Introduction
Enterococci have been recognized as an important
cause of nosocomial infection in the past few decades (1). Clinical infections caused by
Enterococcus species include urinary tract infections,
bacteremia, bacterial endocarditis and diverticulitis (2). Moreover, vancomycin-resistant
isolates of Enterococcus faecium (E. faecium) and
Enterococcus faecalis (E. faecalis) have been
described (3). Three genetic
mechanisms of enterococcal resistance against glycopeptides have
been described (4). Overall,
vancomycin resistance in enterococci has been associated with more
frequent episodes of recurrent bacteremia, persistent isolation of
enterococci from primary infection sites, increased frequency of
endovascular infection, and increased mortality (5).
Due to the rapid increase in the incidence of
vancomycin-resistant enterococci (VRE) infections, particularly of
those acquired in hospitals, the Centers for Disease Control and
Prevention published recommendations for preventing the spread of
vancomycin resistance in 1995 (6).
These recommendations included a policy for deciding the time of
removal of patients from isolation; that is, VRE-negative results
on at least 3 consecutive occasions (≥1 week apart) in all cultures
from multiple body sites. This recommendation has generally been
accepted, but the rationale of this policy seems to stem from an
empirical basis.
The conventional phenotypic method has been the gold
standard for VRE surveillance. However, a previous report suggested
that the molecular identification of VRE is more sensitive and
accurate than the conventional culture method (7). Thus, the molecular method can be used
for a limited population, such as VRE-colonized patients (8).
In Korea, VRE infections have recently been
increasing, with a clinical isolation rate of up to 25% (9). Most of the cases of VRE infections
are caused by E. faecium (10,11).
A previous study on E. faecium isolates from 10 teaching
hospitals showed that all isolated VRE harboured the vanA
gene (12).
The aim of the present study was to determine the
clinical usefulness of real-time PCR for the detection of the
vanA gene over the conventional culture method in patients
confirmed as being VRE-colonized, and to re-evaluate the optimal
number of negative tests before deciding a patient’s removal from
isolation.
Materials and methods
Evaluation stage
We compared the results of real-time PCR for the
detection of the vanA gene, for which 4 preparations (direct
specimen, isolate cultured in blood agar plate for 24 h, isolate
cultured in broth for 24 h and isolate cultured in blood agar plate
for 24 h, as well as in broth for the previous 24 h) of identical
clinical specimens were used, with the results of the conventional
culture method. A total of 100 consecutive routine clinical
specimens were collected for 180 days at the Seoul National
University Bundang Hospital. All specimens were simultaneously
confirmed as VRE-positive or vancomycin-susceptible enterococci
(VSE)-positive through the conventional method. Screening and
identification of isolates were performed by plating in brain heart
infusion agar with 6 μg vancomycin and via MicroScan WalkAway
(Siemens Healthcare Diagnostics, West Sacramento, CA, USA),
respectively. Additionally, antimicrobial susceptibility test
profiles and minimum inhibitory concentraion (MIC) levels were
determined by the disc diffusion method and E-test (bioMérieux,
Marcy l’Etoile, France). Specimens used in the present study were
extracted from the gastrointestinal tract (56 isolates),
genitourinary tract (16 isolates), respiratory tract (8 isolates)
and fluid (4 isolates); 16 isolates were from other sources.
Application stage
Between May 2008 and December 2009, a total of 1,115
clinical consecutive specimens from 82 previously established
VRE-infected or VRE-colonized patients at the Seoul National
University Bundang Hospital were evaluated for VRE surveillance.
Specimens were obtained from 21 blood (8 patients), 16 fluid (5
patients), 636 gastrointestinal tract (79 patients), 320
genitourinary tract (48 patients), 98 other (16 patients) and 24
respiratory tract (7 patients) samples. All specimens were
incubated in broth for 24 h.
For both the evaluation and the application, nucleic
acid extractions were performed on each pre-treated specimen. In
the direct specimen culture, a few colonies grown on blood agar
plate were transferred to phosphate-buffered saline (PBS) via a
cotton swab. The amount of 1 ml of mixture or broth was centrifuged
at 15,000 rpm for 10 min. The supernatant was discarded and the
pellet was washed with 1 ml PBS through centrifugation. After
mixing the pellet with 20 μl of Chelex solution, the tube was
heated at 100°C for 20 min and cooled subsequently. After
centrifugation at 15,000 rpm for 3 min, 100 μl of the supernatant
was obtained for real-time PCR.
