Introduction
Nasopharyngeal carcinoma (NPC) is a tumor arising
from the epithelial cells that cover the surface and line the
nasopharynx (1). NPC differs
significantly from other cancers of the head and neck in its
occurrence, causes, clinical behavior and treatment. It is vastly
more common in certain regions of East Asia and Africa than
elsewhere, with viral, dietary and genetic factors implicated in
its causation (2,3). Although rare, NPC accounts for one
third of childhood nasopharyngeal neoplasms in the USA (4).
Chemotherapy with anti-neoplastic (or cytotoxic)
drugs is one of the most extensively adopted practices for managing
cancers (5). Apart from radiation
therapy and surgical resection, chemotherapy should be considered
for patients with metastatic disease or local recurrence that is no
longer amenable to surgery or radiation therapy (6–8). For
NPC therapy, it is most common to use a combination of two or more
chemotherapeutic drugs, known as combination chemotherapy. Using
two or more drugs together is often more effective than using one
drug.
Two of the main drugs used in the treatment of NPC
are cisplatin and 5-fluorouracil (5-FU) (9,10).
Cisplatin is a DNA-binding agent that is widely used to treat
different types of cancer. At the centre of this drug is a platinum
metal atom. The drug forms DNA crosslinks via the platinum which
damages the cancer cells. 5-FU is one of the most commonly used
drugs to treat cancer. It is part of a group of chemotherapeutic
drugs known as the anti-metabolites. In cancer cells, 5-FU is
converted intracellularly into three cytotoxic metabolites:
fluorodeoxyuridine monophosphate (FdUMP), fluorodeoxyuridine
triphosphate (FdUTP) and fluorouridine triphosphate (FUTP). These
metabolites inhibit the formation and repair of DNA in cancer cells
(11,12).
Despite the widespread clinical use of cisplatin and
5-FU for over 40 years and the aberrant expression of
protein-coding genes observed after chemotherapy (13–15),
the regulatory mechanisms invovled remain poorly understood. We
hypothesized that microRNAs (miRNAs), a class of
post-transcriptional gene regulators, may play an important role in
the cisplatin-and 5-FU-induced alteration of gene expression in
cancer cells. miRNAs are a group of small (20–22 nt) endogenous
non-protein-coding RNA molecules that negatively regulate gene
expression (16,17). miRNAs are involved in multiple
biological processes and metabolic regulation, including cell
proliferation, differentiation and programmed cell death. As the
dysregulation of these processes is a hallmark of cancer, miRNAs
are viewed as major contributors to the pathogenesis of cancer,
including the initiation and progression of cancer (18). A growing body of evidence has
suggested the importance of miRNAs in managing the efficiency of
chemotherapy in several human cancers (19,20).
Although a number of studies have shown that
chemotherapy drugs alter miRNA expression in many cancer cells,
there is no report on the effect of cisplatin or 5-FU on miRNAs in
human NPC (21,22). In this study, we used a label-free
high-throughput miRNA array technology, the stacking-hybridized
universal tag (SHUT) assay (23),
to investigate the effect of cisplatin and/or 5-FU exposure on the
global expression profile of miRNAs in the CNE human NPC cell
line.
Materials and methods
Cell line and cell culture
All the cell culture reagents were purchased from
Invitrogen, Inc. (Carlsbad, CA, USA). The CNE human NPC cell line
was obtained from the Cell Bank of the Chinese Academy of Sciences
(CAS). Cells were cultured in Dulbecco’s modified Eagle’s medium
(DMEM) supplemented with 10% fetal bovine serum (FBS). They were
maintained at 37°C in a humidified incubator with 5%
CO2.
