Introduction
Schistosoma japonicum (S. japonicum)
is an extremely harmful pathogen, which infects humans and causes
severe public health problems. To date, effective therapeutic drugs
for this pathogen are lacking. Scientists are diligently searching
for anti-infectious vaccines against S. japonicum; however,
all of the vaccine candidates reported are not effective enough to
protect individuals from the infection of S. japonicum.
Therefore, effective methods for curing S. japonicum-related
diseases are needed.
S. japonicum causes disease mainly through
its egg granuloma in hosts, which is followed by host hepatic and
intestinal fibrosis (1).
Therefore, a suitable strategy may be to target the maturity of
eggs and reproduction of S. japonicum. The eggshell protein
gene (SjESG) of S. japonicum encodes the major
component of its egg yolk (2). It
is known that this gene is required for the maturity and egg
deposition of female S. japonicum (3). Sequence analyses show that over 85%
of the SjESG sequences are conserved among members in the
family (4).
Ribozymes, a class of RNA molecules with
endonuclease activities, were found to be able to bind to their
targeting mRNAs and digest them specifically (5). Ribozymes bind to target RNAs
containing the NUH sequence (N stands for any base; H stands for C,
U or A) via specific cleavage sites downstream of the NUH triplet.
The ribozyme-mediated mRNA cleavage results in inhibition or
blockage of expression of the target genes (6,7).
Hammerhead ribozymes have been found to control many
types of diseases (8,9). In this study, we designed and
synthesized 3 ribozymes targeting the SjESG gene and
evaluated their cleavage abilities in vitro and in
vivo. We found that the in vitro cleavage abilities of
two of these 3 designed ribozymes were correlated with their
abilities to inhibit the maturity of eggs and reproduction of S.
japonicum in mice.
Materials and methods
DNA preparation
Snails infected with S. japonicum cercarie
were purchased from the Hunan Institute of Schistosomiasis
Prevention and Treatment. Genomic DNA was isolated from the
collected worms (S. japonicum) according to the methods
described previously (10). The
designed ribozyme DNA oligonucleotides were synthesized by
Invitrogen and dissolved in TE buffer (pH 8.0). The
oligonucleotides (A chain and B chain) (Table I) for each ribozyme were mixed
equivalently and annealed by denaturation for 5 min at 95°C,
followed by slow cooling to room temperature.
 | Table IOligonucleotides annealed for making
Rz1, Rz2 and Rz3. |
Table I
Oligonucleotides annealed for making
Rz1, Rz2 and Rz3.
| Rzl | A chain:
5′AGCTTCACACCCTGATGAGTCCGTGAGGACGAAACCTCCG3′
B chain: 5′GATCCGGAGGTTTCGTCCTCACGGACTCATCAGGGTGTGA3′ |
| Rz2 | A chain:
5′AGCTTCACCATCTGATGAGTCCGTGAGGACGAAACCGCG3′
B chain: 5′GATCCGCGGTTTCGTCCTCACGGACTCATCAGATGGTCA3′ |
| Rz3 | A chain:
5′AGCTTTCACCATCTGATGAGTCCGTGAGGACGAAACCACCG3′
B chain: 5′GATCCAGTGGTTTCGTCCTCACGGACTCATCAGATGGTGAA3′ |
Plasmid constructs
The SjESG gene fragment was amplified from
the genomic DNA of S. japonicum by PCR, using primers
(5′-CCAAGCTTATGTACCCACCATCATCC-3′ and
5′-CGGGATCCTCAATAATAGGAGGGTGCA-3′). For constructing the plasmid
pcDNA3.1(+)/SjESG, the SjESG PCR products were cloned
into the HindIII and BamHI sites of the vector
pcDNA3.1(+), which were purchased from Promega. The annealed
ribozyme dsDNAs (Rz1, Rz2, Rz3) were cloned into the sites of
HindIII and BamHI of pcDNA3.1(+), resulting in
plasmids pcDNA3.1(+)/Rz1, pcDNA3.1(+)/Rz2 and pcDNA3.1(+)/Rz3. The
ligation products were transformed into E. coli JM109, and
positive colonies were selected.