Real-time PCR for the detection of the vanA
gene was performed with 3 μl of template DNA, 0.6 μl of each primer
(vanA 3F, 5′-CTG TGA GGT CGG TTG TGC G-3′; vanA 3R,
5′-TTT GGT CCA CCT CGC CA-3′), 2.4 μl of MgCl2, and 2 μl
of LightCycler FastStart DNA Master HybProbe (vanA 3P,
FAM-5′-CAA CTA ACG CGG CAC TGT TTC CCA AT-3′-TAMRA; Roche
Diagnostics GmbH, Germany). The final volume was adjusted to 20 μl
with distilled water. PCR was performed under the following
conditions: 95°C for 3 min; 40 cycles of 95°C for 10 sec, 58°C for
20 sec and 72°C for 20 sec; followed by a final extension at 40°C
for 30 sec. As an internal control, real-time PCR for 16S ribosomal
RNA (rRNA) was performed under the same conditions.
Results
Evaluation stage
Between March 2007 and September 2007, a total of
100 clinical isolates of E. faecium were obtained
consecutively from 19 patients at the Seoul National University
Bundang Hospital. All 100 isolates were identified as E.
faecium. A total of 52 of these isolates were resistant and 48
were susceptible to vancomycin. Clinical specimens with VRE
included fluid (n=4), gastrointestinal tract (n=30), genitourinary
tract (n=9), respiratory tract (n=6) and others (n=3).
The results of real-time PCR for the detection of
vanA in 52 isolates, for which 4 preparation methods were
used, are shown in Table I. For
isolates prepared via incubation in broth for 24 h, the PCR results
were concordant with those of the conventional method. The results
of real-time PCR for the detection of vanA in isolates
prepared by the other 3 preparation methods showed relatively low
sensitivity.
 | Table IResults of real-time PCR for the
detection of vanA in 4 preparations of specimens from
VRE-infected or VRE-colonized patients. |
Table I
Results of real-time PCR for the
detection of vanA in 4 preparations of specimens from
VRE-infected or VRE-colonized patients.
| Method |
|---|
|
|
|---|
| Culture | Real-time PCR |
|---|
|
|
|
|---|
| Specimen type | Broth and BAP | Direct | BAP | Broth | Broth and BAP |
|---|
| Othera | 3 | 2 | 1 | 3 | 3 |
| Fluid | 4 | 4 | 3 | 4 | 4 |
| Gastrointestinal | 31 | 16 | 18 | 31 | 26 |
| Genitourinary | 9 | 4 | 5 | 9 | 9 |
| Respiratory | 5 | 4 | 4 | 5 | 5 |
| Total | 52 | 30 | 31 | 52 | 47 |
The results of real-time PCR for the detection of
vanA in isolates prepared by 24-h incubation in broth were
compared with the results of the conventional culture method. The
results obtained from the total isolates and from those from the
most dominant isolation site are summarized in Table II. Both specimen groups had 100%
sensitivity. On the other hand, the specificity was 71.4% in the
total number of specimens and 50% in the gastrointestinal
specimens.
 | Table IIComparison of real-time PCR for the
detection of vanA and conventional culture for total and
gastrointestinal specimens. |
Table II
Comparison of real-time PCR for the
detection of vanA and conventional culture for total and
gastrointestinal specimens.