Drug treatment and
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay
5-FU and cisplatin were purchased from Sigma-Aldrich
(St. Louis, MO, USA) and stored at 4°C, protected from lighting and
moisture. For the cell treatments, 5-FU and cisplatin were freshly
prepared in DMEM complete medium in 5-fold series dilution
concentrations, with final starting concentrations of 48.05 and
1.04 mM. The cells were seeded 10,000 cells/well (100 μl) in a
96-well plate (Corning Costar, #3599) and treated with the freshly
prepared 5-FU or cisplatin in duplicate for 48 h. The half maximal
inhibitory concentration (IC50) (concentration of the
two drugs to produce half maximal cell inhibition) values were
determined by the MTT method and their 10% inhibitory concentration
(IC10) values were calculated. Briefly, after 48 h of
drug incubation, 90 μl fresh medium and 10 μl MTT-Dulbecco’s
phosphate-buffered saline (DPBS) solution were added to each well,
and the plate was incubated in the dark for 4 h at 37°C. The medium
was removed by inverting and tapping the plate, and 100 μl/well of
dimethyl sulfoxide (DMSO) were added and shaken for 5 min to
dissolve the formazan crystals. The absorbance at 570 nm was read
on a microplate spectrophotometer (BioTek) and the results were
expressed as a percentage of the control.
RNA isolation
Cells were seeded at 300,000 cells/well in a
flat-bottom 6-well plate in 2 ml of complete medium. After 18 h,
the medium was replaced with fresh medium. The cells were dived
into four groups: control group (CNE-normal), IC10 of
5-FU (CNE-5-FU), IC10 of cisplatin (CNE-Cis), or both
IC10 values of 5-FU and cisplatin (CNE-Cis + FU) in
complete medium, respectively. After 48 h, the cells were collected
and total RNA was prepared using TRIzol (Invitrogen, Inc.)
following the manufacturer’s instructions, except that the
precipitation was allowed to proceed for 12 h at −20°C, which
efficiently recovers all RNA species, including miRNAs. RNA
concentration was determined using a nanodrop spectrophotometer.
RNA integrity was determined by TAE agarose gel electrophoresis and
the relation of the 28S, 18S and 5S ribosomal RNA (rRNA) bands.
Microarray miRNA profiling
The SHUT assay, recently presented by Duan et
al (23), is an efficient
technique for high-throughput miRNA profiling using microarrays.
Being a label-free approach, it eliminates the cost and potential
biases of the fluorescent-labeling of miRNAs, while offering good
specificity and sensitivity. Following the strategies presented by
the authors, we designed a full-spectrum microarray targeting of
each of the 1,733 mature human miRNAs listed in miRBase release 17.
This customized microarray design was ordered from Agilent
Technologies Inc. (Santa Clara, CA, USA) via its eArray online
service. The final product is an in situ synthesized
high-definition DNA microarray of the format 8×15 k (arrays per
slide × probes per array; Agilent G2509F). For each array, 5.0 μg
total RNA extracted from each cell line and 200 nM (final
concentration) Cy3-labeled universal tag DNA oligonucleotides (UT,
of the sequence AGGTCGCA; synthesized by Integrated DNA
Technologies, Inc., Coralville, IA, USA) were dissolved in
hybridization buffer (5X SSC with 0.2% SDS in nuclease-free water)
to make a 40-μl sample solution. In a typical experiment, the four
cell samples along with a blank buffer were applied respectively to
one of the eight arrays on the same microarray slide, which was
part of the SureHyb hybridization assembly (with G2534-60014 and
G2534A; Agilent Technologies, Inc.). The assembly was placed in a
hybridization oven (Agilent 2545A) at 42°C for 22 h with a constant
rotation speed of 20 rpm. After hybridization, the slides were
washed in 5X SSC and 0.1% SDS at 30°C for 6 min, and then washed
for 3 min twice at room temperature in 0.2X SSC. The slides were
immediately dried and scanned.
Image scanning and data analysis
Following hybridization and washing, the slides were
scanned using a GenePix 4100A Microarray Scanner (Molecular
Devices, Inc., USA) at constant power and photomultiplier (PMT)
gain settings through a single-color channel (532 nm wavelength).
The raw pixel intensities were extracted using the GenePix Pro 7.0
software (Molecular Devices, Inc.). Data processing (including
filtration, background-correction, transformation and
normalization) and subsequent statistical analysis were performed
using the methods provided by Duan et al (23). Finally, the expression levels of
all mature miRNAs were plotted on a heatmap.
