Introduction
Non-alcoholic fatty liver disease (NAFLD) is the
most common type of chronic liver disease in Western Europe,
America, Australia and Japan, and is secondary to the growing
epidemic of metabolic syndrome (MetS) (1,2).
Non-alcoholic steatohepatitis (NASH) is a progressive form of
NAFLD; this subset of the population exhibits hepatic steatosis
accompanied by inflammation and fibrosis (3). NASH is the hepatic manifestation of
MetS associated with obesity, non-insulin-dependent diabetes and
hypertriglyceridemia (4,5).
To control the development of NAFLD, it is
significant to determine the precise mechanism of regulation of
lipid accumulation in the human liver. Nakamuta et al
(6) investigated fatty acid
synthesis in NAFLD and reported that the de novo synthesis
of fatty acids, including acetyl-CoA carboxylase 1 (ACC1), ACC2,
glycerol-3-phosphate acyltransferase (GPAT), stearoyl-Co A
desaturase-1 (SCD) and fatty acid synthase (FAS), were upregulated
in NAFLD patients. The expression of these lipogenic genes was
regulated by sterol regulatory element binding protein-1 (SREBP-1)
at the transcriptional level in response to insulin and glucose.
SREBPs are transcription factors that belong to the basic
helix-loop-helix leucine zipper family. SREBP-1, a SREBP isoform,
is mostly expressed in the liver and regulates fatty acid synthesis
(7,8). SREBP1 transgenic mice also manifest
pronounced NASH (9). These
findings indicate that SREBP1 is important in the development of
NAFLD.
SREBP1 is degraded in a Fbxw7-dependent manner
(10). Fbxw7 (also known as Fbw7,
SEL-10, hCdc4 and hAgo) is a member of the F-box protein family
which determines the substrate specificity of the SCF-type
ubiquitin ligase (E3) complex. SCFFbxw7 is a type of E3
that belongs to the ubiquitin-proteasome system (UPS).
SCFFbxw7 consists of the RING-finger protein ring-box 1
(Rbx1; also known as Roc1 and Hrt1), the scaffold protein cullin 1
(Cul1) and the adaptor protein S-phase kinase-associated protein 1
(Skp1), in addition to an F-box protein (11). Fbxw7 targets various mammalian
proteins for degradation, including cyclin E, c-Myc, c-Jun, SREBPs,
mammalian target of rapamycin (mTOR) and PPAR-γ coactivator-1α
(PGC-1α) (11).
In light of the above findings, we hypothesized that
there may be a reduction of Fbxw7 in NAFLD and an induced
accumulation of SREBP-1. To investigate this hypothesis, we used a
mouse model of NAFLD and initially detected the expression of Fbxw7
in the liver tissues by reverse transcription-polymerase chain
reaction (RT-PCR), immunohistochemistry and western blot analysis,
and then evaluated its correlation with the expression of SREBP-1
at the protein level.
Materials and methods
Animals and diets
C57BL/6J mice (n=40; age, 5 weeks) purchased from
the Centre of Laboratory Animals, The Medical College of Xi’an Jiao
Tong University (Xi’an, China) were acclimatized to a vivarium
(temperature, 23–25°C; 12 h light/dark; 50–60% humidity) for 1
week. All animal protocols were approved by the Institutional
Animal Care and Use Committee. Following acclimatization, animals
were weighed and divided into two groups (n=20 each). The mice in
the experimental group were fed HF diets (45% calories as fat),
while the mice in the control group were maintained on a low-fat
diet (10% calories as fat) for 8 weeks. The mice had access to food
and water ad libitum. Mouse chow was purchased from Research
Diets Inc. (New Brunswick, NJ, USA) (12).
Measurement of triglyceride and total
cholesterol levels in the liver
The frozen liver tissue was homogenized,
triglyceride and total cholesterol were extracted from the
homogenate with chloroform/methanol (2:1; vol/vol), and then dried
and resuspended in 2-propanol. The amounts of triglyceride and
total cholesterol in the extract were measured using Lipidos liquid
and Cholescolor liquid kits, respectively (Toyobo, Osaka,
Japan).
Histopathological and biochemical
analyses
Tissue samples excised from the liver were fixed
with 4% paraformaldehyde before processing for histological
analyses by conventional methods. Step sections (5 μm) of the liver
were obtained and stained with hematoxylin and eosin (H&E).
Frozen sections were stained with Oil Red O (Nakalai Tesque Inc.,
Kyoto, Japan), according to the manufacturer’s instructions, to
examine the extent of lipid accumulation in the hepatocytes.
