Introduction
Esophageal cancer occurs frequently, with an
incidence ranked 8th among all tumor types. This form of cancer is
associated with a high degree of malignancy and a poor 5-year
survival rate of <10% (1–2). The
prevalence of esophageal cancer, in particular esophageal squamous
cell carcinoma (ESCC), is extremely high in China, with ~250,000
new cases arise annually, accounting for 50% of all cases worldwide
(3).
However, despite the frequent occurrence and high
mortality of esophageal cancer, its pathogenesis remains unknown.
Esophageal cancer is generally accepted to be a complex polygenic
disease caused by genetic and environmental factors. Studies have
demonstrated that defects in DNA damage repair lead to accumulated
DNA damage, which confers a higher risk of cancer (4). The most important DNA damage repair
mechanism in humans is nucleotide excision repair (NER) (5). NER is performed by a number of
proteins, for deficiencies that cause human diseases, including
Xeroderma pigmentosum and Cockayne syndrome. A key NER protein,
Xeroderma pigmentosum group D (XPD), a subunit of
transcription factor IIH, has ATP-dependent 5′→3′ helicase activity
and is involved in gene transcription with the action of RNA
polymerase II (6,7). Additionally, XPD interacts with p53
to co-regulate cell apoptosis (8).
Thus, XPD is essential to DNA repair capacity and complete
XPD gene deletion results in embryonic death (9).
XPD, also known as excision repair
cross-complementing, maps to a 5.4-kb region of human
chromosome 9. Eight single nucleotide polymorphisms (SNPs) have
been found in the coding region thus far and among these,
variations in codons 312 (G→A) and 751 (A→C) are the most common
(10). Numerous studies have
investigated potential associations between XPD
polymorphisms and various types of cancer, including head and neck
squamous, lung squamous and basal cell carcinomas and breast and
gastric cancers (11–15). However, a limited number of studies
have explored the relationship between XPD polymorphisms and
ESCC and have yielded inconsistent results (16–19).
Therefore, this case-control study assessed whether SNPs in codons
312 and 751 of XPD are associated with ESCC in a Chinese
population.
Materials and methods
Study subjects
The present study was a population-based
case-control study. Between January 2008 and December 2011, 400
patients with ESCC (case group) whose diagnoses were confirmed by
pathological biopsy of tissues removed by endoscopic or surgical
excision were recruited. Healthy individuals (control group) were
confirmed by endoscopic census to be free of esophageal cancer and
precancerous lesions over the same period in the same geographical
area. Information on gender, age, smoking status and alcohol
consumption of case and control group individuals was obtained. The
400 healthy individuals were gender- and age-matched with the case
group. Smoking was defined as ≥1 cigarette/day continuously for
>6 months. Alcohol consumption was defined as at least once/week
continuously for >6 months. The study was approved by the First
People’s Hospital of Yancheng City Ethics Committee (Jiangsu,
China) and all subjects provided written informed consent.
Specimen collection
Peripheral venous blood samples were obtained from
all participants in a fasting state and stored at −20°C for
subsequent use.
Extraction of genomic DNA
Wizard genomic DNA extraction kits (Promega
Corporation, Madison, WI, USA) were used to extract genomic DNA
according to the manufacturer’s instructions.
Concentration and purity of genomic
DNA
ND-1000 UV/VIS spectrophotometer (Nanodrop
Corporation, Wilmington, DE, USA) was used to perform quantitative
determination of the DNA samples. Template DNA concentration was
adjusted to 25–50 ng/μl.