Results
Effects of 5-FU and cisplatin on the CNE
human NPC cell line
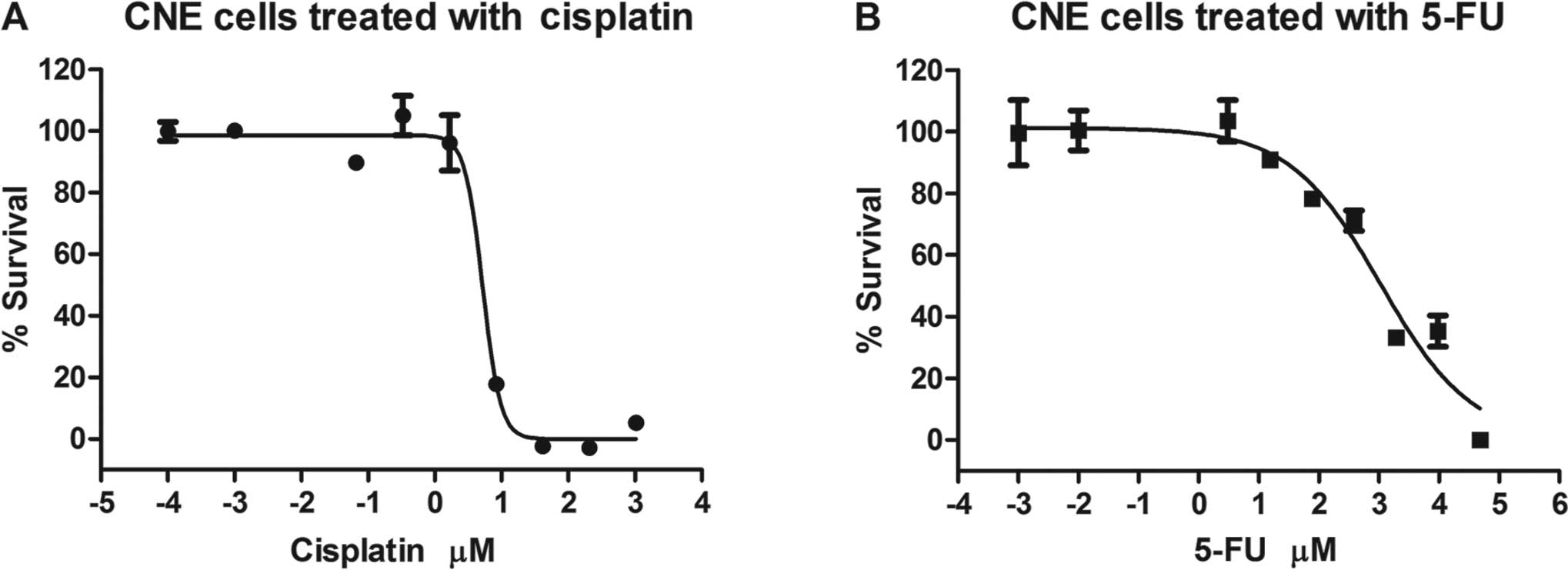
To determine the sensitivity of CNE cells to 5-FU or
cisplatin, MTT assay was performed and dose-response curves were
generated to determine the IC50 values. As shown in
Fig. 1, the IC50 of
5-FU and cisplatin was approximately 1,058 and 5.237 μM,
respectively. Using a formula from GraphPad {ECF =
IC50 × [F/(100-F)]1/H}, the IC10
values of these two drugs were calculated to be approximately 22.91
and 2.67 μM, respectively. To avoid genetic changes as much as
possible, which are more sensitive as compared to cellular changes,
low concentrations (IC10) of these two drugs were
selected to determine their effects on the global expression levels
of miRNAs in the CNE cells. Furthermore, as the combination of
these two drugs is used as a popular first-line therapy (22.91 μM
of 5-FU and 2.67 μM of cisplatin), we also investigated their
combined effect on the global miRNA expression levels.