Stained sections were evaluated (light microscopy; magnification,
×40) by a pathologist (Department of Pathology, The First
Affiliated Hospital of the Medical College of Xi’an Jiao Tong
University, Xi’an, China) in a blinded analysis. Plasma levels of
alanine transaminase (ALT), aspartate transaminase (AST), alkaline
phosphatase (AP), γ-glutamyl transpeptidase (GGT), free fatty acids
(FFA), low-density lipoprotein-cholesterol (LDL-c), high-density
lipoprotein-cholesterol (HDL-c), triglycerides (TG) and total
cholesterol (TC) were measured using a standard clinical
autoanalyzer.
RT-PCR
The frozen specimens for RT-PCR were stored at
−80°C. The RNAfast 200 purification kit was obtained from Shanghai
Fastagen Biotech Ltd. Co., Shanghai, China; the PrimeScript RT
reagent kit was purchased from Takara Bio Inc., Shiga, Japan and
the 2X Taq Master mix was obtained from Beijing CoWin Biotech.,
Beijing, China. The Fbxw7 primers for RT-PCR, designed by Beacon
Designer software (Premier Biosoft International, Palo Alto, CA,
USA), were: sense, 5′-CACAGGCCTTCAAGAGTGGC-3′; and antisense,
5′-TTGCATCATATGCTTCACTTGTGT-3′. The PCR product was ~117 bp.
β-actin, a housekeeping gene, served as an internal control to
ensure that an equal amount of mRNA was analyzed from each sample.
The upstream primer sequence for β-actin was
5′-TGATGACATCAAGAAGGTGGTGAA-3′ and the downstream sequence was
5′-TCCTTGGAGGCCAT GTAGGCCAT-3′, which was predicted to produce a
239 bp PCR product.
The total RNA of the samples was extracted using the
RNAfast 200 purification kit, according to the manufacturer’s
instructions. The optical density of RNA at A260/280 nm was between
1.7 and 1.9. The integrity of the RNA was confirmed by the presence
of intact 18S and 28S bands on 2% agarose gels. The total RNA was
then reverse-transcribed at 37°C for 15 min, and the cDNA was
incubated at 85°C for 5 sec to inactivate the reverse
transcriptase. For PCR amplification, the cDNA was used as the
template to amplify specific PCR products of the Fbxw7 and GAPDH
genes. The PCR reaction was performed in a 25 μl system, starting
with denaturation for 5 min at 94°C; then 35 cycles of denaturation
for 30 sec at 94°C; annealing at 62°C for Fbxw7 and GAPDH for 30
sec; and extension for 30 sec at 72°C, followed by a final
extension for 7 min at 72°C. The PCR products were separated by 2%
agarose gel electrophoresis and stained with ethidium bromide. The
electrophoresis bands were analyzed using the Gene Genius Gel
Imaging System. The gene expression intensity (relative
coefficient) was quantitatively analyzed by the ratio of the
expression intensity of the electrophoresis bands to β-actin.
Immunohistochemical staining
The primary mouse anti-Fbxw7 antibody (ab74054) was
purchased from Abcam (Hong Kong, China). The primary mouse
anti-SREBP-1 antibody (2A4) was purchased from Lab Vision &
Neomarkers (Fremont, CA, USA).
Immunohistochemistry was performed on
paraformaldehyde-fixed paraffin sections. The sections were
dewaxed, dehydrated, rehydrated and antigen retrieval was performed
in citrate buffer. Endogenous peroxidase activity was blocked for
10 min using 3.0% hydrogen peroxide, the sections were blocked for
30 min using 10% goat plasma, and then separately incubated with
the primary antibodies directed against Fbxw7 and SREBP-1 at 4°C
overnight. The primary antibody was detected using biotinylated
secondary antibodies (Beijing Zhongshan Goldenbridge Biotechnology
Ltd., Co., Beijing, China) according to the manufacturer’s
instructions. The staining of the sections was performed using
streptavidin-HRP conjugates for Fbxw7 and SREBP-1 (SP method). The
sections were visualized with diaminobenzidine, counterstained with
hematoxylin, then dehydrated in alcohol and xylene and mounted onto
glass slides.