| Total no. of
specimens | Gastrointestinal
specimens |
|---|
|
|
|
|---|
| real-time PCR for
vanA | VRE detected by
conventional culture n (%) | No VRE detected by
conventional culture n (%) | VRE detected by
conventional culture n (%) | No VRE detected by
conventional culture n (%) |
|---|
| Positive | 52 (52.0) | 20 (20.0) | 30 (53.6) | 13 (23.2) |
| Negative | 0 | 28 (28.0) | 0 | 13 (23.2) |
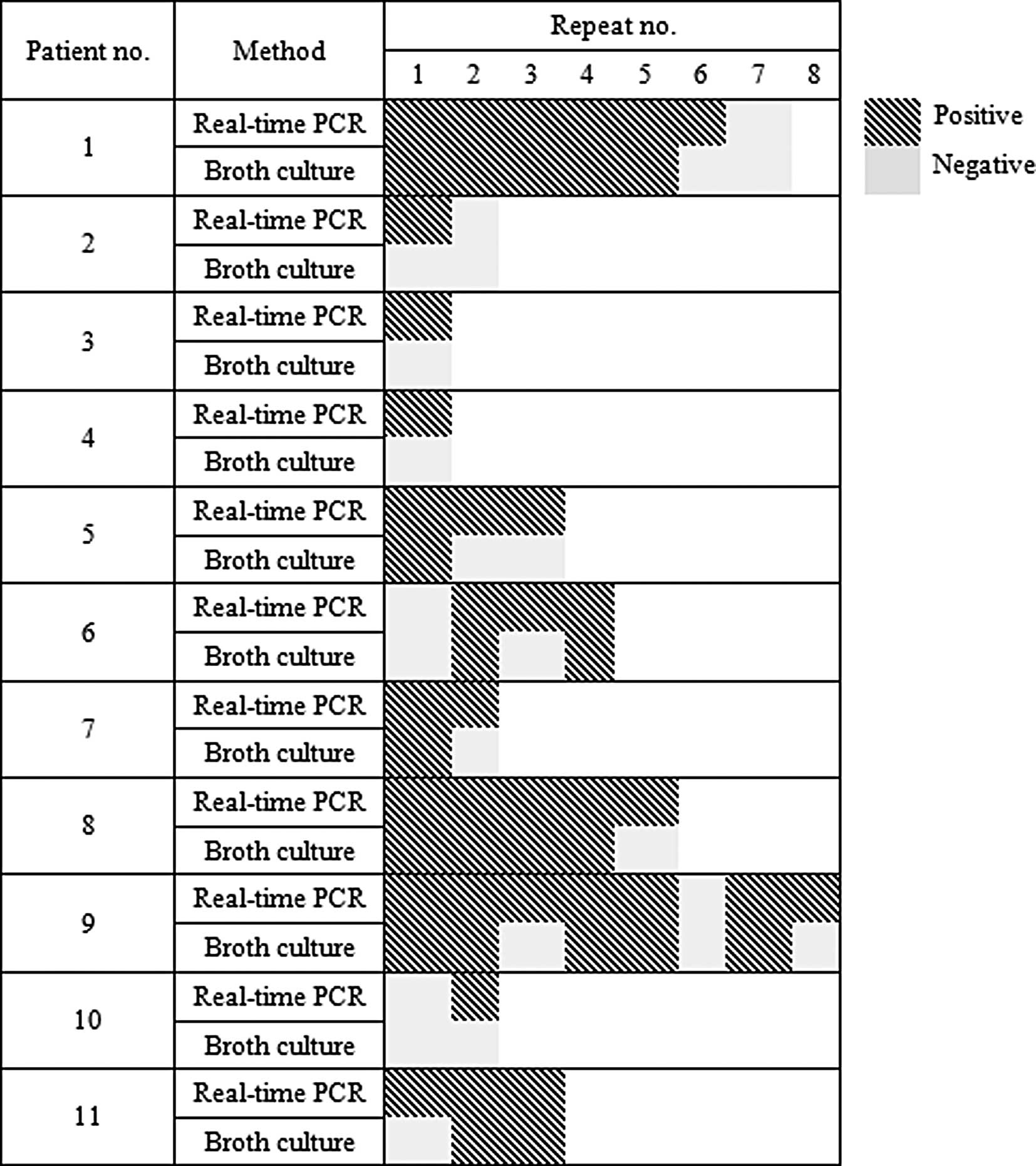
Disagreements between the results of real-time PCR
for the detection of vanA and the conventional method were
observed in 11 of 19 patients (Fig.
1). More than 1 discordant result was detected in 2 patients
(patient nos. 5 and 9).
Application stage
For the preparation, we applied the 24-h broth
incubation method to a total of 1,117 clinical follow-up isolates
from 82 VRE-infected or VRE-colonized patients between June 2008
and December 2009. Of the 1,117 isolates, 372 were resistant to
vancomycin as detected via real-time PCR for the detection of
vanA. The most prevalent collection site was the
gastrointestinal tract (including stool), with up to 636 isolates
(56.9%).