Polymerase chain reaction (PCR)
Genotypes at the 312 and 751 loci of XPD were
determined using PCR-restriction fragment length polymorphism
(RFLP). Primers were synthesized by Sangon Biological Engineering
Technology Co., Ltd. (Shanghai, China). For codon 312, the primer
sequences used were: upstream, 5′-CTGTTGGTGGGTGCCCGTATCTGTT
GGTCT-3′; downstream, 5′-TAATATCGGGGCTCACCCT GCAGCACTTCCT-3′. PCR
mix (20 μl) containing 100 ng DNA template, 2.0 μl 10X PCR buffer
solution (with 15 mM MgCl2), 0.4 μl dNTPs (10 mM), 1.5
μl each primer (150 mM), 1 unit Taq DNA polymerase and 16.5
μl ddH2O. Reaction conditions were as follows: 94°C for
5 min; 35 cycles of 94°C for 45 sec, 69°C for 45 sec and 72°C for
45 sec; and 72°C for 7 min. PCR products were digested using the
StyI restriction enzyme (New England Biolabs, Ipswich, MA,
USA) at 37°C for 16 h. For codon 751, primer sequences used were:
upstream, 5′-GCCCGCTCTGGATTATACG-3′ downstream, 5′-CTATC
ATCTCCTGGCCCCC-3′. PCR mix (20 μl) containing 100 ng DNA template,
2.0 μl 10Χ PCR buffer solution (Mg2+-free), 1.2 μl
Mg2+ solution (25 mM MgCl2), 0.5 μl dNTPs (10
mM), 1.5 μl each primer (150 mM) and 25 units Taq DNA
polymerase. Reaction conditions were as follows: 94°C for 5 min; 35
cycles of 94°C for 45 sec, 63°C for 45 sec and 72°C for 45 sec; and
72°C for 7 min. PCR products were digested using the PstI
restriction enzyme (New England Biolabs) at 37°C for 16 h. Codon
products were visualized on 2% agarose gels using a gel imaging