Alteration of miRNA expression profiles
in CNE cells after drug treatment
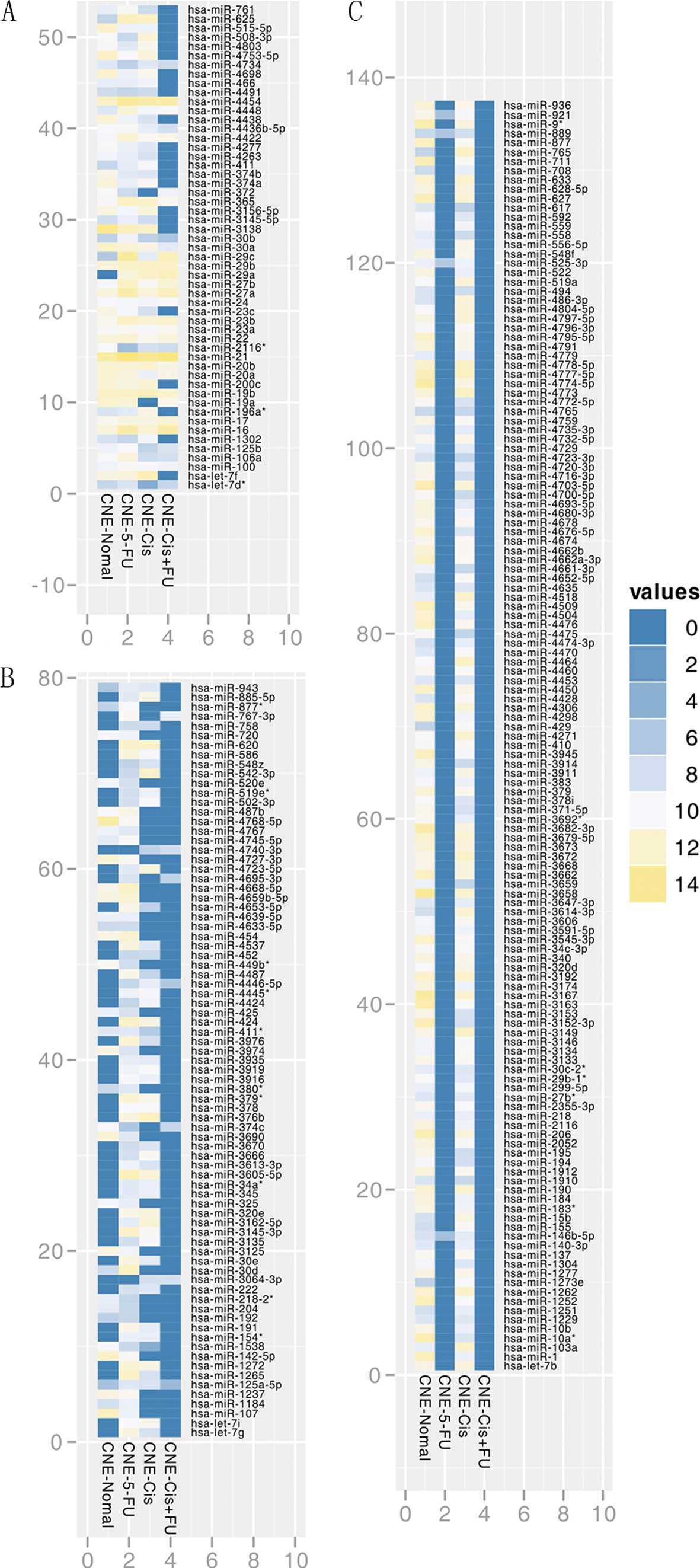
Based on the SHUT assay, the miRNA expression
profiles in CNE cells with different treatments were obtained and
the heatmap of part of the results is shown in Fig. 2. Of the 1,733 human miRNAs
analyzed, a total of 701 were detected to be expressed
significantly with or without low-dose drug treatment, among which
23 miRNAs were detected in all the four groups (Table I and Fig. 2). Apart from the 23 miRNAs commonly
expressed, 431 miRNAs were detected to be significantly expressed
in the control cells, while 184 miRNAs were detected in the
5-FU-treated cells, 336 miRNAs in the cisplatin-treated cells and
13 miRNAs in the cells treated with the combination of both drugs.