Three consecutive negative results from real-time
PCR for the detection of vanA were found in 23 patients; the
median period until removal from isolation was 71 days, and the
median number of tests was 5.5 (Table III). In these 23 patients, there
were only 2 (8.3%) initial specimens from the gastrointestinal
tract, whereas 20 (83.3%) gastrointestinal specimens were submitted
for release from isolation (Table
III). Two out of the 23 patients showed 2 consecutive negative
real-time PCR results. After 1 negative result, the test results
were once again positive in 5 patients. In 7 patients, only one
type of specimen was submitted, whereas in 16, more than 2 types of
specimen were submitted consecutively. Gastrointestinal specimens
were submitted in the case of all patients, except in 1 specimen
labelled ‘other’. A total of 5 of 16 patients with 2 specimen types
showed negative results simultaneously.
 | Table IIIDays and number of tests performed
before release from isolation and specimen types in 23
patients. |
Table III
Days and number of tests performed
before release from isolation and specimen types in 23
patients.
| Patient no. | Days | Repeats | Last specimen
type | Initial specimen
type |
|---|
| 1 | 1 | 6 | Other | Other |
| 2 | 277 | 6 | Gastrointestinal | Gastrointestinal |
| 3 | 51 | 10 | Gastrointestinal | Genitourinary |
| 4 | 52 | 9 | Gastrointestinal | Genitourinary |
| 5 | 163 | 25 | Other | Other |
| 6 | 63 | 10 | Gastrointestinal | Genitourinary |
| 7 | 44 | 8 | Gastrointestinal | Genitourinary |
| 8 | 472 | 12 | Gastrointestinal | Genitourinary |
| 9 | 21 | 4 | Genitourinary | Genitourinary |
| 10 | 47 | 8 | Gastrointestinal | Gastrointestinal |
| 11 | 19 | 3 | Gastrointestinal | N/A |
| 12 | 55 | 9 | Gastrointestinal | Genitourinary |
| 13 | 21 | 3 |
Gastrointestinal/Genitourinary | Genitourinary |
| 14 | 417 | 15 |
Gastrointestinal | N/A |
| 15 | 102 | 16 |
Gastrointestinal | Other |
| 16 | 13 | 3 |
Gastrointestinal/Other | Other |
| 17 | 14 | 3 |
Gastrointestinal/Genitourinary | Blood |
| 18 | 16 | 4 |
Gastrointestinal/Genitourinary | Other |
| 19 | 236 | 10 |
Gastrointestinal | Genitourinary |
| 20 | 19 | 4 |
Gastrointestinal | Genitourinary |
| 21 | 17 | 3 |
Gastrointestinal | Other |
| 22 | 142 | 22 |
Gastrointestinal | Other |
| 23 | 33 | 6 | Genitourinary | Genitourinary |
| Median | 71 | 5.5 | | |
Discussion
Vancomycin resistance is an independent predictor of
mortality among patients with enterococcal bloodstream infection
(13). Considering the clinical
significance of VRE colonization, infection and spread, we
evaluated the usefulness of the real-time PCR method in comparison
to the conventional phenotypic method. It is widely acknowledged
that real-time PCR shows more rapid, sensitive and reproducible
results than conventional PCR. Moreover, it can minimize the
carryover contamination rate (14). Particularly in the clinical
laboratory, contamination can lead to wrong diagnosis and
treatment.
As previously reported (15), enterococcal broth culture is more
effective for VRE detection than enterococcal agar culture.
Similarly, the most efficient preparation method for real-time PCR
is 24-h incubation in enterococcal broth, which produced the same
results as conventional PCR in a previous study (16).
On the other hand, real-time PCR was found to have a
much higher positive rate of VRE detection in all types of samples
than the conventional method (data not shown). This may be due to
the administration of antibiotics or the difference in the limit of
detection. In the clinical laboratory, real-time PCR of isolates
prepared by 24-h broth incubation may be more adequate and easier
to perform, given that broth medium is more convenient and less
time-consuming to use than agar medium.
There are 3 consecutive negative results in VRE
surveillance accepted as the standard period until removal from
isolation (6). As shown in
Fig. 1 (particularly in the cases
of patient nos. 5 and 10), real-time PCR showed a higher detection
rate than the conventional method. However, to be released from
isolation, 3 or more consecutive negative results of real-time PCR
maybe optimal, which remains the current recommendation in the
conventional method. This is supported by the fact that in the
application stage, 2 of the 23 patients removed from isolation
tested positive for VRE after 2 consecutive negative results of
real-time PCR for the detection of vanA. Moreover, as shown
in Fig. 1, real-time PCR was much
more sensitive than conventional culture.
Three consecutive negative results of vanA
real-time PCR were shown in 23 patients, with a median period for
removal from precautionary isolation of 71 days, which is shorter
than that of the previous report (246 days) (16). The median number of tests was 5.5.
