1α gene expression and subsequently in the development of
insulin resistance in type 2 diabetes. It should be noted that the
corresponding methylation pattern of the genes examined in our
study was only undertaken at 18 months. However, Lillycrop et
al(33) reported that the
pattern of methylation in the hepatic PPARα promoter induced by
maternal PR may persist into adulthood. Whether methylation changes
of the PGC-1α promoter in skeletal muscle exhibit the same trend
requires further investigation.
In conclusion, we have shown that a PR diet during
pregnancy leads to epigenetic modulation of PGC-1α in the skeletal
muscles of 18-month-old female offspring, which may be associated
with downregulation of PGC-1α transcription. Perturbations in
PGC-1α and GLUT4 expression in skeletal muscle may contribute to
the insulin resistance in offspring with IUGR. These findings
provide novel insights into the molecular mechanisms of skeletal
dysfunction, indicating that transcription regulation of oxidative
phosphorylation by PGC-1α may be involved in the pathological
process of IUGR through epigenetic factors such as DNA methylation.
This hypothesis requires confirmation by further elucidation of the
signaling pathways leading to DNA methylation of PGC-1α and other
potential genes.
Acknowledgements
This study was supported by a grant from the program
(ZKX09013) of Nanjing Medical Science and Technique Development
Foundation.
References
|
1
|
Barker DJ: The fetal and infant origins of
adult disease. BMJ. 301:11111990. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kanaka-Gantenbein C: Fetal origins of
adult diabetes. Ann N Y Acad Sci. 1205:99–105. 2010. View Article : Google Scholar
|
|
3
|
Hales CN and Barker DJ: The thrifty
phenotype hypothesis. Br Med Bull. 60:5–20. 2001. View Article : Google Scholar
|
|
4
|
Ozanne SE, Martensz ND, Petry CJ, Loizou
CL and Hales CN: Maternal low protein diet in rats programmes fatty
acid desaturase activities in the offspring. Diabetologia.
41:1337–1342. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Thamotharan M, Shin BC, Suddirikku DT,
Thamotharan S, Garg M and Devaskar SU: GLUT4 expression and
subcellular localization in the intrauterine growth-restricted
adult rat female offspring. Am J Physiol Endocrinol Metab.
288:E935–E947. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lisle SJ, Lewis RM, Petry CJ, Ozanne SE,
Hales CN and Forhead AJ: Effect of maternal iron restriction during
pregnancy on renal morphology in the adult rat offspring. Br J
Nutr. 90:33–39. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
de Grauw TJ, Myers RE and Scott WJ: Fetal
growth retardation in rats from different levels of hypoxia. Biol
Neonate. 49:85–89. 1986.PubMed/NCBI
|
|
8
|
Simmons RA, Templeton LJ and Gertz SJ:
Intrauterine growth retardation leads to the development of type 2
diabetes in the rat. Diabetes. 50:2279–2286. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kahn BB: Facilitative glucose
transporters: regulatory mechanisms and dysregulation in diabetes.
J Clin Invest. 89:1367–1374. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sampaio de Freitas M, Garcia De Souza EP,
Vargas da Silva S, et al: Up-regulation of phosphatidylinositol
3-kinase and glucose transporter 4 in muscle of rats subjected to
maternal undernutrition. Biochim Biophys Acta. 1639:8–16.
2003.PubMed/NCBI
|
|
11
|
Ozanne SE, Olsen GS, Hansen LL, et al:
Early growth restriction leads to down regulation of protein kinase
C zeta and insulin resistance in skeletal muscle. J Endocrinol.
177:235–241. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Agote M, Goya L, Ramos S, et al: Glucose
uptake and glucose transporter proteins in skeletal muscle from
undernourished rats. Am J Physiol Endocrinol Metab.
281:E1101–E1109. 2001.PubMed/NCBI
|
|
13
|
Gavete ML, Martin MA, Alvarez C and
Escriva F: Maternal food restriction enhances insulin-induced
GLUT-4 translocation and insulin signaling pathway in skeletal
muscle from suckling rats. Endocrinology. 146:3368–3378. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Selak MA, Storey BT, Peterside I and
Simmons RA: Impaired oxidative phosphorylation in skeletal muscle
of intrauterine growth-retarded rats. Am J Physiol Endocrinol
Metab. 285:E130–E137. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lin J, Handschin C and Spiegelman BM:
Metabolic control through the PGC-1 family of transcription
coactivators. Cell Metab. 1:361–370. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bird A: DNA methylation patterns and
epigenetic memory. Genes Dev. 16:6–21. 2002. View Article : Google Scholar
|
|
17
|
Lillycrop KA, Phillips ES, Jackson AA,
Hanson MA and Burdge GC: Dietary protein restriction of pregnant
rats induces and folic acid supplementation prevents epigenetic
modification of hepatic gene expression in the offspring. J Nutr.