system. Restriction fragments were sent to Beijing Genomics
Institute (Beijing, China) for direct gene sequencing.
Statistical analysis
SPSS 17.0 statistical software was used for
statistical analysis. The two-sample t-test was used to compare
differences between the two groups and the χ2 test was
used to compare age, gender, smoking status, alcohol consumption
and genotype and allele frequencies between the two groups. The
relationship between gene polymorphism and ESCC was analyzed using
logistic regression to obtain odds ratios (OR) and 95% confidence
intervals (CI). Analyses were performed with two-sided tests.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Population characteristics
Age, gender, smoking status and alcohol consumption
of the 400 ESCC cases and 400 healthy controls included in this
study are presented in Table I.
Controls were age- and gender-matched to cases. Age ranges and
gender distributions of the two groups were not identified as
significantly different, although ESCC occurred twice as often in
males. However, statistically significant increases in cigarette
smoking and alcohol consumption were observed in the ESCC patients
(44.0 and 34.3%, respectively) compared with the controls (33.5 and
26%, respectively; P<0.05).
 | Table IComparison of basic characteristics
between ESCC cases and healthy controls. |
Table I
Comparison of basic characteristics
between ESCC cases and healthy controls.
| Variable | Controls (n=400) | Cases (n=400) |
t/χ2a | P-value |
|---|
| Age, years (mean ±
SD) | 63.2±9.6 | 62.9±9.5 | 0.498 | 0.618 |
| Age, years (%) |
| <60 | 137 (34.3) | 150 (37.5) | 0.918 | 0.338 |
| ≥60 | 263 (65.8) | 250 (62.5) | | |
| Gender (%) |
| Male | 272 (68.0) | 257 (64.3) | 1.256 | 0.262 |
| Female | 128 (32.0) | 143 (35.8) | | |
| Smoking status
(%) |
| Non-smoking | 266 (66.5) | 224 (56.0) | | |
| Smoking | 134 (33.5) | 176 (44.0) | 9.290 | 0.002 |
| Alcohol consumer
(%) |
| No | 296 (74.0) | 263 (65.8) | | |
| Yes | 104 (26.0) | 137 (34.3) | 6.467 | 0.011 |
Comparison of genotype and allele
frequencies for SNPs 312 and 751 between ESCC and healthy
individuals
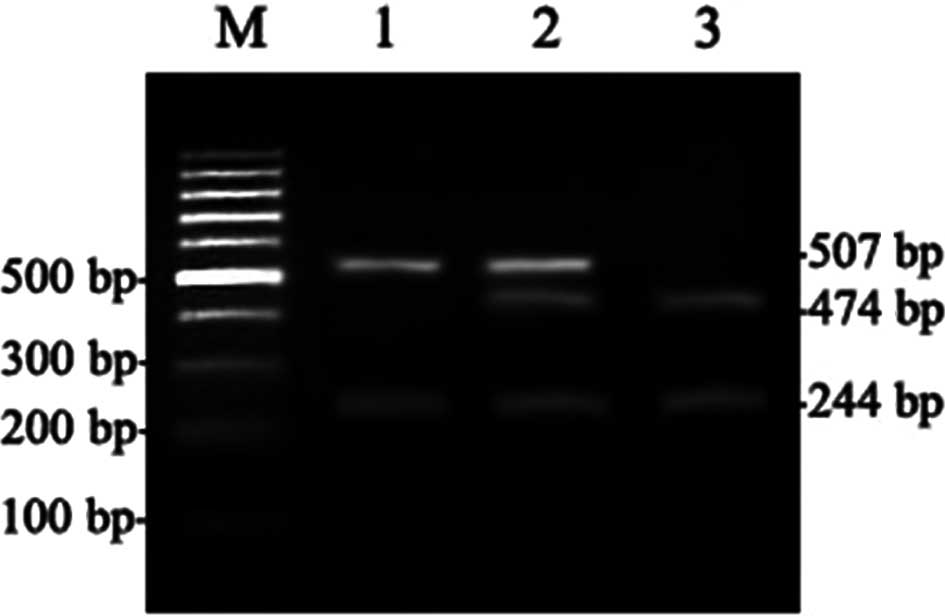
The expected PCR product size for the 312 locus was
751 bp. StyI restriction digestion generated various
products based on the genotype: GG resulted in 2 fragments, 507 and
244 bp; GA resulted in 4 fragments, 507, 474, 244 and 33 bp; and AA
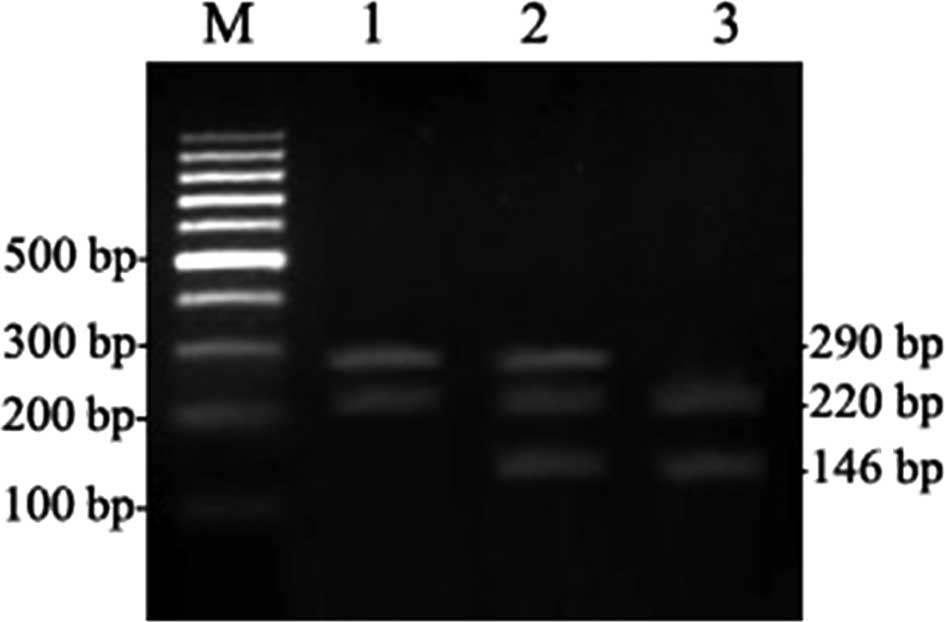
resulted in 3 fragments, 474, 244 and 33 bp (Fig. 1). For the 751 locus, the expected
PCR product size was 436 bp. PstI restriction digestion
generated the following: 2 fragments for the AA genotype, 290 and
146 bp; 4 fragments for AC, 290, 227, 146 and 63 bp; and 3
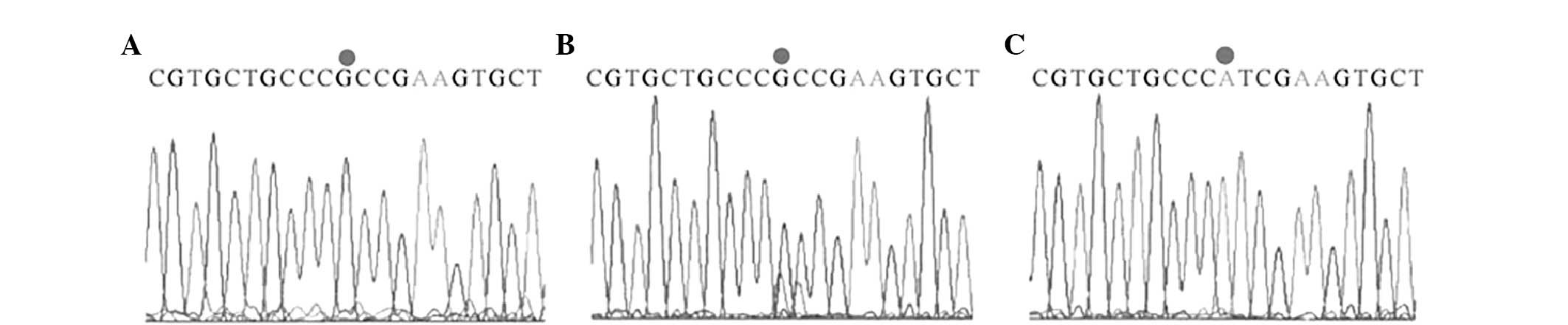
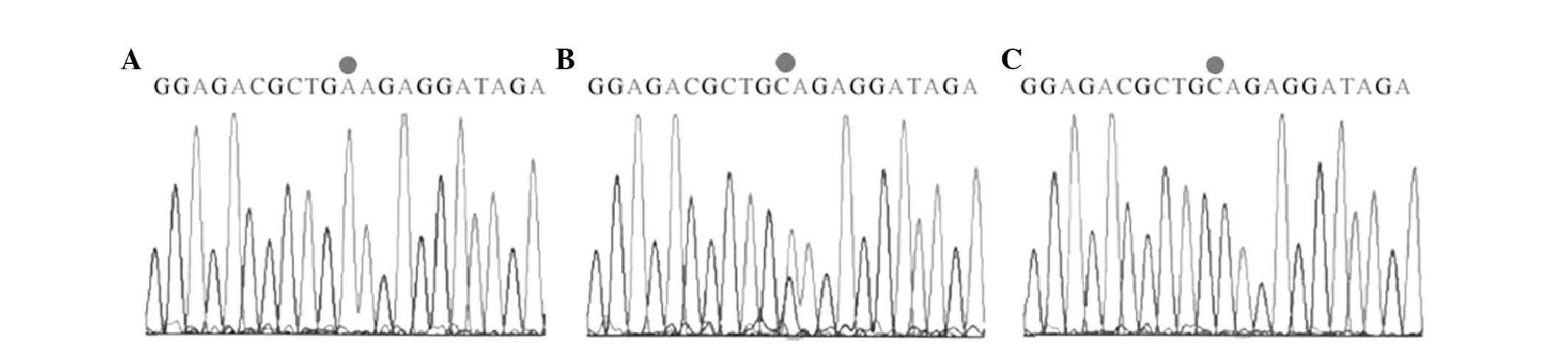
fragments for CC, 227, 146 and 63 bp (Fig. 2). Sequencing of these restriction
products was consistent with the expected sequence (Figs. 3 and 4).