In vitro transcription
The plasmids pcDNA3.1(+)/SjESG,
pcDNA3.1(+)/Rz1, pcDNA3.1(+)/Rz2 and pcDNA3.1(+)/Rz3 were
linearized with BamHI and purified with a gel purification
kit (catalogue no. DV805A; Takara Co.). The in vitro
transcription was performed by using a transcription kit (catalogue
no. L1170; Promega), according to the manufacturer’s instructions.
Briefly, DNA constructs were linearized and used as templates in a
20-μl in vitro transcription reaction containing 4 μl of 5X
buffer, 2 μl of DTT, 1 μl of RNase inhibitor, with or without 2 μl
of digoxin labeling mix, 1 μl of the DNA template, 1 μl of T7 RNA
polymerase, and H2O. The reactions were performed at
37°C for 2 h, followed by addition of stop buffer. The RNAs labeled
with or without digoxin were purified and used for ribozyme
cleavage experiments.
RNA blotting
RNAs were separated on formaldehyde denaturation
agarose gels, transferred to a nylon membrane by a transblotting
system, and then detected by a DIG nucleic acid detection kit
(catalogue no. 11175025910; Roche).
RNA cleavage by ribozymes
The 3 ribozymes (Rz1, Rz2 and Rz3) were respectively
mixed with the substrate (SjESG mRNA, labeled with a digoxin
marker) in a ratio of 1:1 in a 10-μl reaction system, containing 50
mM of Tris-HCl (pH 7.5), 20 mM of MgCl2, 20 mM of NaCl
and 1 μl of RNasin (11). The
mixture was incubated at 95°C for 1 min, rapidly transferred to
ice, incubated at 37°C for 2 h, and followed by addition of the
stop buffer (formamide 960 mM, EDTA 20 mM). The cleaved products
were detected using RNA blotting with a digoxin marker. The bands
were scanned and analyzed with the software AlphaImager 2200
(Beckman). Cleavage efficiency was calculated with the following
formula: CE = [P/(S + P)] × 100%; S, substrate; P, digested
product; CE, cleavage efficiency.
Animal experiments
Forty age-matched (4–6 weeks of age) BALB/c female
mice were infected with S. japonicum cercarie via the vena
caudalis. The infected mice were randomly divided into 5 groups and
injected i.v. with PBS, vector, or the ribozyme constructs (Rz1,
Rz2 or Rz3) as previously described (12). The constructs were diluted to 0.25
μg/μl with PBS to construct a plasmid DNA solution for the mouse
treatments. Each mouse was injected with 200 μl of plasmid DNA
solution containing DNA 50 μg or PBS at the schedule of 14, 21 and
28 days post-infection with S. japonicum.
Measurement of IFN-γ and IL-4 levels in
the serum of the treated mice
Serum samples were obtained from the mice by cutting
the vena caudalis prior to treatment, 2 days after the first
treatment and 2 weeks after the third treatment, respectively.
IFN-γ and IL-4 levels in the serum were measured by ELISA and
analyzed using SPSS software as described (13).
Measurement of quantities of worms and
eggs in the treated mice
The mice were sacrificed by extracting the eyeballs
45 days after the third treatment of ribozymes. Adult worms were
collected by flushing the portal of vein. Livers were extracted and
incubated in 5% of KOH solution for 20 h at 37°C, and then the
worms and eggs in these tissues were counted under a
microscope.
Results
Design of ribozymes targeting the SjESG
mRNA
Computer software (14) was used to analyze the
computer-predicted secondary structure of SjESG mRNA. Six
potential hammerhead ribozyme sites were found in the mRNA
(Table II). After further
analyses, the 76th, 283rd and 160th sites were chosen for ribozyme
cleavage, since they have a lower ΔEr than the sequences on other
sites (Table II). In addition,
the sequences on these 3 sites (76th, 283rd and 160th) and the
sequences around them were conserved among all members of the
SjESG gene family (GenBank nos. M32280, M32281, M59318,
DQ225185 and AB017096).
 | Table IIParameters of the hammerhead
ribozymes. |
Table II
Parameters of the hammerhead
ribozymes.