This may be due to the outlier in the evaluation of the mean and
median values.
A previous study reported urine to be the most
frequent site from which VRE was isolated (17) and the number of cultures counted
was up to 88 out of 240 isolates (36.7%). Similarly, 11 of 19
‘cleared’ patients were primarily identified with a positive urine
culture.
We believe that our study proves the usefulness of
real-time PCR for surveillance purposes. Considering the
geographical difference, real-time PCR may replace the conventional
culture method in VRE surveillance.
Acknowledgements
The authors would like to thank H.S. Park for the
laboratory assistance.
References
|
1
|
Murray B: The life and times of the
Enterococcus. Clin Microbiol Rev. 3:46–65. 1990.PubMed/NCBI
|
|
2
|
Fisher K and Phillips C: The ecology,
epidemiology and virulence of Enterococcus. Microbiology.
155:1749–1757. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Uttley AH, Collins CH, Naidoo J and George
RC: Vancomycin-resistant enterococci. Lancet. 1:57–58. 1988.
View Article : Google Scholar
|
|
4
|
Cetinkaya Y, Falk P and Mayhall CG:
Vancomycin-resistant enterococci. Clin Microbiol Rev. 13:686–707.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Winn W, Allen S, Janda W, et al: Koneman’s
color atlas and textbook of diagnostic microbiology. 6th edition.
Lippincott Williams and Wilkins; Philadelphia, PA: pp. 10122006
|
|
6
|
Centers for Disease Control and
Prevention. Recommendations for preventing the spread of vancomycin
resistance: recommendations of the Hospital Infection Control
Practices Advisory Committee (HICPAC). Am J Infect Control.
23:87–94. 1995. View Article : Google Scholar
|
|
7
|
Sedgley CM, Nagel AC, Shelburne CE,
Clewell DB, Appelbe O and Molander A: Quantitative real-time PCR
detection of oral Enterococcus faecalis in humans. Arch Oral Biol.
50:575–583. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Paule SM, Trick WE, Tenover FC, Lankford
M, Cunningham S, Stosor V, Cordell RL and Peterson LR: Comparison
of PCR assay to culture for surveillance detection of
vancomycin-resistant enterococci. J Clin Microbiol. 41:4805–4807.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hwang K, Sung H, Namgoong S, Yoon N and
Kim M: Microbiological and epidemiological characteristics of
vancomycin-dependent enterococci. Korean J Lab Med. 29:299–306.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Song JH, Ko KS, Oh WS, Park S, Heo ST,
Kwon KT, Ryu SY, Peck KR and Lee NY: High frequency of
vancomycin-resistant Enterococcus faecium isolates with vanB
phenotype and vanA genotype in Korean hospitals. Diagn
Microbiol Infect Dis. 56:401–406. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yoon YK, Sim HS, Kim JY, Park DW, Sohn JW,
Roh KH, Lee SE and Kim MJ: Epidemiology and control of an outbreak
of vancomycin-resistant enterococci in the intensive care units.
Yonsei Med J. 50:637–643. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lee W, Kim Y and Huh J: Glycopeptide and
aminoglycoside resistance of vancomycin-resistant Enterococcus
faecium in Korea. Korean J Clin Microbiol. 6:18–22. 2003.
|
|
13
|
Diaz Granados CA, Zimmer SM, Klein M and
Jernigan JA: Comparison of mortality associated with
vancomycin-resistant and vancomycin-susceptible enterococcal
bloodstream infections: a meta-analysis. Clin Infect Dis.
41:327–333. 2005.PubMed/NCBI
|
|
14
|
Mackay IM: Real-time PCR in the
microbiology laboratory. Clin Microbiol Infect. 10:190–212. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kim S, Sung H, Jeon H, Park S, Park S and
Kim M: Evaluation of a rapid enrichment-PCR method for the
detection of vanA vancomycin-resistant enterococci in fecal
specimens. Korean J Clin Microbiol. 10:44–48. 2007.
|
|
16
|
Byers KE, Anglim AM, Anneski CJ and Farr
BM: Duration of colonization with vancomycin-resistant
Enterococcus. Infect Control Hosp Epidemiol. 23:207–211.
2002. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Furtado G, Martins S, Coutinho A, Soares
GM, Wey SB and Medeiros EA: Incidence of vancomycin-resistant
Enterococcus at a university hospital in Brazil. Rev Saúde
Pública. 39:41–46. 2005.
|