The distributions of genotypes for XPD codons
312 and 751 were consistent with Hardy-Weinberg equilibrium for
cases and controls (P>0.05). No statistically significant
differences in genotype or allele frequencies for codon 312 were
observed between cases and controls (Table II). However, statistically
significant differences in genotype and allele frequencies for
codon 751 were observed between cases and controls (P<0.05;
Table III).
 | Table IIComparison of genotype and allele
frequencies for XPD codon 312 between groups, no. (%). |
Table II
Comparison of genotype and allele
frequencies for XPD codon 312 between groups, no. (%).
| | Genotype | Allele |
|---|
| |
|
|
|---|
| Group | No. | GG | GA | AA | G | A |
|---|
| Controls | 400 | 351 (87.8) | 47 (11.8) | 2 (0.5) | 749 (93.6) | 51 (6.4) |
| Cases | 400 | 342 (85.5) | 56 (14.0) | 2 (0.5) | 740 (92.5) | 60 (7.5) |
| χ2 | | | 0.903 | | 0.784 | 0.784 |
| P-value | | | 0.637 | | 0.376 | 0.376 |
 | Table IIIComparison of genotype and allele
frequencies for XPD codon 751 between groups, no. (%). |
Table III
Comparison of genotype and allele
frequencies for XPD codon 751 between groups, no. (%).
| | Genotype | Allele |
|---|
| |
|
|
|---|
| Group | No. | AA | AC | CC | A | C |
|---|
| Controls | 400 | 321 (80.3) | 73 (18.3) | 6 (1.5) | 715 (89.4) | 85 (10.6) |
| Cases | 400 | 283 (70.8) | 105 (26.3) | 12 (3.0) | 671 (83.9) | 129 (16.1) |
| χ2 | | | 10.144 | | 10.444 | 10.444 |
| P-value | | | 0.006 | | 0.001 | 0.001 |
Relationship between genotype and
ESCC
Results of the logistic regression analysis
indicated that gender was a confounding factor for ESCC (P=0.039),
while age, smoking status and alcohol consumption had no
correlation with the occurrence of ESCC (P>0.05). Excluding the
effect of gender resulted in no correlation between SNP in codon
312 and occurrence of ESCC (P>0.05). However, a correlation was
observed between codon 751 genotype and ESCC occurrence. No
significant increase in ESCC risk was observed for cases with the
AC genotype compared with those with the AA genotype (P=0.137).
ESCC risk in cases with the CC genotype was increased by 1.6-fold
(OR, 1.600; 95% CI; 1.137–2.253; P=0.007; Table IV).
 | Table IVAssociation between XPD
polymorphism and ESCC. |
Table IV
Association between XPD
polymorphism and ESCC.