| Rz | 5′ΔE
(kcal/mol) | 3′ΔE
(kcal/mol) | ΔEs (kcal/mol) | ΔEr (kcal/mol) |
|---|
| Rzl (76th
site) | −11.9 | −10.7 | 1.2 | 4.2 |
| Rz2 (283rd
site) | −10.4 | −9.5 | 2.7 | 2.6 |
| Rz3 (160th
site) | −11.8 | −11.8 | 0.0 | 1.6 |
| Rz4 (35th
site) | −11.8 | −11.6 | 0.0 | 5.5 |
| Rz5 (67th
site) | −11.9 | −11.0 | 1.2 | 4.2 |
| Rz6 (109th
site) | −11.9 | −12.4 | 0.0 | 4.2 |
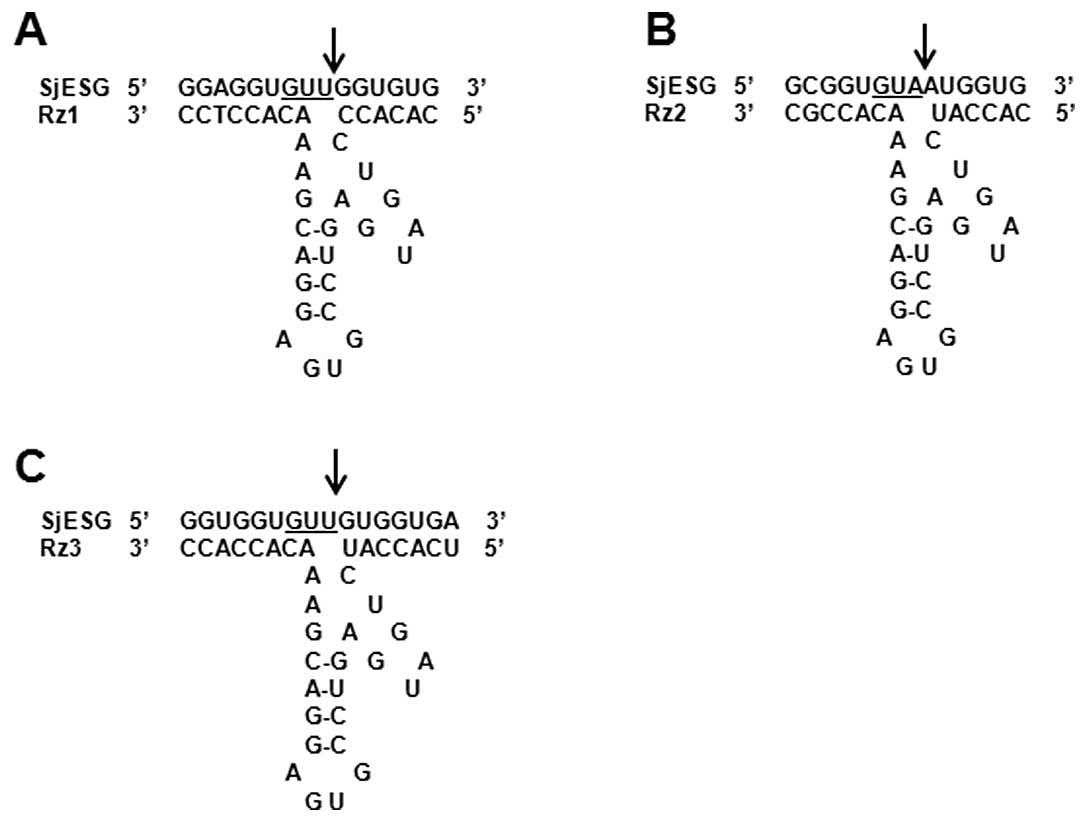
The ribozymes targeting the 76, 283 and 160 sites
were named Rz1, Rz2 and Rz3, respectively. The sequence of the
catalytic center was designed according to the hammerhead structure
model introduced by Symons et al (15), and the sequences on both sides of
the ribozyme were complementary to corresponding substrate
(16). Restricted enzyme sequences
were added to the 5′ side and 3′ side for easy cloning. The
sequences of Rz1, Rz2 and Rz3 targeting the 76, 283 and 160-bp
sites, respectively, are shown in Fig.
1.