Protein expression revealed by western
blot analysis
For western blot analysis, ~200 mg of liver samples
were homogenized in 1 ml of lysis buffer (20 mM Tris, 145 mM NaCl,
10% glycerol, 5 mM EDTA, 1% Triton-X, 0.5% Nonidet P-40, 100 M
phenylmethylsulfonyl fluoride, 50 M NaF and 1 mM sodium
orthovanadate). Lysates were centrifuged at 8850 × g at 4°C for 10
min. The supernatant was collected, and the protein concentration
was determined using the Bradford method; bovine serum albumin was
used as a standard. Samples (1.5 g/mm gel thickness) were heated
for 5 min under reducing conditions and loaded onto sodium dodecyl
sulfate-polyacrylamide gels. Proteins from the gels were then
transferred onto nitrocellulose membranes. Electroblots were
blocked in Tris-buffered NaCl-Tween (TBST) containing 5% skimmed
milk powder at room temperature. Western blot analysis was then
conducted using specific antibodies for Fbxw7 and SREBP-1. Blots
were incubated with antibodies of interest (in TBST buffer and 5%
bovine serum albumin) and agitated overnight at 4°C. Following
washing with TBST, the blots were incubated with horseradish
peroxidase-labeled anti-rabbit antibody (Cell Signaling Technology,
Inc., Beverly, MA, USA) in skimmed milk for 1 h at room
temperature. Immunoreactivity was detected by enhanced
chemiluminescence. Bands were quantified by scanning densitometry
and expressed as arbitrary units.
Statistical analysis
Data were shown as the mean ± standard deviation
(SD) and were analyzed using the two-tailed Student’s t-test.