| Variable | Controls
(n=400) | Cases (n=400) | OR (95% CI) | P-value |
|---|
| Age |
| <60 | 137 (34.3) | 150 (37.5) | Reference | |
| ≥60 | 263 (65.8) | 250 (62.5) | 0.866
(0.644–1.164) | 0.341 |
| Gender |
| Male | 272 (68.0) | 257 (64.3) | Reference | 0.039 |
| Female | 128 (32.0) | 143 (35.8) | 1.391
(1.016–1.905) | |
| Smoking status |
| Non-smoking | 266 (66.5) | 224 (56.0) | Reference | |
| Smoking | 134 (33.5) | 176 (44.0) | 1.507
(0.985–2.305) | 0.059 |
| Alcohol
consumer |
| Never consume
alcohol | 296 (74.0) | 263 (65.8) | Reference | |
| Consume
alcohol | 104 (26.0) | 137 (34.3) | 1.177
(0.748–1.850) | 0.481 |
| Codon |
| 312 GG | 351 (87.8) | 342 (85.5) | Reference | |
| 312 GA | 47 (11.8) | 56 (14.0) | 1.166
(0.762–1.783) | 0.480 |
| 312 AA | 2 (0.5) | 2 (0.5) | 1.013
(0.138–7.441) | 0.990 |
| 751 AA | 321 (80.3) | 283 (70.8) | Reference | |
| 751 AC | 73 (18.3) | 105 (26.3) | 2.149
(0.785–5.884) | 0.137 |
| 751 CC | 6 (1.5) | 12 (3.0) | 1.600
(1.137–2.253) | 0.007 |
Discussion
Previous studies on the association between
XPD polymorphisms and ESCC are controversial. However, the
present study provides additional confirmation of a correlation
between a SNP in XPD, specifically within codon 751 and
ESCC. No significant correlation was observed between the SNP in
codon 312 and occurrence of ESCC, consistent with results of
previous studies on the relationship between polymorphisms of
XPD codon 312 and risk of esophageal cancer. Specifically,
Zhou et al(17) and Wu
et al(19) reported no
correlation between polymorphisms at this locus and occurrence of
ESCC in Chinese populations. Similarly, Tse et al(20) found no such correlation in a
predominantly Caucasian population. By contrast, we report that a
polymorphism in codon 751 of XPD is significantly correlated
with occurrence of ESCC and that individuals with the CC genotype
(vs. AA) have a 1.6-fold higher risk of ESCC. Thus, the CC genotype
confers increased susceptibility to ESCC, consistent with specific
previous studies. In particular, researchers in the Taiwan Province
of China reported that the XPD codon 751 SNP is correlated
with risk of esophageal cancer (21); Ye et al(16) found a similar correlation in a
Swedish population. However, an earlier study in northern China
identified no correlation between XPD gene polymorphisms and
ESCC (19), indicating that
XPD polymorphisms may have different effects on genetic
susceptibility to ESCC in various populations. A previous study
indicated that, compared with the AA genotype, CC individuals have
significantly reduced repair capacity of chromatid-type DNA
aberrations (22). The structure
of codon 751 is highly evolutionarily conserved, indicating a role
in maintaining function of XPD. Therefore, mutation at this locus
may affect the repair ability of XPD, as well as affecting its
transcriptional activity and role in apoptosis, thereby promoting
tumorigenesis.
In short, XPD codon 751 polymorphism is
markedly associated with ESCC susceptibility and the CC genotype
may be a risk factor for ESCC. Therefore, detection of XPD
polymorphism at this locus may aid in prevention and early
diagnosis of ESCC. Corresponding functional studies are required to
reveal molecular mechanisms that promote ESCC in individuals
carrying XPD variants.
References
|
1
|
Lambert R and Hainaut P: The
multidisciplinary management of gastrointestinal cancer.
Epidemiology of oesophagogastric cancer. Best Pract Res Clin
Gastroenterol. 21:921–945. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Holmes RS and Vaughan TL: Epidemiology and
pathogenesis of esophageal cancer. Semin Radiat Oncol. 17:2–9.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lu S: Research progress on etiology of
esophageal cancer. A specially invited set of reports for Annual
Meeting Seminar of the Tenth China Association for Science and
Technology. 129–132. 2008.
|
|
4
|
Matullo G, Palli D, Peluso M, et al:
XRCC1, XRCC3, XPD gene polymorphisms, smoking and 32P-DNA adducts
in a sample of healthy subjects. Carcinogenesis. 22:1437–1445.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ishikawa T, Zhang SS, Qin X, Takahashi Y,
Oda H, Nakatsuru Y and Ide F: DNA repair and cancer: lessons from
mutant mouse modle. Cancer Sci. 95:112–117. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Egly JM: The 14th Datta Lecture. TFIIH:
from transcription to clinic. FEBS Lett. 498:124–128. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lehmann AR: The xeroderma pigmentosum
group D (XPD) gene: one gene, two functions, three diseases. Genes
Dev. 15:15–23. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang XW, Vermeulen W, Coursen JD, et al:
The XPB and XPD DNA helicases are components of the p53-mediated
apoptosis pathway. Genes Dev. 10:1219–1232. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
de Boer J, Donker I, de Wit J, Hoeijmakers
JH and Weeda G: Disruption of the mouse xeroderma pigmentosum
group D DNA repair/basal transcription gene results in
preimplantation lethality. Cancer Res. 58:89–94. 1998.