Confirmation of the plasmid
construction
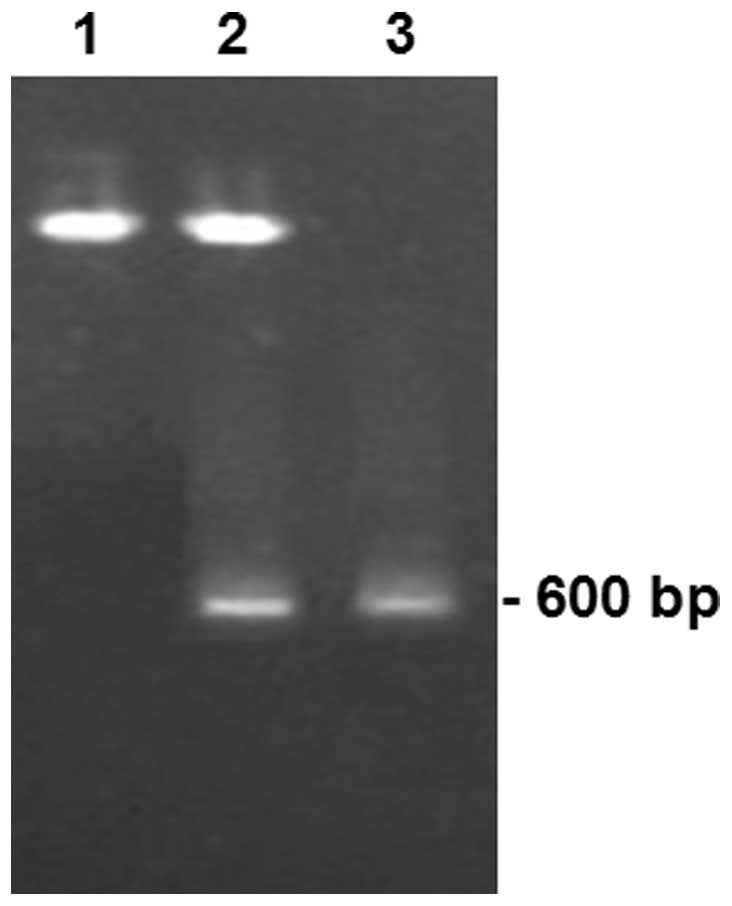
To confirm whether the plasmid constructs were
cloned correctly, the pcDNA3.1(+)/SjESG plasmid was digested
with HindIII and BamHI. The digested products were
separated on agarose gels along with the PCR product of the
SjESG gene. As shown in Fig.
2, a DNA fragment ~600 bp was dropped off from the
pcDNA3.1(+)/SjESG, but not from the vector plasmid. The
dropped fragments had a similar size with the PCR product amplified
from the SjESG gene. In addition, the 3 ribozyme constructs
[pcDNA3.1(+)/Rz1, pcDNA3.1(+)/Rz2 and pcDNA3.1(+)/Rz3] were
confirmed by DNA sequencing. Therefore, these results suggest that
the constructs were constructed correctly.
In vitro transcription of SjESG mRNA and
ribozyme RNAs
To obtain the SjESG mRNAs and the 3 ribozyme
RNAs, in vitro transcription was performed using the
enzyme-linearized pcDNA3.1(+)/SjESG, pcDNA3.1(+)/Rz1,
pcDNA3.1(+)/Rz2, or pcDNA3.1(+)/Rz3, respectively, as templates.