Additionally, the Spearman’s rank test was used to analyze the
correlation between the Fbxw7 protein expression and SREBP-1.
P<0.05 was considered statistically significant. SPSS 13.0
software package was used for all statistical analyses (SPSS Inc.,
Chicago, IL, USA)
Results
Mouse model for NAFLD
The HF diet increased body weight and adiposity in
C57BL/6J mice (Fig. 1A). Body
weight and liver weight (wet) in the HF group were significantly
higher than those of the control mice (45.10±2.56 vs. 29.40±2.10;
5.16±0.28 vs. 2.56±0.25, respectively; P<0.05) (Fig. 1A). Elevated plasma AST, ALT, AP and
GGT blood biomarkers were associated with liver dysfunction in
human NAFLD (Fig. 1B). The HF diet
increased plasma FFA to an average of 39 mg/l in the HF group,
which was higher than that in the control group, but not
statistically significant (P=0.063). No significant changes were
observed in plasma TG concentrations. However, clear
hypercholesterolemia was observed in the HF group, mostly due to an
increase in LDL-c, although no significant alterations occurred in
HDL-c (Fig. 1C). TG levels were
significantly increased in the livers of the HF group compared with
those of the control group (P<0.05). The concentration of TC was
not affected in the HF group (Fig.
1D).
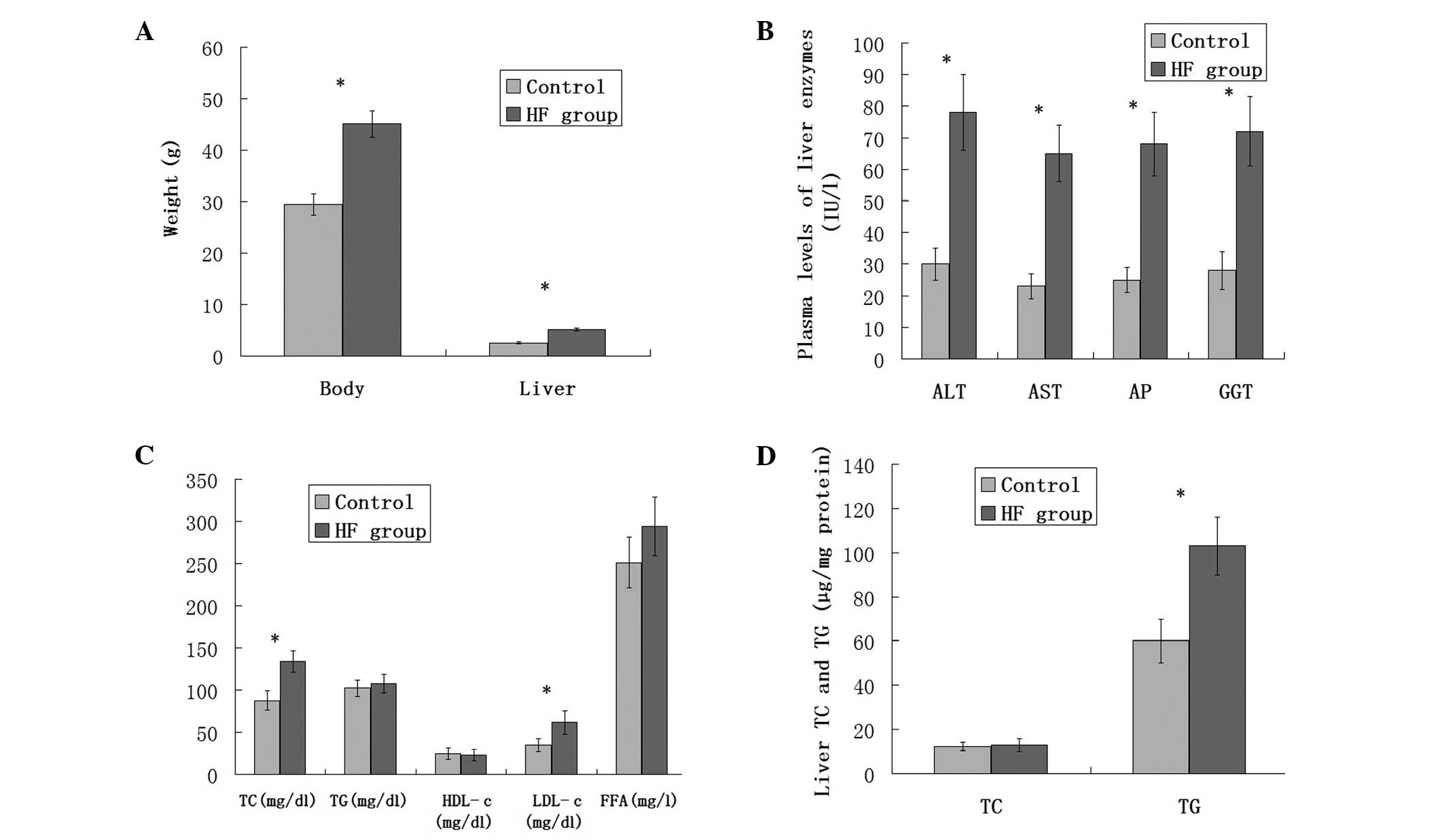 | Figure 1(A) Body and liver weight gain in
C57BL/6J mice in the control or HF diet group fed for 8 weeks
(n=20). (B) Plasma levels of the liver enzymes in the C57BL/6J mice