The above results showed that all the treated cells expressed a
lower number of miRNAs as compared to the control cells.
 | Table IComparison of miRNA expression profile
between the four groups. |
Table I
Comparison of miRNA expression profile
between the four groups.
| miRNA category | No. | miRNAs |
|---|
| Expressed
significantly only in the CNE-normal, CNE-Cis and CNE-5-FU
group | 24 | let-7f, miR-1302,
miR-200c, miR-23c, miR-30b, miR-3145-5p, miR-3156-5p, miR-374a,
miR-374b, miR-411, miR-4263, miR-4277, miR-4438, miR-4491, miR-466,
miR-4698, miR-4753-5p, miR-4803, miR-508-3p, miR-515-5p, miR-625,
miR-196a*, miR-3138, miR-761 |
| Expressed
significantly only in the CNE-normal, CNE-Cis and CNE-Cis + FU
group | 3 | let-7d*,
miR-19a, miR-372 |
| Expressed
significantly only in the CNE-normal, CNE-5-FU and CNE-Cis + FU
group | 1 |
miR-2116* |
| Expressed
significantly only in the CNE-Cis, CNE-5-FU and CNE-Cis + FU
group | 2 | miR-29c, miR-29a |
| Expressed in all the
four groups | 23 | miR-100, miR-106a,
miR-125b, miR-16, miR-17, miR-19b, miR-20a, miR-20b, miR-21,
miR-22, miR-23a, miR-23b, miR-27a, miR-27b, miR-29b, miR-30a,
miR-365, miR-4422, miR-4448, miR-24, miR-4436b-5p, miR-4454,
miR-4734 |
When compared by group, the control cells
collectively expressed 185, 80 and 28 miRNAs significantly with the
cisplatin-, the 5-FU- and the combination-treated cells,
respectively (Table II and
Fig. 2). Furthermore, only 137
miRNAs were found in the control and the cisplatin-treated cells,
30 miRNAs in the control and the 5-FU-treated cells and only one
miRNA, hsa-miR-374c, was found in the control and the
combination-treated cells. The cisplatin-treated and 5-FU-treated
cells exclusively expressed 44 miRNAs. As indicated in Table II and Fig. 2, there were only two miRNAs,
hsa-miR-29c and hsa-miR-29a, expressed in the drug-treated groups,
but not in the control. Three miRNAs, hsa-let-7d*,
hsa-miR-19a and hsa-miR-372, were found to be expressed in all
groups, except the cisplatin-treated group. One miRNA,
hsa-miR-2116*, was expressed in all the groups, except
for the 5-FU-treated group. Notably, a total of 235 miRNAs were
exclusively expressed in the control cells, while 116 miRNAs were
expressed in the cisplatin-treated cells, 109 miRNAs in the
5-FU-treated group and only two miRNAs (hsa-miR-195* and
hsa-miR-3714) in the combination-treated group. Therefore, it can
be concluded that 5-FU, cisplatin and their combination treatment
significantly altered the miRNA expression profile in the CNE human
NPC cell line.
 | Table IIComparison of miRNAs expressed
significantly in only two of the four groups. |
Table II
Comparison of miRNAs expressed
significantly in only two of the four groups.
| miRNA category | No. | miRNAs |
|---|
| Expressed
significantly only in the CNE-normal and CNE-Cis group | 30 | miR-142-5p, miR-192,
miR-204, miR-30d, miR-3125, miR-325, miR-380*, miR-3974,
miR-411*, miR-4633-5p, miR-4639-5p, miR-4659b-5p,
miR-4668-5p, miR-4727-3p, miR-4768-5p, miR-487b, miR-520e, miR-720,
miR-107, miR-1184, miR-1237, miR-1538, miR-218-2*,
miR-3690, miR-425, miR-449b*, miR-4745-5p, miR-4767,
miR-877* |
| Expressed
significantly only in the CNE-normal and CNE-5-FU group | 137 | let-7b, miR-1,
miR-10a*, miR-10b, miR-1252, miR-1262, miR-1277,