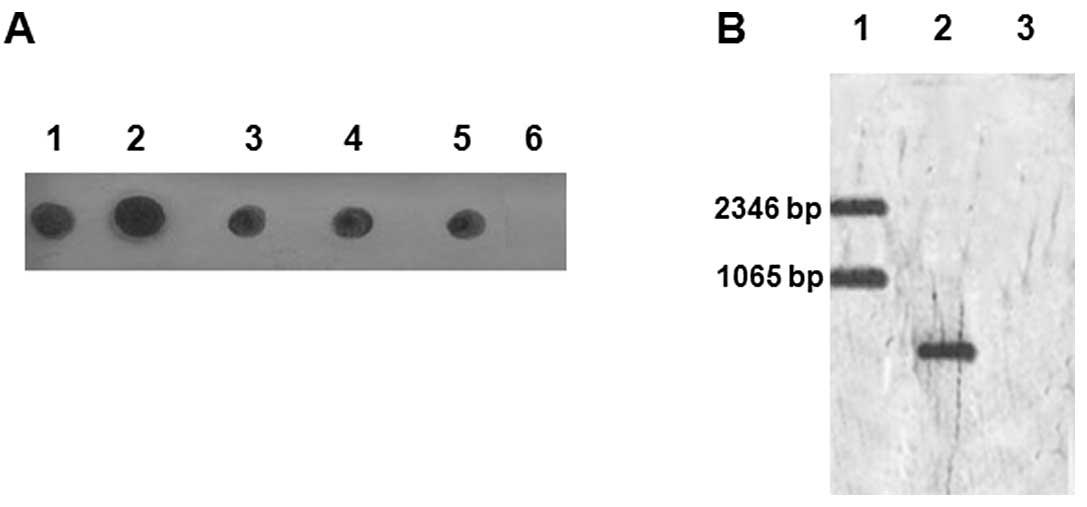
After in vitro transcription reactions were completed, the
DNA templates in the reaction system were digested using DNase, and
the synthesized SjESG mRNA and the ribozyme RNA were gel
purified. The digoxin-labeled SjESG mRNA and ribozyme RNAs
were confirmed using RNA blotting (Fig. 3A). The synthesized SjESG
mRNA products, which were labeled with a digoxin marker, were also
confirmed by the RNA blotting as shown in Fig. 3B. The SjESG mRNA and
ribozyme RNAs labeled with or without digoxin were prepared and
used for the following in vitro cleavage experiments.
The ribozymes efficiently cleave SjESG
mRNA
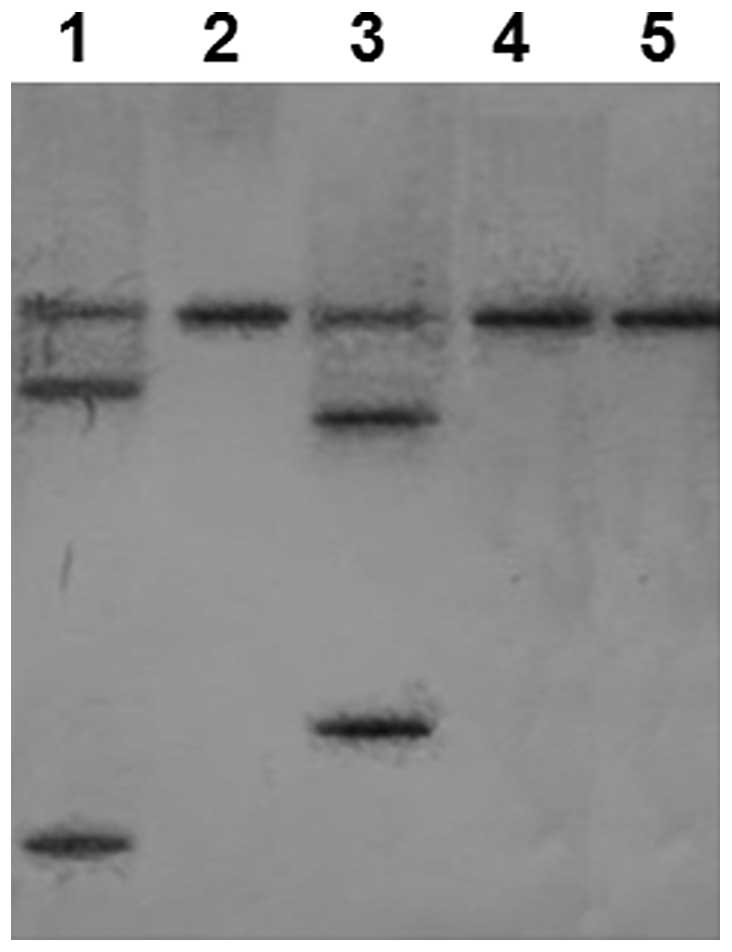
To determine whether the 3 ribozymes (Rz1, Rz2 and
Rz3) were able to cleave the SjESG mRNA, in vitro
cleavage experiments using digoxin-labeled SjESG mRNA and
unlabeled ribozyme RNAs were performed. The cleaved,
digoxin-labeled SjESG mRNA products were detected using RNA
blotting (the unlabeled ribozymes were undetectable on the blots).
As shown in Fig. 4, Rz1 cleaved
the SjESG mRNA into two smaller fragments: 561 and 76 bp.