in the control or HF diet groups fed for 8 weeks (n=20). (C) Plasma
levels of TC, TG, HDL-c, LDL-c and FFA in the C57BL/6J mice in the
control or HF diet groups fed for 8 weeks (n=20). (D) Triglyceride
and total cholesterol concentrations in the livers of C57BL/6J mice
fed the control or HF diet group fed for 8 weeks. Values are the
mean ± standard error. *P<0.05. HF, high fat; TC,
total cholesterol; TG, triglycerides; HDL-c, high-density
lipoprotein-cholesterol; LDL-c, low-density
lipoprotein-cholesterol; FFA, free fatty acids; ALT, alanine
transaminase; AST, aspartate transaminase; AP, alkaline
phosphatase; GGT, γ-glutamyl transpeptidase. |
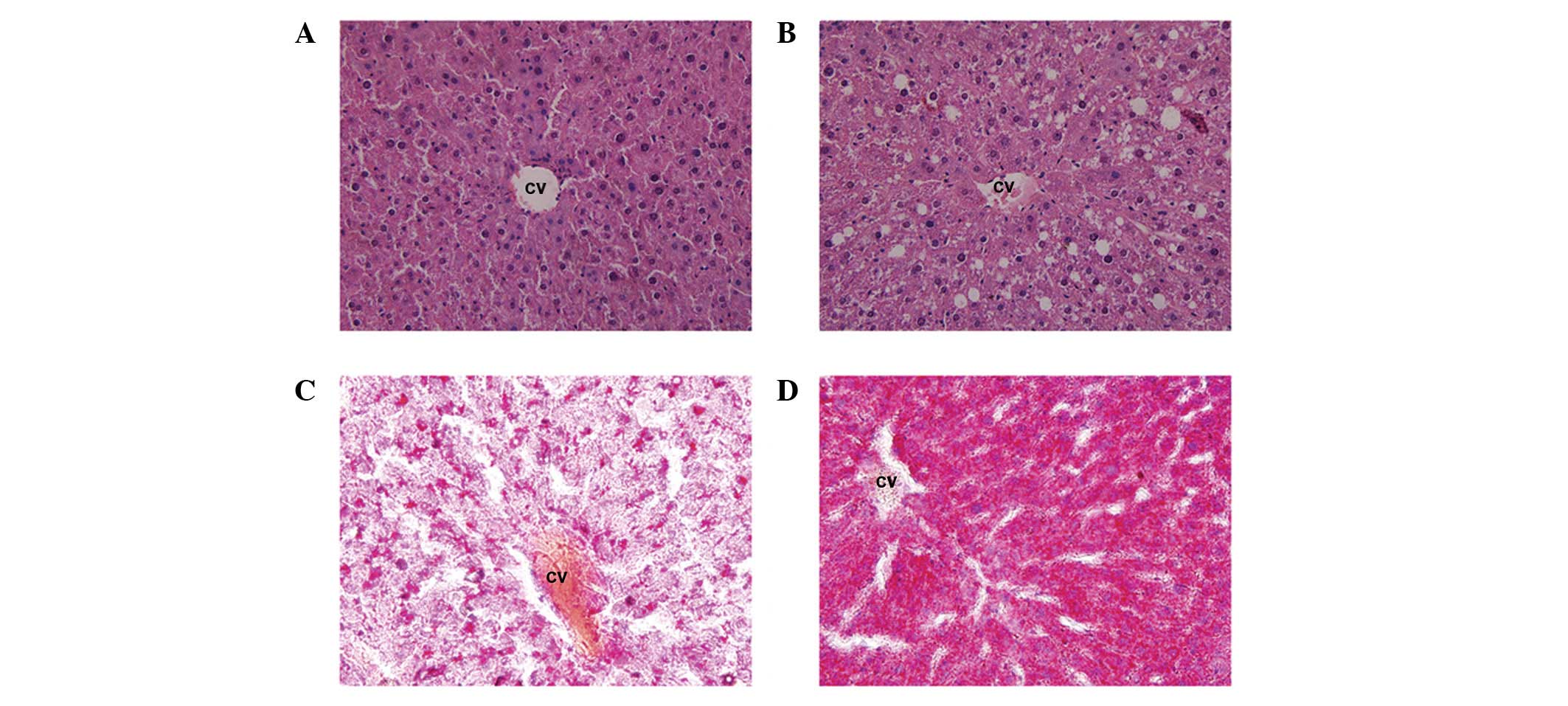
Liver histology of the control and HF groups was
evaluated to investigate the occurrence of NAFLD. Representative
images of liver H&E- and Oil Red O-stained sections from the
control and HF diet groups were captured. Fig. 2A shows sections of the liver from
the control animals demonstrating no steatosis, inflammation or
fibrosis. Macrovesicular and microvesicular steatosis was observed
in the HF group (Fig. 2B). Oil Red
O staining demonstrated an accumulation of triglyceride molecules
in the HF group (Fig. 2D), which
was significantly higher than that in the control group (Fig. 2C).
Expression of Fbxw7 in the control and HF
groups
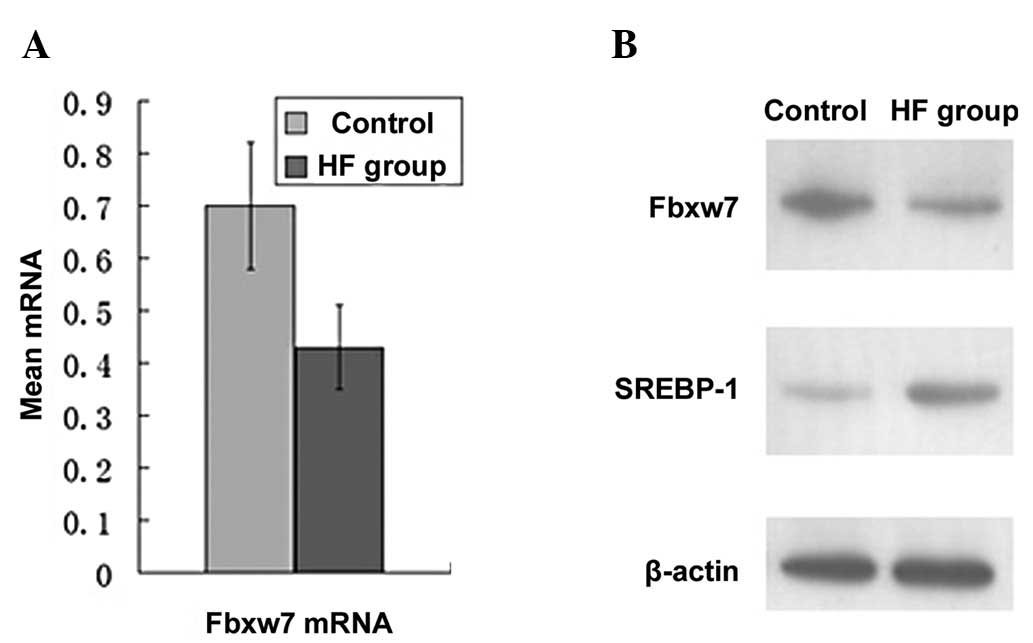
Following analysis of the RT-PCR results, the
expression of Fbxw7 mRNA in the HF group was found to be
significantly lower than that in the control group (0.43±0.12 vs.