miR-1304, miR-137, miR-146b-5p, miR-155, miR-15b,
miR-183*, miR-184, miR-190, miR-194, miR-195, miR-2052,
miR-206, miR-2116, miR-218, miR-2355-3p, miR-27b*,
miR-299-5p, miR-29b-1*, miR-30c-2*, miR-3133,
miR-3134, miR-3146, miR-3149, miR-3152-3p, miR-3163, miR-3167,
miR-3174, miR-320d, miR-340, miR-3545-3p, miR-3591-5p, miR-3606,
miR-3614-3p, miR-3647-3p, miR-3658, miR-3659, miR-3662, miR-3668,
miR-3672, miR-3673, miR-3682-3p, miR-371-5p, miR-378i, miR-379,
miR-383, miR-3914, miR-3945, miR-410, miR-4271, miR-429, miR-4306,
miR-4450, miR-4453, miR-4460, miR-4464, miR-4475, miR-4476,
miR-4504, miR-4509, miR-4635, miR-4652-5p, miR-4661-3p,
miR-4662a-3p, miR-4662b, miR-4678, miR-4680-3p, miR-4693-5p,
miR-4703-5p, miR-4720-3p, miR-4729, miR-4735-3p, miR-4759,
miR-4765, miR-4772-5p, miR-4773, miR-4774-5p, miR-4777-5p,
miR-4778-5p, miR-4779, miR-4791, miR-4795-5p, miR-4796-3p,
miR-4797-5p, miR-4804-5p, miR-494, miR-519a, miR-522, miR-548f,
miR-556-5p, miR-558, miR-559, miR-592, miR-617, miR-627,
miR-628-5p, miR-633, miR-708, miR-765, miR-877, miR-889,
miR-9*, miR-936, miR-103a, miR-1229, miR-1251,
miR-1273e, miR-140-3p, miR-1910, miR-1912, miR-3153, miR-3192,
miR-34c-3p, miR-3679-5p, miR-3692*, miR-3911, miR-4298,
miR-4428, miR-4470, miR-4474-3p, miR-4518, miR-4674, miR-4676-5p,
miR-4700-5p, miR-4716-3p, miR-4723-3p, miR-4732-5p, miR-486-3p,
miR-525-3p, miR-711, miR-921 |
| Expressed
significantly only in the CNE-normal and CNE-Cis + FU group | 1 | miR-374c |
| Expressed
significantly only in the CNE-Cis and CNE-5-FU group | 44 | let-7g, let-7i,
miR-1265, miR-154*, miR-222, miR-30e, miR-3145-3p,
miR-320e, miR-34a*, miR-3613-3p, miR-3666, miR-376b,
miR-378, miR-379*, miR-3919, miR-3976, miR-424,
miR-4424, miR-4445*, miR-4487, miR-452, miR-4653-5p,
miR-519e*, miR-542-3p, miR-548z, miR-586, miR-620,
miR-758, miR-885-5p, miR-125a-5p, miR-1272, miR-191, miR-3135,
miR-3162-5p, miR-345, miR-3605-5p, miR-3670, miR-3916, miR-3935,
miR-4446-5p, miR-4537, miR-4723-5p, miR-502-3p, miR-943 |
| Expressed
significantly only in the CNE-Cis and CNE-Cis + FU group | 2 | miR-4695-3p,
miR-767-3p |
| Expressed
significantly only in the CNE-5-FU and CNE-Cis + FU group | 2 | miR-3064-3p,
miR-4740-3p |
Discussion
miRNAs are an important class of gene regulators for
a variety of human cancers. Each miRNA has the ability to regulate
the expression of hundreds of target genes, including oncogenes and
tumor suppressor genes (16,17).
Extensive research is currently being focused on identifying miRNAs
that may play an important role in cancer therapy. Thus, a study of
the possible effects of chemotherapeutic drugs on the expression
profile of miRNAs is of prime importance for cancer therapy and
resistance.
NPC is a special type of squamous cell carcinoma of
the head and neck. Clinically, poorly differentiated squamous cell
carcinoma accounts for 98% of NPC cases in Southern China, where
this disease is particularly prevalent (3). For NPC therapy, cisplatin and 5-FU
are two of the main drugs that are widely used (9,10).
The present study focused on the alteration of miRNA expression in
the CNE cell line, with or without cisplatin and/or 5-FU treatment.
We applied a label-free high-throughput microRNA array technology,
the SHUT assay, to study the miRNA profiling between untreated and
treated CNE cells.