Rz3 cleaved the SjESG mRNA into 477- and 160-bp fragments.
However, no Rz2-mediated cleavage products were detectable.
Densitometry analyses indicated that Rz1 and Rz3 cleaved the
SjESG mRNAs with an efficiency of 68.9 and 69.6%,
respectively.
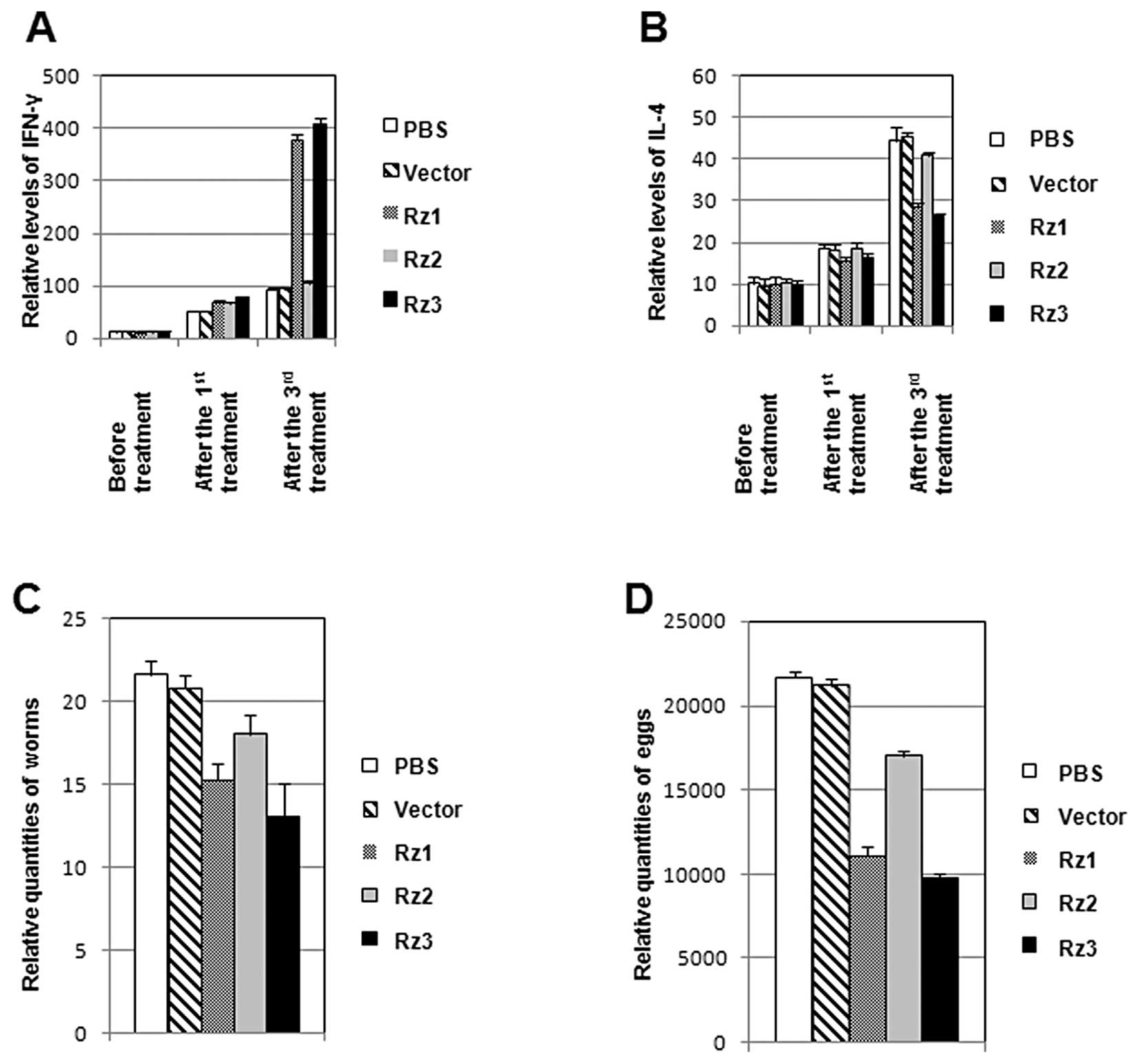
IFN-γ and IL-4 levels in the serum of
mice
Since the in vitro ribozyme cleavage results
found that two of the 3 designed ribozymes cleaved SjESG
mRNA efficiently, these ribozymes were investigated in vivo.