|
|
10
|
Manuguerra M, Saletta F, Karagas MR,
Berwick M, Veglia F, Vineis P and Matullo G: XRCC3 and XPD/ERCC2
single nucleotide polymorphisms and the risk of cancer: a HuGe
review. Am J Epidemiol. 164:297–302. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sturgis EM, Zheng R, Li L, Castillo EJ,
Eicher SA, Chen M, Strom SS, Spitz MR and Wei Q: XPD/ERCC2
polymorphism and risk of head and neck cancer: a case-control
analysis. Carcinogenesis. 21:2219–2223. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Butkiewicz D, Rusin M, Enewold L, Shields
PG, Chorazy M and Harris CC: Genetic polymorphisms in DNA repairs
genes and risk of lung cancer. Carcinogenesis. 22:593–597. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dybdahl M, Vogel U, Frentz G, Wallin H and
Nexø BA: Polymorphisms in the DNA repair gene XPD: correlations
with risk and age at onset of basal cell carcinoma. Cancer
Epidemiol Biomarkers Prev. 8:77–81. 1999.PubMed/NCBI
|
|
14
|
Tang D, Cho S, Rundle A, Chen S, Phillips
D, Zhou J, Hsu Y, Schnabel F, Estabrook A and Perera FP:
Polymorphisms in the DNA repair enzyme XPD are associated with
increased levels of PAH-DNA adducts in a case-control study of
breast cancer. Breast Cancer Res Treat. 75:159–166. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ruzzo A, Canestrari E, Maltese P, et al:
Polymorphisms in genes involved in DNA repair and metabolism of
xenobiotics in individual susceptibility to sporadic diffuse
gastric cancer. Clin Chem Lab Med. 45:822–828. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ye W, Kumar R, Bacova G, Lagergren J,
Hemminki K and Nyrén O: The XPD 751Gln allele is associated with an
increased risk for esophageal adenocarcinoma: a population-based
case-control study in Sweden. Carcinogenesis. 27:1835–1841. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhou R, Li Y, Wang N, et al: Association
study on mononucleotide polymorphism of XPD gene and risk of
esophageal squamous cell carcinoma, gastric cardiac adenocarcinoma.
Tumor. 27:118–123. 2007.
|
|
18
|
Chen M, Wang J and Guo G: The correlation
of repair gene XPD Lys751Gln for DNA damage, XRCC1 Arg399Gln
polymorphism mononucleotide with genetic susceptibility of
esophageal cancer. Fudan University (Med Sci). 35:273–278.
2008.
|
|
19
|
Wu X, Wang P, Yun Y, et al: The
relationship between esophageal squamous cell carcinoma in Henan
Han Chinese and XPD gene polymorphism. Public Health of China.
28:446–449. 2012.(In Chinese).
|
|
20
|
Tse D, Zhai R, Zhou W, Heist RS, Asomaning
K, Su L, Lynch TJ, Wain JC, Christiani DC and Liu G: Polymorphisms
of the NER pathway genes, ERCCI and XPD are associated with
esophageal adenocarcinoma risk. Cancer Causes Control.
19:1077–1083. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lee JM, Lee YC, Yang SY, Yang PW, Luh SP,
Lee CJ, Chen CJ and Wu MT: Genetic polymorphisms of XRCC1 and risk
of the esophageal cancer. Int J Cancer. 95:240–246. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lunn RM, Helzlsouer KJ, Parshad R, Umbach
DM, Harris EL, Sanford KK and Bell DA: XPD polymorphisms: effects
on DNA repair proficiency. Carcinogenesis. 21:551–555. 2000.
View Article : Google Scholar : PubMed/NCBI
|