The results from our study revealed that all the
treated cells expressed a lower number of miRNAs as compared to the
control cells. In total, 454 miRNAs were detected to be expressed
in the control cells, over 200 in the 5-FU- or cisplatin-treated
cells, while only 36 miRNAs were detected in the 5-FU and cisplatin
combination-treated cells. Such a sharp decrease indicated that
most of the miRNAs related to the regulation of cell growth and
proliferation were suppressed by the combination treatment, which
may be the main reason for the improved therapeutic effectiveness
of the combination treatment. Furthermore, many of the miRNAs that
were significantly detected only in the cisplatin-treated cells
(109 miRNAs) or 5-FU-treated cells (116 miRNAs), as shown by our
microarray analysis, have been shown to have tumor suppressive and
anti-migration/anti-metastatic roles in cancer. Two miRNAs,
hsa-miR-29c and hsa-miR-29a, were detected to be signigicantly
expressed in the drug-treated groups, but not in the control group,
and have been shown to induce apoptosis by targeting p85α and
CDC42, both of which negatively regulate p53 (24). Hsa-let-7d*, hsa-miR-19a
and hsa-miR-372 were found to be significantly expressed in all the
groups, except for the cisplatin-treated group.
Hsa-miR-2116* was significantly expressed in all the
groups, except for the 5-FU-treated group. The regulation of
hsa-let-7d* and hsa-miR-19a has been reported to be
related to inflammation (25,26).
Hsa-miR-372 has been reported to take part in tumorigenesis
(27). However, the function of
hsa-miR-2116* has not yet been reported. Also, several
miRNAs with unknown function were detected significantly in some of
the four groups. Hsa-miR-4695-3p and hsa-miR-767-3p were
significantly expressed in both the cisplatin- and the
combination-treated cells. Hsa-miR-3064-3p and hsa-miR-4740-3p were
significantly expressed in both the 5-FU- and the
combination-treated cells. Hsa-miR-195* and hsa-miR-3714
were significantly detected only in the combination-treated cells.
The function of these miRNAs was not fully elucidated and we
believe that these miRNAs may also play a primary role in
cancer.
In conclusion, in the present study, we performed a
comparative miRNA expression profile analysis of the two drugs,
cisplatin and 5-FU, and their combination, across the CNE cell line
panel. A number of miRNAs were uncovered, which may form a basis
for the rational clinical use of these drugs as anticancer agents
and may correlate drug response with genomic characteristics. Our
data provide insights into the cytotoxic mechanisms of anticancer
drugs, as well as new clues for the treatment of NPC.
Acknowledgements
The authors would like to thank Kexiao Zheng and
Jiong Li from the Suzhou Institute of Nano-Tech and Nano-Bionics,
Chinese Academy of Sciences, for providing the miRNA microarray
assay support.
References
|
1
|
Brennan B: Nasopharyngeal carcinoma.
Orphanet J Rare Dis. 1:232006.
|
|
2
|
Chang ET and Adami HO: The enigmatic
epidemiology of nasopharyngeal carcinoma. Cancer Epidemiol
Biomarkers Prev. 15:1765–1777. 2006.
|
|
3
|
Fang W, Li X, Jiang Q, et al:
Transcriptional patterns, biomarkers and pathways characterizing
nasopharyngeal carcinoma of Southern China. J Transl Med.
6:322008.
|
|
4
|
Young JL Jr and Miller RW: Incidence of
malignant tumors in U.S. Children. J Pediatr. 86:254–258. 1975.
|
|
5
|
Joensuu H: Systemic chemotherapy for
cancer: from weapon to treatment. Lancet Oncol. 9:3042008.
|
|
6
|
Al-Sarraf M: Head and neck cancer:
chemotherapy concepts. Semin Oncol. 15:70–85. 1988.
|
|
7
|
Jacobs C, Lyman G, Velez-García E, Sridhar
KS, Knight W, Hochster H, Goodnough LT, Mortimer JE, Einhorn LH, et
al: A phase III randomized study comparing cisplatin and
fluorouracil as single agents and in combination for advanced
squamous cell carcinoma of the head and neck. J Clin Oncol.
10:257–263. 1992.
|
|
8
|
Foo KF, Tan EH, Leong SS, Wee JT, Tan T,
Fong KW, Koh L, Tai BC, Lian LG and Machin D: Gemcitabine in
metastatic nasopharyngeal carcinoma of the undifferentiated type.