135:1382–1386. 2005.PubMed/NCBI
|
|
18
|
Bock C, Reither S, Mikeska T, Paulsen M,
Walter J and Lengauer T: BiQ Analyzer: visualization and quality
control for DNA methylation data from bisulfite sequencing.
Bioinformatics. 21:4067–4068. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu XM, Kong J, Song WW and Lu Y: Glucose
metabolic and gluconeogenic pathways disturbance in the
intrauterine growth restricted adult male rats. Chin Med Sci J.
24:208–212. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Blondeau B, Avril I, Duchene B and Breant
B: Endocrine pancreas development is altered in foetuses from rats
previously showing intra-uterine growth retardation in response to
malnutrition. Diabetologia. 45:394–401. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Phipps K, Barker DJ, Hales CN, Fall CH,
Osmond C and Clark PM: Fetal growth and impaired glucose tolerance
in men and women. Diabetologia. 36:225–228. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sadiq HF, Das UG, Tracy TF and Devaskar
SU: Intra-uterine growth restriction differentially regulates
perinatal brain and skeletal muscle glucose transporters. Brain
Res. 823:96–103. 1999. View Article : Google Scholar
|
|
23
|
Raychaudhuri N, Raychaudhuri S,
Thamotharan M and Devaskar SU: Histone code modifications repress
glucose transporter 4 expression in the intrauterine
growth-restricted offspring. J Biol Chem. 283:13611–13626. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Boloker J, Gertz SJ and Simmons RA:
Gestational diabetes leads to the development of diabetes in
adulthood in the rat. Diabetes. 51:1499–1506. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ozanne SE, Jensen CB, Tingey KJ, Storgaard
H, Madsbad S and Vaag AA: Low birthweight is associated with
specific changes in muscle insulin-signalling protein expression.
Diabetologia. 48:547–552. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jaquet D, Vidal H, Hankard R, Czernichow P
and Levy-Marchal C: Impaired regulation of glucose transporter 4
gene expression in insulin resistance associated with in utero
undernutrition. J Clin Endocrinol Metab. 86:3266–3271.
2001.PubMed/NCBI
|
|
27
|
Ling C, Del Guerra S, Lupi R, et al:
Epigenetic regulation of PPARGC1A in human type 2 diabetic islets
and effect on insulin secretion. Diabetologia. 51:615–622. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mootha VK, Lindgren CM, Eriksson KF, et
al: PGC-1alpha-responsive genes involved in oxidative
phosphorylation are coordinately downregulated in human diabetes.
Nat Genet. 34:267–273. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ling JQ and Hoffman AR: Epigenetics of
long-range chromatin interactions. Pediatr Res. 61:R11–R16. 2007.
View Article : Google Scholar
|
|
30
|
Thompson JD, Higgins DG and Gibson TJ:
CLUSTAL W: improving the sensitivity of progressive multiple
sequence alignment through sequence weighting, position-specific
gap penalties and weight matrix choice. Nucleic Acids Res.
22:4673–4680. 1994. View Article : Google Scholar
|
|
31
|
Gemma C, Sookoian S, Alvarinas J, et al:
Maternal pregestational BMI is associated with methylation of the
PPARGC1A promoter in newborns. Obesity (Silver Spring).
17:1032–1039. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Brons C, Jacobsen S, Nilsson E, et al:
Deoxyribonucleic acid methylation and gene expression of PPARGC1A
in human muscle is influenced by high-fat overfeeding in a
birth-weight-dependent manner. J Clin Endocrinol Metab.
95:3048–3056. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lillycrop KA, Phillips ES, Torrens C,
Hanson MA, Jackson AA and Burdge GC: Feeding pregnant rats a
protein-restricted diet persistently alters the methylation of
specific cytosines in the hepatic PPAR alpha promoter of the
offspring. Br J Nutr. 100:278–282. 2008. View Article : Google Scholar : PubMed/NCBI
|