0.70±0.18; P<0.05) (Fig. 3A).
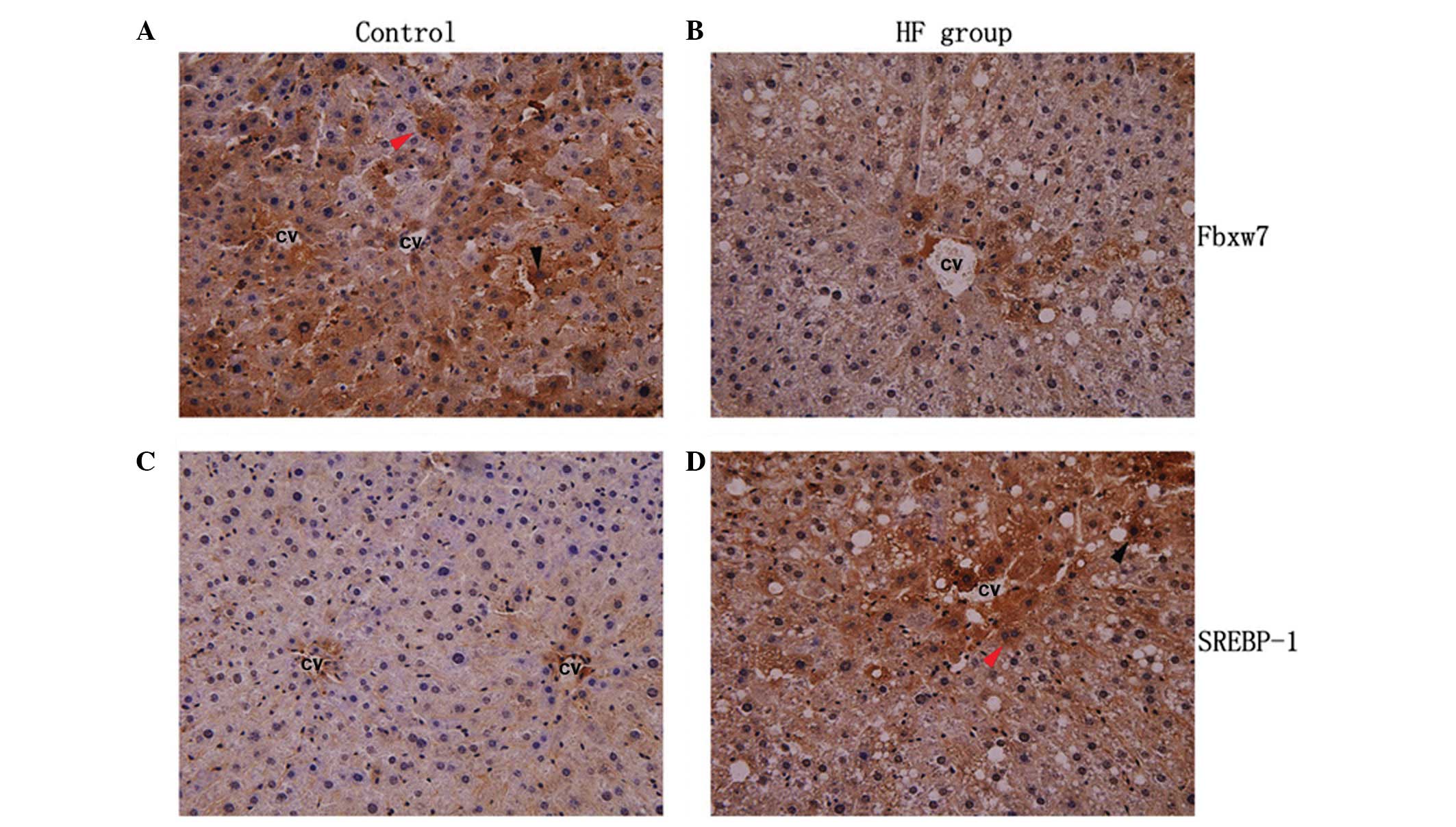
Immunohistochemistry results revealed that the majority of the
positive liver cells demonstrated diffuse cytoplasmic staining of
Fbxw7, while nuclear staining was identified in few cells (Fig. 4). Results of the western blot
analysis indicated that the expression of the Fbxw7 protein was
also significantly lower in the HF group compared to the control
group (0.44±0.04 vs. 0.82±0.08; P<0.05) (Fig. 3B). Additionally, the results were
in accordance with those at the mRNA level.