Mice infected with S. japonicum received injections of Rz1,
Rz2 or Rz3 in PBS at several time points. The serum samples were
collected from mice to measure the levels of two important
cytokines, IFN-γ and IL4, using ELISA. As shown in Fig. 5A, after the 1st treatment, the
levels of IFN-γ in all 5 groups (treated with PBS, vector, Rz1,
Rz2, or Rz3) increased very slightly. After the 3rd treatment,
IFN-γ levels in the 3 groups (treated with PBS, vector, and Rz2)
still increased slightly, while IFN-γ levels in the groups treated
with Rz1 and Rz3 increased by up to 4-fold, when compared to the
levels in the groups treated with PBS and vector. Notably, when
compared with the highly up-regulated IL-4 levels (Fig. 5B) in the groups treated with PBS,
vector, and Rz2, the IL-4 levels in the groups treated with Rz1 and
Rz3 increased much less. These results suggest that Rz1 and Rz3
treatments induce similar effects on IFN-γ and IL-4 levels in mice,
which is different from the groups treated with PBS, vector, or
Rz2.
Anti-reproduction contribution of the
ribozymes
In order to investigate whether the ribozyme
treatments affect the amount of worms and eggs, the infected mice
in the animal experiments (Fig. 5A and
B) were sacrificed 45 days after the 3rd treatment of
ribozymes. Adult worms were collected by flushing portal of vein
and the amounts of the worm eggs in the mouse livers were counted
under a microscope. As shown in Fig.
5C, Rz1 and Rz3 caused more marked decreases in the amounts of
adult worms when compared with the groups treated with PBS, vector,
or Rz2. Similarly, Rz1 and Rz3 also decreased the amounts of eggs
more effectively than Rz2 and the vector controls (Fig. 5D). Rz3 was more effective than Rz1,
resulting in a 39.82% reduction in amounts of worms and a 54.95%
decrease in the egg reduction rate in the liver. Altogether, these
results suggest the in vitro SjESG RNA cleavage
mediated by Rz1 and Rz3 may be related to the regulation of the
levels of the cytokines, IFN-γ and IL-4, consequently decreasing
the amounts of adult worms and eggs.
Discussion
S. japonicum infects humans, livestock and
snails, resulting in severe diseases. No effective therapeutic
strategies are available to date. In this study, we designed 3
hammerhead ribozymes targeting the SjESG gene of S.
japonicum. We studied their cleavage activity in vitro
and their roles in inhibiting the reproduction of S.
japonicum. Our results suggest that there is a correlation
between the in vitro cleavage abilities of Rz1 and Rz3 and
their roles in reproduction inhibition of S. japonicum. To
our knowledge, this is the first preliminary study of specific
hammerhead ribozymes targeting SjESG as a possible method to
treat S. japonicum-related diseases.
After in vitro mRNA transcription of
SjESG and ribozymes from the constructs, we conducted in
vitro cleavage experiments. The results showed that Rz1 and Rz3
cleaved their targeted mRNAs at specific sites, but Rz2 did not.
These results indicated that the interaction between ribozyme and
it’s substrate mRNA as predicted by computer analyses may not be
real. In addition, the cleavage efficiency between the different
ribozymes was varied.
Since RNA is degraded easily in vivo, in the
animal experiments we used the corresponding expression vectors to
express RNA rather than the synthesized ribozyme RNAs. Secondly, we
injected ribozymes frequently through the vena caudalis to maintain
the concentration of ribozymes in the mice. In addition, the
secondary structure of the ribozymes and the substrates had
features to avoid degradation.
Notably, the IFN-γ level in the serum of the Rz
groups was higher than that of the control groups, while the IL-4
level in the serum of the Rz groups was lower. One reasonable
explanation may be that the Th1 cytokines of IFN-γ are correlated
with S. japonicum granuloma formation and vigor, and the Th2
cytokines of IL-4 play an important role in down-regulating egg
granuloma reaction at chronic schistosomiasis by inhibiting the Th1
cytokines.