Ann Oncol. 13:150–156. 2002.
|
|
9
|
Rosenberg B, Vancamp L, Trosko JE and
Mansour VH: Platinum compounds: a new class of potent antitumour
agents. Nature. 222:385–386. 1969.
|
|
10
|
Heidelberger C, Chaudhuri NK, Danneberg P,
et al: Fluorinated pyrimidines, a new class of tumour-inhibitory
compounds. Nature. 179:663–666. 1957.
|
|
11
|
Santi DV, McHenry CS and Sommer H:
Mechanism of interaction of thymidylate synthetase with
5-fluorodeoxyuridylate. Biochemistry. 13:471–481. 1974.
|
|
12
|
Sommer H and Santi DV: Purification and
amino acid analysis of an active site peptide from thymidylate
synthetase containing covalently bound 5-fluoro-20-deoxyuridylate
and methylenetetrahydrofolate. Biochem Biophys Res Commun.
57:689–695. 1974.
|
|
13
|
Maxwell PJ, Longley DB, Latif T, et al:
Identification of 5-fluorouracil-inducible target genes using cDNA
microarray profiling. Cancer Res. 63:4602–4606. 2003.
|
|
14
|
Mauritz R, van Groeningen CJ, Smid K, et
al: Thymidylate synthase and dihydropyrimidine dehydrogenase mRNA
expression after administration of 5-fluorouracil to patients with
colorectal cancer. Int J Cancer. 120:2609–2612. 2007.
|
|
15
|
Shah MY, Pan X, Fix LN, Farwell MA and
Zhang B: 5-Fluorouracil drug alters the microRNA expression
profiles in MCF-7 breast cancer cells. J Cell Physiol.
226:1868–1878. 2011.
|
|
16
|
Zhang BH, Pan XP, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007.
|
|
17
|
Zhang BH, Stellwag EJ and Pan XP:
Large-scale genome analysis reveals unique features of microRNAs.
Gene. 443:100–109. 2009.
|
|
18
|
Koturbash I, Zemp FJ, Pogribny I and
Kovalchuk O: Small molecules with big effects: the role of the
microRNAome in cancer and Carcinogenesis. Mutat Res. 722:94–105.
2011.
|
|
19
|
Meng F, Henson R, Lang M, et al:
Involvement of human micro-RNA in growth and response to
chemotherapy in human cholangiocarcinoma cell lines.
Gastroenterology. 130:2113–2129. 2006.
|
|
20
|
Xia L, Zhang D, Du R, et al: miR-15b and
miR-16 modulate multidrug resistance by targeting BCL2 in human
gastric cancer cells. Int J Cancer. 123:372–379. 2008.
|
|
21
|
Kovalchuk O, Filkowski J, Meservy J, et
al: Involvement of microRNA-451 in resistance of the MCF-7 breast
cancer cells to chemotherapeutic drug doxorubicin. Mol Cancer Ther.
7:2152–2159. 2008.
|
|
22
|
Rossi L, Bonmassar E and Faraoni I:
Modification of miR gene expression pattern in human colon cancer
cells following exposure to 5-fluorouracil in vitro. Pharmacol Res.
56:248–253. 2007.
|
|
23
|
Duan D, Zheng KX, Shen Y, et al:
Label-free high-throughput microRNA expression profiling from total
RNA. Nucleic Acids Res. 39:e1542011.
|
|
24
|
Park SY, Lee JH, Ha M, et al: miR-29
miRNAs activate p53 by targeting p85 alpha and CDC42. Nat Struct
Mol Biol. 16:23–29. 2009.
|
|
25
|
Reina Ortiz M, Schreiber F, Benitez S, et
al: Effects of chronic ascariasis and trichuriasis on cytokine
production and gene expression in human blood: a cross-sectional
study. PLoS Neql Trop Dis. 5:e11572011.
|
|
26
|
Xie YF, Shu R, Jiang SY, et al: Comparison
of microRNA profiles of human periodontal diseased and healthy
gingival tissues. Int J Oral Sci. 3:125–134. 2011.
|
|
27
|
Tian RQ, Wang XH, Hou LJ, et al:
MicroRNA-372 is down-regulated and targets cyclin-dependent kinase
2 (CDK2) and cyclin A1 in human cervical cancer, which may
contribute to tumorigenesis. J Biol Chem. 286:25556–25563.
2011.
|