Expression of SREBP-1 in the control and
HF groups
The protein expression of SREBP-1 was assessed by
immunohistochemical and western blot analysis. SREBP-1 protein was
localized in the cytoplasm and nucleus (Fig. 4) and was significantly elevated in
the HF group (P<0.05) (Fig.
3B).
Correlation between Fbxw7 and SREBP-1
protein expression in the HF group
As mentioned above, the Fbxw7 protein expression in
the HF group was significantly lower compared to the control group.
However, the results showed SREBP-1 protein to be higher in the
mouse liver tissues in the HF group compared to in the control
group. Spearman’s rank test analysis revealed a significant
negative correlation between Fbxw7 and SREBP-1 at the protein level
in the HF group (r=−0.584, P<0.05).
Discussion
The abundance of cell proteins is regulated in a
coordinated manner at the levels of synthesis and degradation
(13). In particular,
intracellular proteolysis is important to cell survival and
self-repair. The UPS is responsible for the specific degradation of
proteins and contains 3 enzymes: a ubiquitin-activating enzyme
(E1), a ubiquitin-conjugating enzyme (E2) and E3, and a 26S
proteasome (14). Ubiquitin is
first bound and activated via the C-terminal adenylation by E1. E1
catalyzes the transfer of charged ubiquitin to a cysteine residue
of E2, E3 recognizes the substrate of the UPS, and then the
ubiquitylated substrates are recognized and degraded by the 26S
proteasome (15–16). UPS regulates a number of
significant cell processes, including differentiation, apoptosis
and proliferation (17).
F-box proteins determine the substrate specificity
of the SCF-type E3 complex (11).
Fbxw7 is a member of the F-box family, and was first identified as
a negative regulator of Notch-mediated (LIN-12-mediated) signaling
in Caenorhabditis elegans by genetic analysis (18). The Fbxw7 target proteins contain
conservative phosphorylation amino-acid residues, known as CDC4
phosphodegrons (CPDs) (19).
Phosphorylated CPDs bind to the WD40 repeat domains of Fbxw7 and
target proteins are recognized and degraded in a sequential manner.
Results of our study showed that the Fbxw7 mRNA and protein
expression in the HF group was lower than that in the control group
(P<0.05), indicating that Fbxw7 may be closely correlated with
the pathogenesis of NAFLD.
SREBPs, specific substrates of Fbxw7, are
significant for the onset and development of NASH (20). SREBPs have 3 isozymes: SREBP-1
(SREBP-1a and SREBP-1c) and SREBP-2. SREBP-1 mainly regulates
enzymes involving triglycerides and fatty acid synthesis, including
ACC-1, ACC-2, GPAT, SCD1 and FAS. SREBP-2 regulates cholesterol
synthesis enzymes, including hydroxy-methyl-glutaryl coenzyme A
(HMG-CoA) and HMG-CoA reducing enzymes. Our results indicated that
SREBP-1 was accumulated in the HF group, and was also negatively
correlated with the expression of Fbxw7.
Recently, Onoyama et al (13) demonstrated that mice with the
liver-specific ablation of Fbxw7 developed clinicopathological
features similar to those of NAFLD or NASH in humans. These authors
also found that an abundance of nuclear SREBP1 was increased in
mice with liver-specific null mutations of Fbxw7. Our results
combined with the above findings suggest that the Fbxw7-SREBP-1
axis plays a key physiological role in the regulation of lipid
metabolism in the liver as well as a pathological role in the
development of NAFLD. One possible mechanism is that Fbxw7
expression is reduced during the development of NAFLD, which
prevents UPS from recognizing and degrading SREBP-1, leading to the
accumulation of SREBP-1 which could sequentially upregulate
lipogenic gene transcription and induce NAFLD. However, due to the
complex pathogenesis of NAFLD, more studies are required to
precisely identify the potential mechanisms of occurrence and
development of NAFLD.
Acknowledgements
This study was supported by a grant from the
National Natural Science Foundation of China (No. 81071897).