The results in this study demonstrated that the
hammerhead ribozymes for SjESG play a significant role in
the inhibition of the reproduction of S. japonicum by up to
39.82% in worm reduction in mice and 54.95% in egg reduction in the
mouse liver. Further modification of hammerhead ribozymes for
SjESG and the screening of more ribozyme candidates for
other genes of S. japonicum appears to be a promising novel
therapeutic strategy in this field.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (no. 30972576), the Scientific Research
Fund of Hunan Provincial Scientific and Technological Department
(no. 2010SK3038), and supported by Hunan Provincial Innovation
Foundation for Postgraduate (no. CX2009B190).
References
|
1
|
D SymmersPathogenesis of liver cirrhosis
in schistosomiasisJ Am Med
Assoc147304305195110.1001/jama.1951.0367021001600514873476
|
|
2
|
Q ZengJH XiaoZG WanAcquiring and analysis
of EST and new genes of Schistosoma japonicumZhongguo Ji
Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi2386892005(In
Chinese).
|
|
3
|
SP WangMB ZhouXF ZengH LiXY YiY
ChenSchistosoma japonicum antifemale fecundity and
anti-embryonation immunityChin J Schistosom Control107121998
|
|
4
|
KJ HenkleGA CookLA FosterDM EngmanLA
BobekGD CainJE DonelsonThe gene family encoding eggshell proteins
of Schistosoma japonicumMol Biochem
Parasitol426982199010.1016/0166-6851(90)90114-22172818
|
|
5
|
Q VicensMA AllenSD GilbertB ReznikAR
GoodingRT BateyThe cech symposium: a celebration of 25 years of
ribozymes, 10 years of TERT, and 60 years of
TomRNA14397403200818203922
|
|
6
|
M Asif-UllahM LevesqueG
RobichaudDevelopment of ribozyme-based gene-inactivations; the
example of the hepatitis delta virus ribozymeCurr Gene
Ther7205216200710.2174/15665230778085900817584038
|
|
7
|
HS ZaherPJ UnrauSelection of an improved
RNA polymerase ribozyme with superior extension and
fidelityRNA1310171026200710.1261/rna.54880717586759
|
|
8
|
XM FengLJ CaoFY WangXC ZhangXQ
LuInhibition of pyruvate kinase mRNA expression in Giardia
lamblia by specific hammerhead ribozymeZhongguo Ji Sheng Chong
Xue Yu Ji Sheng Chong Bing Za Zhi282572602010(In Chinese).
|
|
9
|
B KumarM KhannaP KumarV SoodR VyasAC
BanerjeaNucleic acid-mediated cleavage of M1 gene of influenza a
virus is significantly augmented by antisense molecules targeted to
hybridize close to the cleavage siteMol
Biotechnol9110201121744034
|
|
10
|
J SambrookWR DavidPT HuangJX WangHC
ZhuMolecular Cloning: A Laboratory Manual3th editionScience
PressBeijing2008
|
|
11
|
S YangJH XiaoYK ZhangQ ZengQL YangCA LiuKG
WangK YinR TangConstruction and identification of adult worm cDNA
library of Japanese Schistosoma japonicumPract Prev
Med95775792002
|
|
12
|
R KumarV DammaiPK YadavaS KleinauGene
targeting by ribozyme against TNF-a mRNA inhibits autoimmune
arthritisGene
Therapy1214861493200510.1038/sj.gt.330258316034454
|
|
13
|
H XuZ ZhangC XiaoDynamics of liver tissue
immune pathology and IFN-γ/IL-4 cytokine levels in mice infected
with Schistosoma japonicum in different stagesJ Trop Dis
Parasitol87274201019756744
|
|
14
|
CD LuAN ChenGR QiComputer analyses of the
in vitro cleavage of the target RNA by ribozymeBiochem Biophy
J282792851996
|
|
15
|
RH SymonsCJ HutchinsAC ForsterPD RathjenP
KeeseJE VisvaderSelf-cleavage of RNA in the replication of viroids
and virusoidsJ Cell SciSuppl
7S303318198710.1242/jcs.1987.Supplement_7.212460475
|
|
16
|
AC ForsterRH SymonsSelf-cleavage of
virusoid RNA is performed by the proposed 55-nucleotide active
siteCell50916198310.1016/0092-8674(87)90657-X3594567
|