References
|
1
|
Hamaguchi E, Takamura T, Sakurai M, et al:
Histological course of nonalcoholic fatty liver disease in Japanese
patients: tight glycemic control, rather than weight reduction,
ameliorates liver fibrosis. Diabetes Care. 33:284–286. 2010.
View Article : Google Scholar
|
|
2
|
Vanni E, Bugianesi E, Kotronen A, De
Minicis S, Yki-Järvinen H and Svegliati-Baroni G: From the
metabolic syndrome to NAFLD or vice versa? Dig Liver Dis.
42:320–330. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ogawa T, Fujii H, Yoshizato K and Kawada
N: A human-type nonalcoholic steatohepatitis model with advanced
fibrosis in rabbits. Am J Pathol. 177:153–165. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ota T, Takamura T, Kurita S, Matsuzawa N,
Kita Y and Uno M: Insulin resistance accelerates a dietary rat
model of nonalcoholic steatohepatitis. Gastroenterology.
132:282–293. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bugianesi E, Moscatiello S, Ciaravella MF
and Marchesini G: Insulin resistance in nonalcoholic fatty liver
disease. Curr Pharm Des. 16:1941–1951. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nakamuta M, Kohjima M, Morizono S, et al:
Evaluation of fatty acid metabolism-related gene expression in
nonalcoholic fatty liver disease. Int J Mol Med. 16:631–635.
2005.PubMed/NCBI
|
|
7
|
Foufelle F and Ferré P: New perspectives
in the regulation of hepatic glycolytic and lipogenic genes by
insulin and glucose: a role for the transcription factor sterol
regulatory element binding protein-1c. Biochem J. 366:377–391.
2002. View Article : Google Scholar
|
|
8
|
Hagen RM, Rodriguez-Cuenca S and
Vidal-Puig A: An allostatic control of membrane lipid composition
by SREBP1. FEBS Lett. 584:2689–2698. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nakayama H, Otabe S, Ueno T, et al:
Transgenic mice expressing nuclear sterol regulatory
element-binding protein 1c in adipose tissue exhibit liver
histology similar to nonalcoholic steatohepatitis. Metabolism.
56:470–475. 2007. View Article : Google Scholar
|
|
10
|
Sundqvist A, Bengoechea-Alonso MT, Ye X,
et al: Control of lipid metabolism by phosphorylation-dependent
degradation of the SREBP family of transcription factors by
SCFFbw7. Cell Metab. 1:379–391. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Welcker M and Clurman BE: FBW7 ubiquitin
ligase: a tumour suppressor at the crossroads of cell division,
growth and differentiation. Nat Rev Cancer. 8:83–93. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kirpich IA, Gobejishvili LN, Bon Homme M,
et al: Integrated hepatic transcriptome and proteome analysis of
mice with high-fat diet-induced nonalcoholic fatty liver disease. J
Nutr Biochem. 22:38–45. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Onoyama I, Suzuki A, Matsumoto A, et al:
Fbxw7 regulates lipid metabolism and cell fate decisions in the
mouse liver. J Clin Invest. 121:342–354. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livnat-Levanon N and Glickman MH:
Ubiquitin-proteasome system and mitochondria-reciprocity. Biochim
Biophys Acta. 1809:80–87. 2011. View Article : Google Scholar
|
|
15
|
Chan NC, Salazar AM, Pham AH, et al: Broad
activation of the ubiquitin-proteasome system by Parkin is critical
for mitophagy. Hum Mol Genet. 20:1726–1737. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Willis MS, Townley-Tilson WH, Kang EY,
Homeister JW and Patterson C: Sent to destroy: the ubiquitin
proteasome system regulates cell signaling and protein quality
control in cardiovascular development and disease. Circ Res.
106:463–478. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bashir T and Pagano M: Aberrant
ubiquitin-mediated proteolysis of cell cycle regulatory proteins
and oncogenesis. Adv Cancer Res. 88:101–144. 2003.PubMed/NCBI
|
|
18
|
Sundaram M and Greenwald I: Suppressors of
a lin-12 hypomorph define genes that interact with both lin-12 and
glp-1 in Caenorhabditis elegans. Genetics. 135:765–783.
1993.PubMed/NCBI
|
|
19
|
Nash P, Tang X, Orlicky S, et al:
Multisite phosphorylation of a CDK inhibitor sets a threshold for
the onset of DNA replication. Nature. 414:514–521. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bengoechea-Alonso MT and Ericsson J: SREBP
in signal transduction: cholesterol metabolism and beyond. Curr
Opin Cell Biol. 19:215–222. 2007. View Article : Google Scholar : PubMed/NCBI
|